Biological Activities of Lactic Acid Bacteria Isolated from Chinese Traditional Cheese and the Application in Antioxidant Foods
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
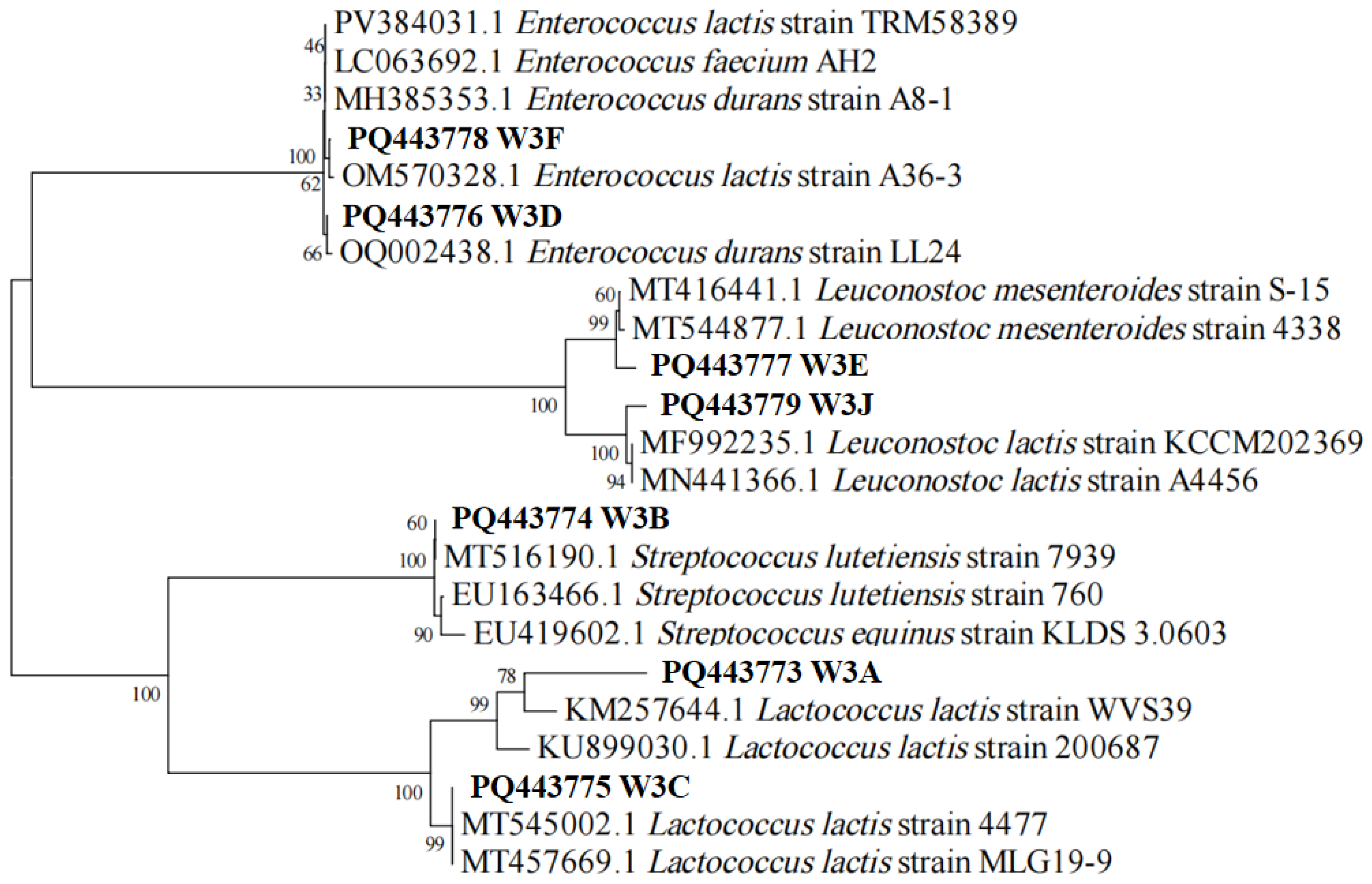
2.2. Isolation and Identification of LAB Strains
2.3. Safety
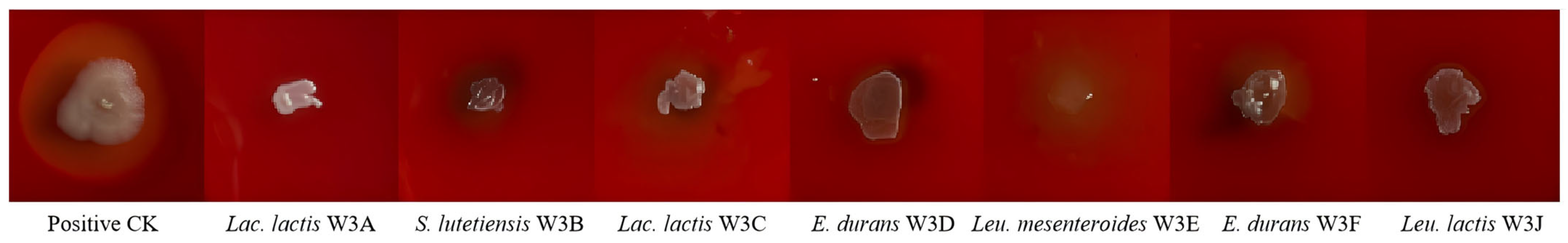
2.3.1. Hemolytic Assay
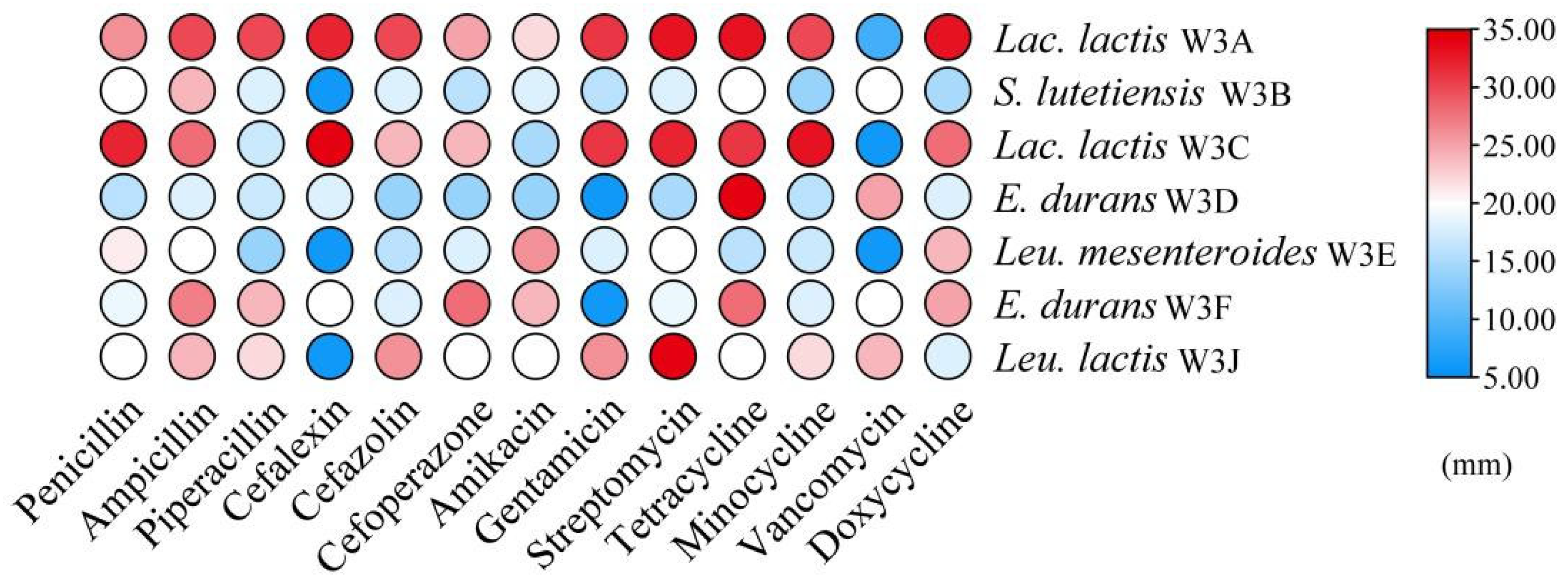
2.3.2. Antibiotic Susceptibility
2.4. The Preparation of Cell-Free Supernatant (CFS) of LAB
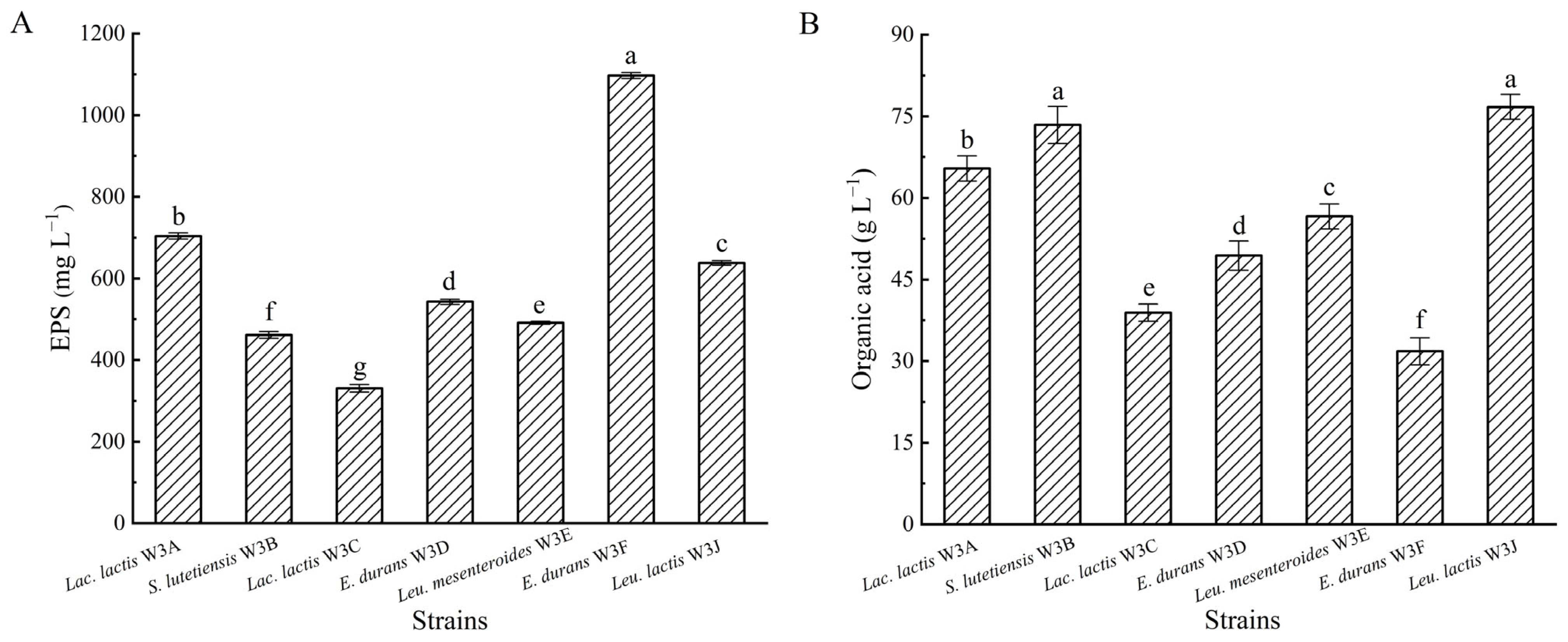
2.5. Exopolysaccharide Production
2.6. Organic Acid Production
2.7. Antioxidant Activity Assay
2.7.1. DPPH Radical Scavenging Activity
2.7.2. ABTS+ Radical Scavenging Activity
2.7.3. Hydroxyl Radical (·OH) Scavenging Activity
2.7.4. Superoxide Anion Radical (·O2−) Scavenging Activity
2.7.5. Ferrous Ion Chelating Capacity (FICC)
2.7.6. Ferric Reducing Antioxidant Power (FRAP)
2.8. Antibacterial Assay
2.9. Antioxidant Effect of Fermented Goat Milk
2.10. Statistical Analysis
3. Results
3.1. Isolation of LAB Strains and Identification by 16S rRNA Sequencing
3.2. Safety Evaluation Through Hemolytic Activity and Antibiotic Susceptibility
3.3. EPS and Organic Acid Production
3.4. Antioxidant Activity
3.5. Antibacterial Activity
3.6. PCA and EW-TOPSIS
3.7. Antioxidant Abilities of Fermented Goat Milk
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- AlKalbani, N.S.; Turner, M.S.; Ayyash, M.M. Isolation, identification, and potential probiotic characterization of isolated lactic acid bacteria and in vitro investigation of the cytotoxicity, antioxidant, and antidiabetic activities in fermented sausage. Microb. Cell Fact. 2019, 18, 188. [Google Scholar] [CrossRef] [PubMed]
- Łepecka, A.; Szymański, P.; Okoń, A.; Zielińska, D. Antioxidant activity of environmental lactic acid bacteria strains isolated from organic raw fermented meat products. LWT 2023, 174, 114440. [Google Scholar] [CrossRef]
- Khalil, E.S.; Abd Manap, M.Y.; Mustafa, S.; Alhelli, A.M.; Shokryazdan, P. Probiotic properties of exopolysaccharide-producing Lactobacillus strains isolated from tempoyak. Molecules 2018, 23, 398. [Google Scholar] [CrossRef]
- Megur, A.; Daliri, E.B.; Balnionyte, T.; Stankeviciute, J.; Lastauskiene, E.; Burokas, A. In vitro screening and characterization of lactic acid bacteria from Lithuanian fermented food with potential probiotic properties. Front. Microbiol. 2023, 14, 1213370. [Google Scholar] [CrossRef]
- Tawab, F.I.A.; Elkadr, M.H.A.; Sultan, A.M.; Hamed, E.O.; El-Zayat, A.S.; Ahmed, M.N. Probiotic potentials of lactic acid bacteria isolated from Egyptian fermented food. Sci. Rep. 2023, 13, 16601. [Google Scholar] [CrossRef]
- Kavitake, D.; Tiwari, S.; Shah, I.A.; Devi, P.B.; Delattre, C.; Reddy, G.B.; Shetty, P.H. Antipathogenic potentials of exopolysaccharides produced by lactic acid bacteria and their food and health applications. Food Control 2023, 152, 109850. [Google Scholar] [CrossRef]
- Feng, T.; Wang, J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef]
- Ramalho, J.B.; Soares, M.B.; Spiazzi, C.C.; Bicca, D.F.; Soares, V.M.; Pereira, J.G.; Silva, W.P.d.; Sehn, C.P.; Cibin, F.W.S. In vitro probiotic and antioxidant potential of Lactococcus lactis subsp. cremoris LL95 and its effect in mice behaviour. Nutrients 2019, 11, 901. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.Y.; Jeong, Y.; Kang, C.-H. Antioxidant activity and probiotic properties of lactic acid bacteria. Fermentation 2022, 8, 29. [Google Scholar] [CrossRef]
- Haghshenas, B.; Kiani, A.; Mansoori, S.; Mohammadi-Noori, E.; Nami, Y. Probiotic properties and antimicrobial evaluation of silymarin-enriched Lactobacillus bacteria isolated from traditional curd. Sci. Rep. 2023, 13, 10916. [Google Scholar] [CrossRef]
- Feng, S.; Wang, H.; Lin, X.; Liang, H.; Zhang, S.; Chen, Y.; Ji, C. Probiotic properties of Lactobacillus plantarum and application in prebiotic gummies. LWT 2023, 174, 114357. [Google Scholar] [CrossRef]
- da Silva, M.N.; Tagliapietra, B.L.; do Amaral Flores, V.; dos Santos Richards, N.S.P. In vitro test to evaluate survival in the gastrointestinal tract of commercial probiotics. Curr. Res. Food Sci. 2021, 4, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kuda, T.; Takahashi, H.; Kimura, B. Bacterial and fungal microbiota of spontaneously fermented Chinese products, Rubing milk cake and Yan-cai vegetable pickles. Food Microbiol. 2018, 72, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Madjirebaye, P.; Xiao, M.; Mahamat, B.; Xiong, S.; Mueed, A.; Wei, B.; Huang, T.; Peng, F.; Xiong, T.; Peng, Z. In vitro characteristics of lactic acid bacteria probiotics performance and antioxidant effect of fermented soymilk. Food Biosci. 2022, 49, 101952. [Google Scholar] [CrossRef]
- Lu, J.; Mao, Y.; Ma, T.; Liu, X.; Cheng, X.; Bai, Y.; Li, S. Screening and genome analysis of lactic acid bacteria with high exopolysaccharide production and good probiotic properties. Food Biosci. 2023, 56, 103211. [Google Scholar] [CrossRef]
- CLSI. Performance standards for antimicrobial susceptibility testing: 21st informational supplement. In CLSI Document M100-S21; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Lappa, I.K.; Kachrimanidou, V.; Alexandri, M.; Papadaki, A.; Kopsahelis, N. Novel probiotic/bacterial cellulose biocatalyst for the development of functional dairy beverage. Foods 2022, 11, 2586. [Google Scholar] [CrossRef]
- Gao, Y.; Zhu, W.; Lou, X.; Zhou, Q.; Tian, T.; Gong, W.; Xiong, J. Protective effects of Bacillus velezensis on texture, physicochemical properties, and lipid oxidation of grass carp fillets during repeated freeze-thaw cycles. J. Sci. Food Agric. 2024, 105, 2497–2505. [Google Scholar] [CrossRef]
- Tian, J.; Mao, Q.; Dong, M.; Wang, X.; Rui, X.; Zhang, Q.; Chen, X.; Li, W. Structural characterization and antioxidant activity of exopolysaccharide from soybean whey fermented by Lacticaseibacillus plantarum 70810. Foods 2021, 10, 2780. [Google Scholar] [CrossRef]
- Shori, A.B. Comparative analysis of Lactobacillus starter cultures in fermented camel milk: Effects on viability, antioxidant properties, and sensory characteristics. Foods 2024, 13, 3711. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Sun, L.; Li, N.; Kwok, L.-Y.; Zhang, H.; Zhang, W. Exopolysaccharide-producing Lacticaseibacillus rhamnosus space mutant improves the techno-functional characteristics of fermented cow and goat milks. J. Agric. Food Chem. 2023, 71, 10729–10741. [Google Scholar] [CrossRef]
- Mohd Yusof, H.; Mohamad, R.; Zaidan, U.H.; Rahman, N.A. Sustainable microbial cell nanofactory for zinc oxide nanoparticles production by zinc-tolerant probiotic Lactobacillus plantarum strain TA4. Microb. Cell Fact. 2020, 19, 10. [Google Scholar] [CrossRef]
- Sakoui, S.; Derdak, R.; Addoum, B.; Pop, O.L.; Vodnar, D.C.; Suharoschi, R.; Soukri, A.; El Khalfi, B. The first study of probiotic properties and biological activities of lactic acid bacteria isolated from Bat guano from Er-rachidia, Morocco. LWT 2022, 159, 113224. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Wu, A.; Ju, C.; Jiang, J.; Chen, J. Multiple-strain Lactobacillus-fermented soymilk with antioxidant capacity and delicate flavour. Int. J. Food Sci. Technol. 2021, 56, 6052–6061. [Google Scholar] [CrossRef]
- Domingos-Lopes, M.F.P.; Lamosa, P.; Stanton, C.; Ross, R.P.; Silva, C.C.G. Isolation and characterization of an exopolysaccharide-producing Leuconostoc citreum strain from artisanal cheese. Lett. Appl. Microbiol. 2018, 67, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Yerlikaya, O. Probiotic potential and biochemical and technological properties of Lactococcus lactis ssp. lactis strains isolated from raw milk and kefir grains. J. Dairy Sci. 2019, 102, 124–134. [Google Scholar] [CrossRef]
- Siroli, L.; Patrignani, F.; Serrazanetti, D.I.; Parolin, C.; Palomino, R.A.Ñ.; Vitali, B.; Lanciotti, R. Determination of antibacterial and technological properties of vaginal Lactobacilli for their potential application in dairy products. Front. Microbiol. 2017, 8, 166. [Google Scholar] [CrossRef]
- Guo, H.; Pan, L.; Li, L.; Lu, J.; Kwok, L.; Menghe, B.; Zhang, H.; Zhang, W. Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 2017, 82, 724–730. [Google Scholar] [CrossRef]
- Mıdık, F.; Tokatlı, M.; Elmacı, S.B.; Özçelik, F. Influence of different culture conditions on exopolysaccharide production by indigenous lactic acid bacteria isolated from pickles. Arch. Microbiol. 2020, 202, 875–885. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, G.; Tian, Y. Biological activities and applications of exopolysaccharides produced by lactic acid bacteria: A mini-review. World J. Microbiol. Biotechnol. 2023, 39, 155. [Google Scholar] [CrossRef]
- Nie, X.; Chen, H.; Xiang, L.; Zhang, Y.; Liu, D.; Zhao, Z. GC-TOF-MS-based non-targeted metabolomic analysis of differential metabolites in Chinese ultra-long-term industrially fermented kohlrabi and their associated metabolic pathways. Metabolites 2022, 12, 991. [Google Scholar] [CrossRef] [PubMed]
- Beltran-Barrientos, L.M.; Garcia, H.S.; Reyes-Diaz, R.; Estrada-Montoya, M.C.; Torres-Llanez, M.J.; Hernandez-Mendoza, A.; Gonzalez-Cordova, A.F.; Vallejo-Cordoba, B. Cooperation between Lactococcus lactis NRRL B-50571 and NRRL B-50572 for aroma formation in fermented milk. Foods 2019, 8, 645. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Zhou, Q.; Jiang, Q.; Lin, L.; Zhu, W.; Mei, X.; Xiong, J.; Gao, Y. Inhibitory effect and mechanism of violacein on planktonic growth, spore germination, biofilm formation and toxin production of Bacillus cereus and its application in grass carp preservation. Int. J. Food Microbiol. 2024, 426, 110917. [Google Scholar] [CrossRef] [PubMed]
- Syromyatnikov, M.Y.; Kokina, A.V.; Solodskikh, S.A.; Panevina, A.V.; Popov, E.S.; Popov, V.N. High-throughput 16S rRNA gene sequencing of butter microbiota reveals a variety of opportunistic pathogens. Foods 2020, 9, 608. [Google Scholar] [CrossRef]
- Sila, A.; Bougatef, A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Gaisawat, M.B.; Iskandar, M.M.; MacPherson, C.W.; Tompkins, T.A.; Kubow, S. Probiotic supplementation is associated with increased antioxidant capacity and copper chelation in C. Difficile-Infected fecal water. Nutrients 2019, 11, 2007. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Measurement of antioxidant activity. J. Funct. Foods 2015, 18, 757–781. [Google Scholar] [CrossRef]
- Saravanan, C.; Kavitake, D.; Kandasamy, S.; Devi, P.B.; Shetty, P.H. Production, partial characterization and antioxidant properties of exopolysaccharide α-d-glucan produced by Leuconostoc lactis KC117496 isolated from an idli batter. J. Food Sci. Technol. 2019, 56, 159–166. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Lorusso, A.; Russo, V.; Pinto, D.; Marzani, B.; Gobbetti, M. Improving the antioxidant properties of quinoa flour through fermentation with selected autochthonous lactic acid bacteria. Int. J. Food Microbiol. 2017, 241, 252–261. [Google Scholar] [CrossRef]
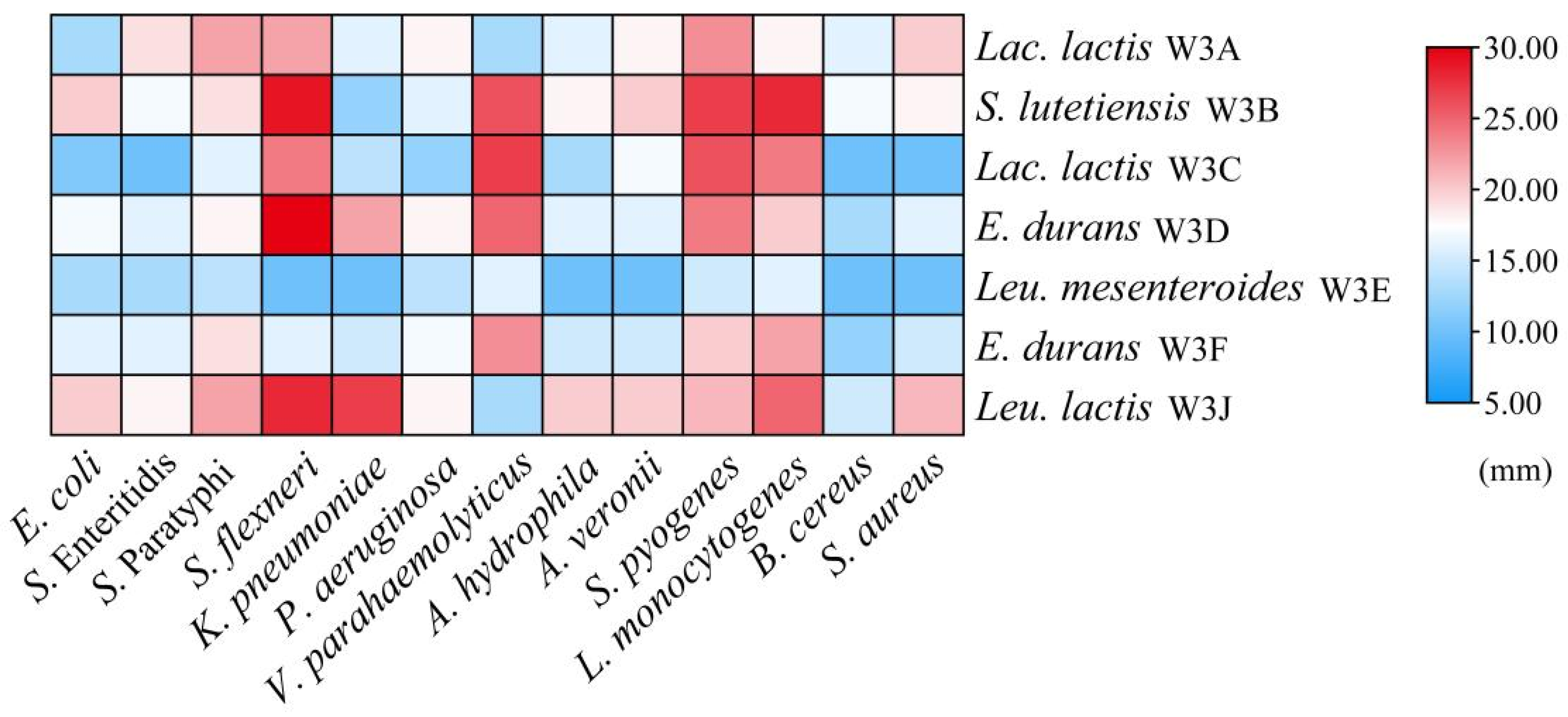
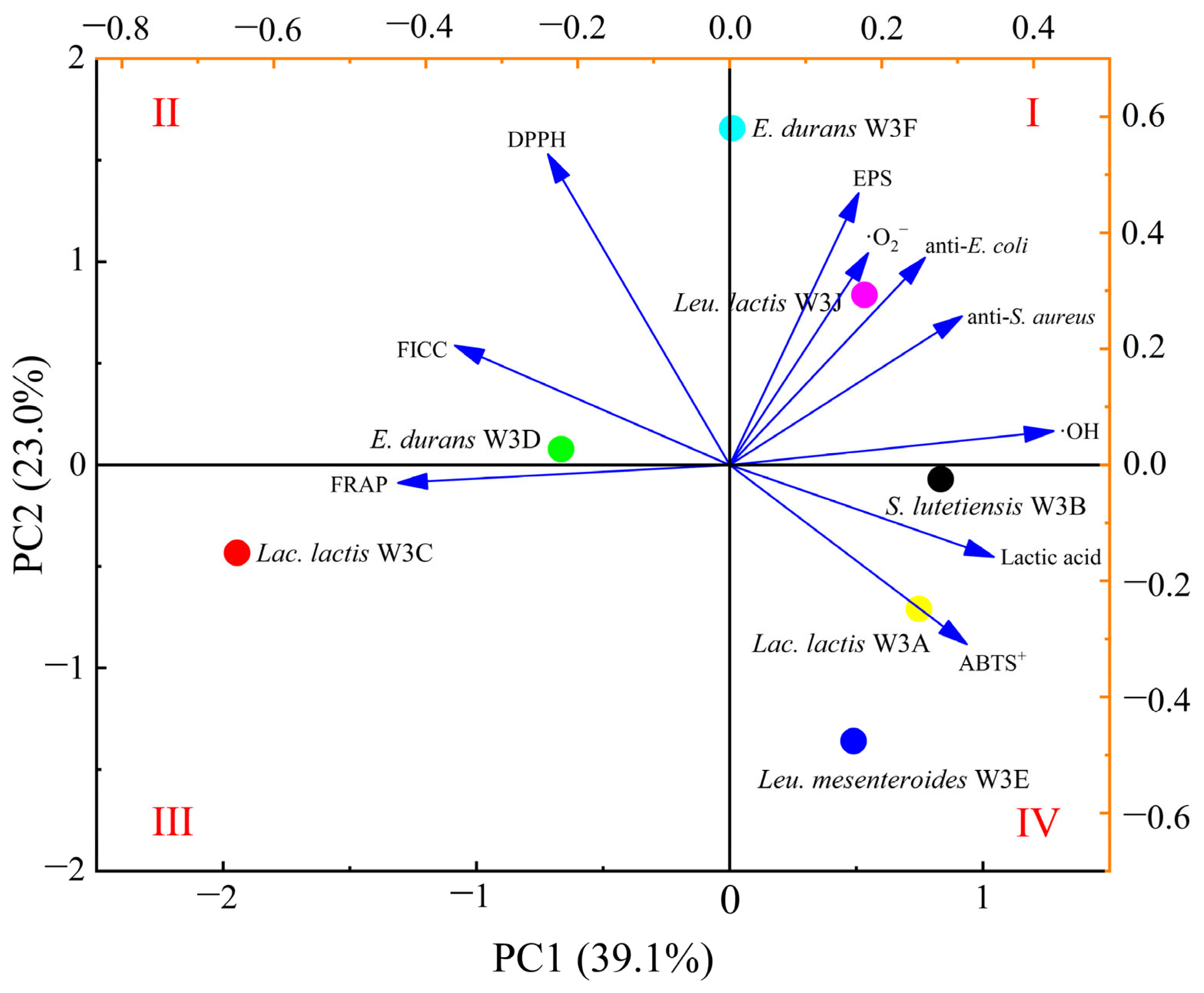
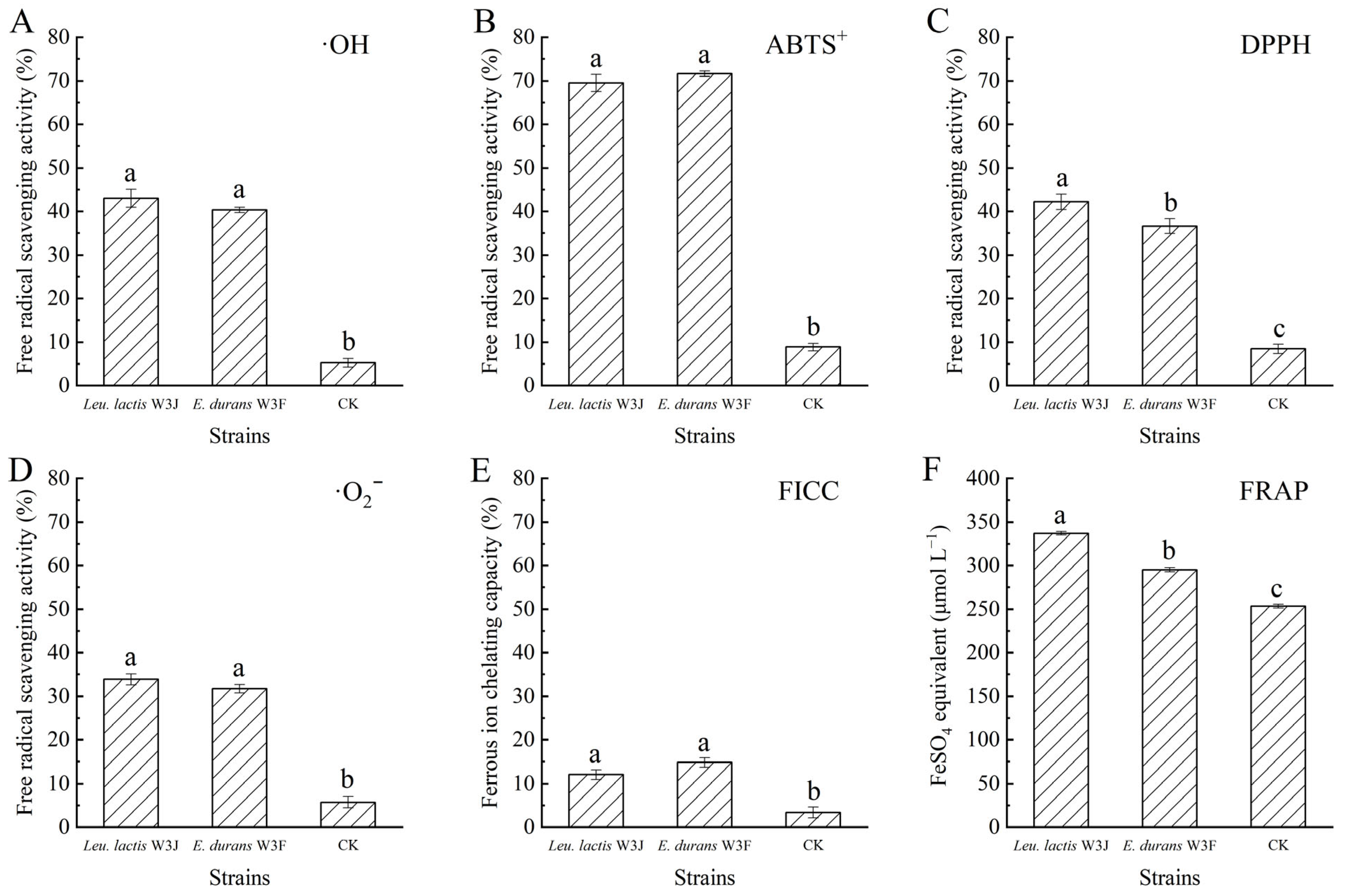
| Strains | ·OH | ABTS+ | DPPH | ·O2− | FICC (%) | FRAP (μmol L−1) |
|---|---|---|---|---|---|---|
| Radical Scavenging Activity (%) | ||||||
| Lac. lactis W3A | 92.69 ± 0.96 c | 98.67 ± 0.75 a | 44.78 ± 1.43 c | 15.99 ± 1.17 d | 28.49 ± 1.17 c | 65.11 ± 0.76 f |
| S. lutetiensis W3B | 95.26 ± 0.38 a | 96.43 ± 0.14 bc | 46.26 ± 0.60 c | 17.24 ± 1.23 d | 25.15 ± 0.95 d | 35.56 ± 1.42 g |
| Lac. lactis W3C | 87.56 ± 0.22 d | 95.86 ± 0.30 c | 51.76 ± 0.38 ab | 11.88 ± 1.27 e | 54.70 ± 1.24 a | 266.83 ± 1.57 b |
| E. durans W3D | 86.06 ± 0.09 e | 96.81 ± 0.29 b | 50.64 ± 1.35 b | 20.26 ± 1.40 c | 25.78 ± 1.16 d | 273.14 ± 1.36 a |
| Leu. mesenteroides W3E | 93.94 ± 0.19 b | 98.09 ± 0.53 a | 37.72 ± 0.58 d | 24.41 ± 1.33 b | 23.98 ± 1.10 d | 98.66 ± 0.17 d |
| E. durans W3F | 93.05 ± 0.03 c | 96.67 ± 0.34 b | 53.13 ± 0.36 a | 31.30 ± 0.41 a | 38.81 ± 1.60 b | 79.53 ± 0.03 e |
| Leu. lactis W3J | 94.44 ± 0.08 b | 96.94 ± 0.65 b | 52.13 ± 1.45 ab | 22.00 ± 1.06 c | 39.44 ± 1.38 b | 141.53 ± 0.62 c |
| Index | Ej | Dj | Wj |
|---|---|---|---|
| ·OH | 0.88 | 0.12 | 7.41% |
| ABTS+ | 0.84 | 0.16 | 9.46% |
| DPPH | 0.90 | 0.10 | 5.76% |
| FRAP | 0.79 | 0.21 | 12.43% |
| FICC | 0.71 | 0.29 | 17.29% |
| ·O2− | 0.86 | 0.14 | 8.41% |
| EPS | 0.83 | 0.17 | 10.35% |
| Lactic acid | 0.86 | 0.14 | 8.46% |
| Anti-S. aureus | 0.81 | 0.19 | 11.33% |
| Anti-E. coli | 0.85 | 0.15 | 9.12% |
| Strains | D+ | D− | Ci | Ranking |
|---|---|---|---|---|
| Lac. lactis W3A | 0.592 | 0.478 | 0.447 | 4 |
| S. lutetiensis W3B | 0.682 | 0.44 | 0.392 | 6 |
| Lac. lactis W3C | 0.662 | 0.58 | 0.467 | 3 |
| E. durans W3D | 0.619 | 0.444 | 0.417 | 5 |
| Leu. mesenteroides W3E | 0.706 | 0.361 | 0.338 | 7 |
| E. durans W3F | 0.496 | 0.558 | 0.53 | 2 |
| Leu. lactis W3J | 0.406 | 0.597 | 0.595 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, X.; Lin, L.; Zhu, W.; Zhang, X.; Xiong, J.; Gao, Y. Biological Activities of Lactic Acid Bacteria Isolated from Chinese Traditional Cheese and the Application in Antioxidant Foods. Microorganisms 2025, 13, 2743. https://doi.org/10.3390/microorganisms13122743
Lou X, Lin L, Zhu W, Zhang X, Xiong J, Gao Y. Biological Activities of Lactic Acid Bacteria Isolated from Chinese Traditional Cheese and the Application in Antioxidant Foods. Microorganisms. 2025; 13(12):2743. https://doi.org/10.3390/microorganisms13122743
Chicago/Turabian StyleLou, Xiangdi, Liping Lin, Wenwu Zhu, Xiaochen Zhang, Jianhua Xiong, and Yanyan Gao. 2025. "Biological Activities of Lactic Acid Bacteria Isolated from Chinese Traditional Cheese and the Application in Antioxidant Foods" Microorganisms 13, no. 12: 2743. https://doi.org/10.3390/microorganisms13122743
APA StyleLou, X., Lin, L., Zhu, W., Zhang, X., Xiong, J., & Gao, Y. (2025). Biological Activities of Lactic Acid Bacteria Isolated from Chinese Traditional Cheese and the Application in Antioxidant Foods. Microorganisms, 13(12), 2743. https://doi.org/10.3390/microorganisms13122743





