Cytotoxic and Antibacterial Angucycline- and Prodigiosin- Analogues from the Deep-Sea Derived Streptomyces sp. SCSIO 11594
Abstract
:1. Introduction
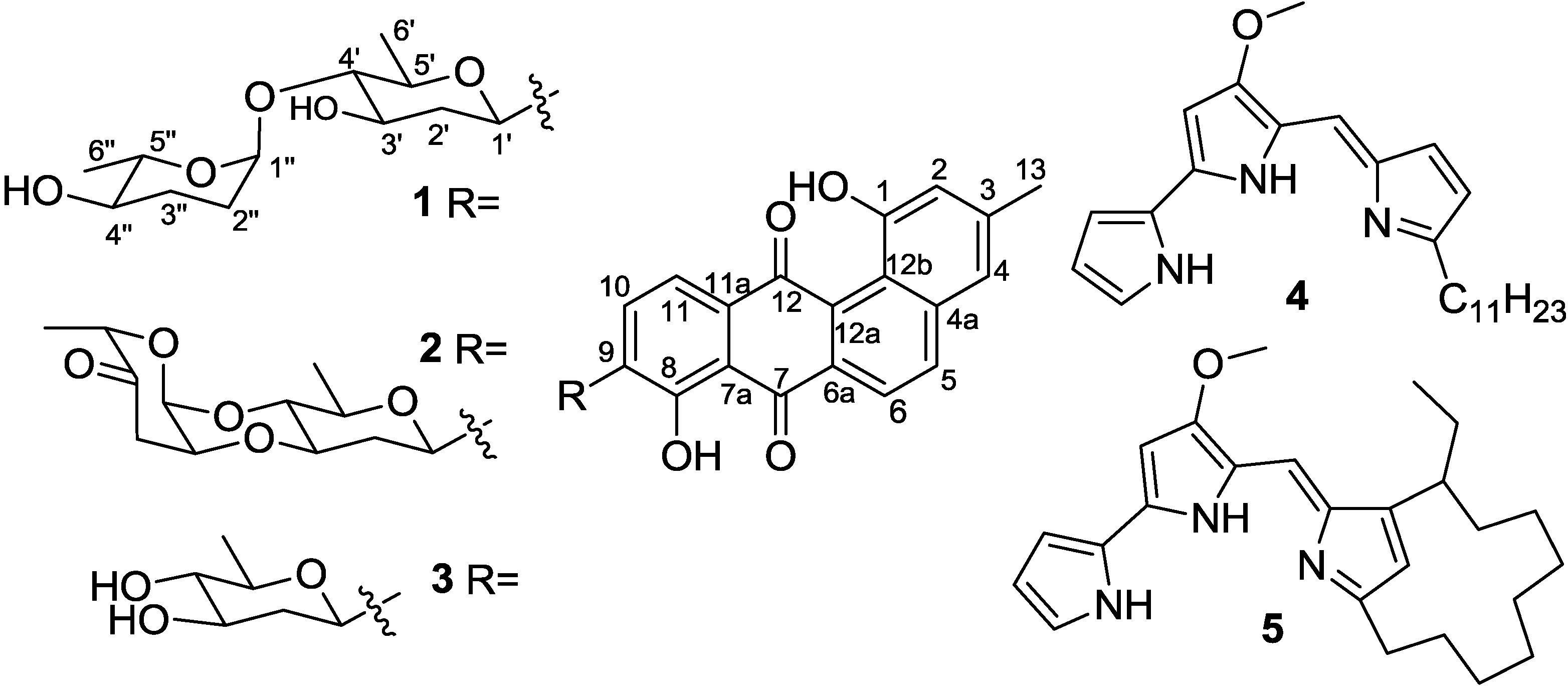
2. Results and Discussion
2.1. Structure Elucidation
| pos. | Marangucycline A (1) | Marangucycline B (2) | ||
|---|---|---|---|---|
| δC | δH, mult. (J in Hz) | δC | δH, mult. (J in Hz) | |
| 1 | 155.6, C | 155.3, C | ||
| 2 | 120.2, CH | 7.12, s | 120.2, CH | 7.15, s |
| 3 | 142.1, C | 142.1, C | ||
| 4 | 121.5, CH | 7.23, s | 121.3, CH | 7.27, s |
| 4a | 132.6, C | 132.5, C | ||
| 5 | 137.7, CH | 8.11, d, J = 8.5 | 137.6, CH | 8.14, d, J = 8.5 |
| 6 | 122.0, CH | 8.29, d, J = 8.5 | 121.8, CH | 8.32, d, J = 8.5 |
| 6a | 135.0, C | 134.8, C | ||
| 7 | 188.3, C | 188.3, C | ||
| 7a | 114.2, C | 114.1, C | ||
| 8 | 158.2, C | 157.8, C | ||
| 9 | 138.6, C | 137.7, C | ||
| 10 | 133.6, CH | 7.90, d, J = 8.0 | 133.6, CH | 7.92, d, J = 8.0 |
| 11 | 121.3, CH | 7.86, d, J = 8.0 | 121.2, CH | 7.88, d, J = 8.0 |
| 11a | 133.6, C | 133.5, C | ||
| 12 | 189.6, C | 189.4, C | ||
| 12a | 139.3, C | 139.2, C | ||
| 12b | 120.2, C | 120.1, C | ||
| 13 | 21.4, CH3 | 2.48, s | 21.3, CH3 | 2.50, s |
| 1-OH | 11.43, br s | 11.38, br s | ||
| 8-OH | 12.62, br s | 12.66, br s | ||
| 1′ | 71.3, CH | 4.90, d, J = 11.2 | 71.5, CH | 5.01, d, J = 11.0 |
| 2′ | 38.8, CH2 | 2.57, m; 1.46, m | 36.6, CH2 | 2.48, m; 1.54, m |
| 3′ | 71.5, CH | 3.87, m | 77.1, CH | 3.84, ddd, J = 11.5, 9.0, 4.5 |
| 4′ | 89.2, CH | 3.07, t, J = 6.5 | 74.5, CH | 3.52, t, J = 9.0 |
| 5′ | 74.7, CH | 3.57, m | 74.6, CH | 3.59, m |
| 6′ | 18.6, CH3 | 1.38, d, J = 6.0 | 17.5, CH3 | 1.43, d, J = 6.0 |
| 1″ | 98.9, CH | 4.92, br s | 91.4, CH | 5.19, d, J = 3.0 |
| 2″ | 27.3, CH2 | 1.93, m; 1.83, m | 71.1, CH | 4.35, q, 3.0 |
| 3″ | 30.1, CH2 | 1.87, m; 1.25, m | 39.9, CH2 | 2.65, m |
| 4″ | 71.8, CH | 3.36, td, J = 10.0, 4.0 | 207.7, C | |
| 5″ | 71.7, CH | 3.91, m | 77.8, CH | 4.75, q, J = 6.5 |
| 6″ | 18.0, CH3 | 1.33, d, J = 6.0 | 16.2, CH3 | 1.39, d, J = 6.5 |
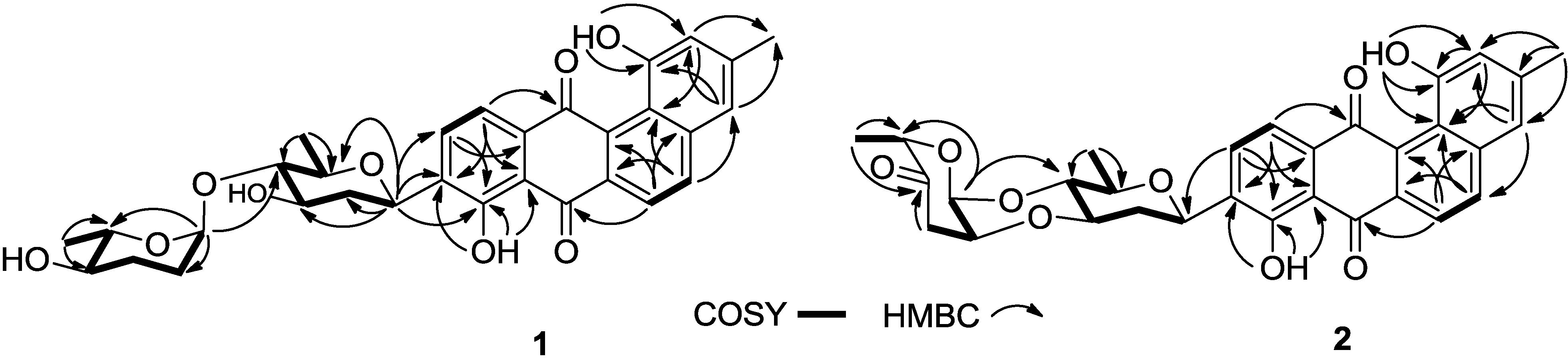
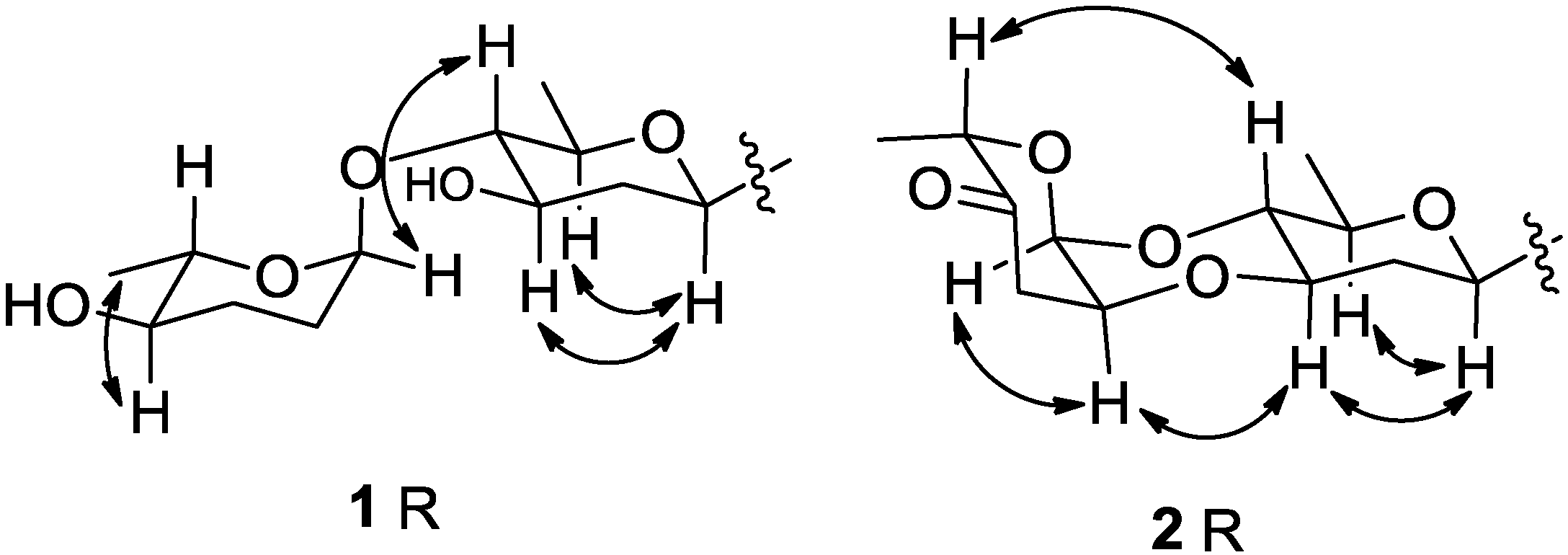
2.2. Cytotoxicities and Antibacterial Activities
| Agent | A549 | CNE2 | MCF-7 | HepG2 | HL7702 | TR |
|---|---|---|---|---|---|---|
| 1 | >50.0 | >50.0 | >50.0 | >50.0 | >50.0 | - |
| 2 | 0.45 ± 0.03 | 0.56 ± 0.02 | 0.24 ± 0.003 | 0.43 ± 0.05 | 3.67 ± 0.07 | 7–15 |
| 3 | 16.40 ± 0.19 | 22.27 ± 0.07 | 23.65 ± 0.09 | 18.81 ± 0.12 | 49.34 ± 0.17 | 2–3 |
| 4 | 0.85 ± 0.01 | 0.28 ± 0.02 | 1.11 ± 0.07 | 4.67 ± 0.09 | 12.47 ± 0.09 | 3–45 |
| 5 | >50.0 | >50.0 | >50.0 | >50.0 | >50.0 | - |
| Cisplatin | 4.56 ± 0.04 | 3.75 ± 0.03 | 5.26 ± 0.07 | 4.14 ± 0.06 | 15.34 ± 0.08 | 3–4 |
2.3. Identification of Strain SCSIO 11594


3. Experimental Section
3.1. General Experimental Procedures
3.2. Fermentation, Extraction and Isolation of the Compounds
3.3. Cytotoxicity Assays
3.4. Antibacterial Activities Assay
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xiong, Z.Q.; Wang, J.F.; Hao, Y.Y.; Wang, Y. Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Eom, S.H.; Kim, Y.M.; Kim, S.K. Marine bacteria: Potential sources for compounds to overcome antibiotic resistance. Appl. Microbiol. Biotechnol. 2013, 97, 4763–4773. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.; Wong, J.H.; Pan, W.L.; Chan, Y.S.; Yin, C.M.; Dan, X.L.; Wang, H.X.; Fang, E.F.; Lam, S.K.; Ngai, P.H.; et al. Antifungal and antiviral products of marine organisms. Appl. Microbiol. Biotechnol. 2014, 98, 3475–3494. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D. Deep-sea natural products. Nat. Prod. Rep. 2008, 25, 1131–1166. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006, 9, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Zotchev, S.B. Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 2012, 158, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Huang, H.; Chen, Y.; Tan, J.; Song, Y.; Zou, J.; Tian, X.; Hua, Y.; Ju, J. Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 2012, 75, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yao, Y.; He, Z.; Yang, T.; Ma, J.; Tian, X.; Li, Y.; Huang, C.; Chen, X.; Li, W.; et al. Antimalarial β-carboline and indolactam alkaloids from Marinactinospora thermotolerans, a deep sea isolate. J. Nat. Prod. 2011, 74, 2122–2127. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, Q.; Liu, X.; Chen, Y.C.; Zhang, Y.; Sun, A.; Zhang, W.; Zhang, J.; Ju, J. Cyclic hexapeptides from the deep South China Sea-derived Streptomyces scopuliridis SCSIO ZJ46 active against pathogenic Gram-positive Bacteria. J. Nat. Prod. 2014, 77, 1937–1941. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, H.; Li, J.; Song, Y.; Jiang, R.; Liu, J.; Zhang, S.; Hua, Y.; Ju, J. New anti-infective cycloheptadepsipeptide congeners and absolute stereochemistry from the deep sea-derived Streptomyces drozdowiczii SCSIO 10141. Tetrahedron 2014, 70, 7795–7801. [Google Scholar] [CrossRef]
- Supong, K.; Thawai, C.; Suwanborirux, K.; Choowong, W.; Supothina, S.; Pittayakhajonwut, P. Antimalarial and antitubercular C-glycosylated benz[α]anthraquinones from the marine-derived Streptomyces sp. BCC45596. Phytochem. Lett. 2012, 5, 651–656. [Google Scholar] [CrossRef]
- Sezaki, M.; Kondo, S.; Maeda, K.; Umezawa, H.; Ono, M. The structure of aquayamycin. Tetrahedron 1970, 26, 5171–5190. [Google Scholar] [CrossRef] [PubMed]
- Kuntsmann, M.P.; Mitscher, L.A. The structural characterization of tetrangomycin and tetrangulol. J. Org. Chem. 1966, 31, 2920–2925. [Google Scholar] [CrossRef] [PubMed]
- Rohr, J.; Thiericke, R. Angucycline group antibiotics. Nat. Prod. Rep. 1992, 9, 103–137. [Google Scholar] [CrossRef] [PubMed]
- Oki, T.; Kitamura, I.; Matsuzawa, Y.; Shibamoto, N.; Ogasawara, T.; Yoshimoto, A.; Inui, T.; Naganawa, H.; Takeuchi, T.; Umezawa, H. Anti-tumor anthracycline antibiotics, aclacinomycin-A and analogs. 2. structural determination. J. Antibiot. 1979, 32, 801–819. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, T.; Ren, X.; Liu, J.; Song, Y.; Sun, A.; Ma, J.; Wang, B.; Zhang, Y.; Huang, C.; et al. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 2012, 75, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Cui, C.B.; Duan, L.; Gu, Q.Q.; Zhu, W.M. Potent in vitro anticancer activity of metacycloprodigiosin and undecylprodigiosin from a sponge-derived actinomycete Saccharopolyspora sp. nov. Arch. Pharm. Res. 2005, 28, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, M.S.; Kharel, M.K.; Hitron, J.A.; Baig, I.; Rohr, J. Moromycins A and B, isolation and structure elucidation of C-glycosylangucycline-type antibiotics from Streptomyces sp. KY002. J. Nat. Prod. 2008, 71, 1569–1573. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Tanaka, H.; Oiwa, R.; Awaya, J.; Masuma, R.; Tanaka, K. New antitumor antibiotics, OS-4742 A1, A2, B1 and B2 produced by a strain of Streptomyces. J. Antibiot. 1977, 30, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Imamura, N.; Kakinuma, K.; Ikekawa, N.; Tanaka, H.; Omura, S. The structure of vineomycin B2. J. Antibiot. 1981, 34, 1517–1518. [Google Scholar] [CrossRef] [PubMed]
- Imamura, N.; Kakinuma, K.; Ikekawa, N.; Tanaka, H.; Omura, S. Biosynthesis of vineomycins A1 and B2. J. Antibiot. 1982, 35, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Morton, G.E.; Barrett, A.G.M. Iterative benzyne-furan cycloaddition reactions: Studies toward the total synthesis of ent-Sch 47554 and ent-Sch 47555. Org. Lett. 2006, 8, 2859–2861. [Google Scholar] [CrossRef] [PubMed]
- Sobti, A.; Kim, K.J.; Sulikowski, G.A. Application of glycosyltetrazoles in oligosaccharide synthesis: Assembly of the C3 trisaccharide component of the antibiotic PI-080. J. Org. Chem. 1996, 61, 6–7. [Google Scholar] [CrossRef]
- Yu, X.; O’Doherty, G.A. De novo asymmetric synthesis and biological evaluation of the trisaccharide portion of PI-080 and vineomycin B2. Org. Lett. 2008, 10, 4529–4532. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.M.; Li, M.S.; O’doherty, G.A. De novo asymmetric approach to the disaccharide portion of Sch-47554. Heterocycles 2011, 82, 1577–1584. [Google Scholar]
- You, Z.Q.; Li, J.; Qin, S.; Tian, X.P.; Wang, F.Z.; Zhang, S. Georgenia sediminis sp. nov. a moderately thermophilic actinobacterium isolated from sediment. Int. J. Syst. Evol. Microbiol. 2013, 63, 4243–4247. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard-8th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009; pp. M07–A8. [Google Scholar]
- Song, Y.; Huang, H.; Chen, Y.; Ding, J.; Zhang, Y.; Sun, A.; Zhang, W.; Ju, J. Cytotoxic and antibacterial marfuraquinocins from the deep South China Sea-derived Streptomyces niveus SCSIO 3406. J. Nat. Prod. 2013, 76, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.; Liu, G.; Li, J.; Huang, H.; Zhang, X.; Zhang, H.; Ju, J. Cytotoxic and Antibacterial Angucycline- and Prodigiosin- Analogues from the Deep-Sea Derived Streptomyces sp. SCSIO 11594. Mar. Drugs 2015, 13, 1304-1316. https://doi.org/10.3390/md13031304
Song Y, Liu G, Li J, Huang H, Zhang X, Zhang H, Ju J. Cytotoxic and Antibacterial Angucycline- and Prodigiosin- Analogues from the Deep-Sea Derived Streptomyces sp. SCSIO 11594. Marine Drugs. 2015; 13(3):1304-1316. https://doi.org/10.3390/md13031304
Chicago/Turabian StyleSong, Yongxiang, Guangfu Liu, Jie Li, Hongbo Huang, Xing Zhang, Hua Zhang, and Jianhua Ju. 2015. "Cytotoxic and Antibacterial Angucycline- and Prodigiosin- Analogues from the Deep-Sea Derived Streptomyces sp. SCSIO 11594" Marine Drugs 13, no. 3: 1304-1316. https://doi.org/10.3390/md13031304
APA StyleSong, Y., Liu, G., Li, J., Huang, H., Zhang, X., Zhang, H., & Ju, J. (2015). Cytotoxic and Antibacterial Angucycline- and Prodigiosin- Analogues from the Deep-Sea Derived Streptomyces sp. SCSIO 11594. Marine Drugs, 13(3), 1304-1316. https://doi.org/10.3390/md13031304








