- Article
Construction of a Eukaryotic Algae-Specific Metagenomic Classification Database and Its Application to Tara Oceans Data
- Xubing Xie,
- Mingjiang Wu and
- Shengqin Wang
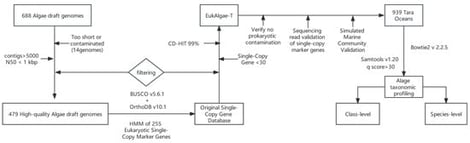
Eukaryotic algae are key contributors to biodiversity and ecosystem functioning in aquatic environments. However, understanding their global diversity patterns and community assembly mechanisms remains limited by the lack of high-resolution, highly specific analytical methods. Here, we present the first eukaryotic algae-specific classification database, EukAlgae-T (Eukaryotic Algae Taxonomic database), constructed based on single-copy orthologous genes. The database integrates 50,581 non-redundant marker genes from 479 high-quality genomes and was applied to analyze 939 marine metagenomic samples from the Tara Oceans project. Our results reveal that the genomically represented fraction of the global algal community is dominated by a widely distributed core taxonomic group, comprising 125 of the 230 detected species. Redundancy analysis indicated that community structure is primarily regulated by latitude and iron concentration on a global scale. In contrast, Mantel tests revealed strong regional heterogeneity, with temperature, salinity, and iron concentration acting as universal local drivers, albeit with varying effect sizes and combinations across ocean basins. Co-occurrence network analysis further demonstrated predominantly cooperative interactions among taxa, forming a highly modular and stable network structure, and identified key hub taxa characterized by low abundance but high connectivity. Together, this study provides a dedicated framework for eukaryotic algae metagenomic analysis and demonstrates that algal community assembly is driven by multi-scale environmental filtering: broadly constrained by climate zones and iron limitation at the global scale and regionally reshaped by local oceanographic processes (e.g., thermohaline structure). Cooperative coexistence and niche differentiation among taxa jointly underpin the maintenance of global algal diversity.
9 February 2026