Abstract
The increasing demand for industrial enzymes calls for cost-effective and sustainable production strategies. This study investigates the potential of industrial wastewater as an alternative fermentation medium for enzyme synthesis, aligning with the principles of the circular bioeconomy. Four wastewater types from Québec, Canada—beverage wastewater (BW), pulp and paper mill activated sludge (PPMS), food industry wastewater (FIW), and starch industry wastewater (SIW)—were evaluated for their potential to support protease, amylase, and lipase production using Bacillus licheniformis, Bacillus amyloliquefaciens, and Bacillus megaterium. Initial screening identified SIW as optimal for amylase production with B. amyloliquefaciens, and PPMS for protease production with B. megaterium. Optimization using the Box–Behnken design was then performed, followed by scale-up experiments in 5 L bioreactors. B. amyloliquefaciens achieved 5.73 ± 0.01 U/mL of amylase at 48 h under 40 g/L total solids, 30 °C, and a 2% inoculum size, while B. megaterium produced the highest protease of 55.41 ± 3.54 U/mL at 24 h. Lipase production remained negligible across all media and strains. These findings demonstrate the feasibility of the potential of wastewater-based enzyme production, reducing reliance on expensive synthetic substrates, mitigating environmental burdens, and contributing to the transition to a circular bioeconomy.
1. Introduction
One of the pressing environmental concerns today is the increasing volume of waste in both solid and liquid phases. This issue stems from the traditional linear economic model and the rapidly expanding global population [1,2]. Waste originates from various sectors and can be broadly classified into industrial production and household consumption [3,4]. The continuous rise in organic waste production highlights the urgent need for a sustainable waste management system to reduce environmental pollution and greenhouse gas emissions [5,6,7].
Recently, a roadmap for achieving a circular and sustainable bioeconomy has been introduced to address these grand challenges by leveraging innovative technologies to convert bioresources into renewable energy and high-value-added products [3,8]. The success of the transition to a bioeconomy depends on advancements in technology, cost-effectiveness, and the availability of sustainable biomass [9]. In this context, industrial wastewater and sludge—such as starch wastewater, beverage wastewater, and pulp and paper mill activated sludge, etc., which are rich in diverse nutrients and components—have emerged as valuable renewable substrates for ensuring the availability of sustainable biomass [10].
In contrast to conventional treatment plants, which primarily treat wastewater to reduce organic and suspended solid loads for discharge compliance, various wastewater sources can instead be valorized into value-added products such as fertilizers, biofuels, biopesticides and enzymes, etc. [10,11]. However, in practice, effectively addressing the heterogeneity of biowaste and the presence of unwanted chemicals to maximize productivity requires further comprehensive studies adopting a sector-specific approach.
The valorization of organic wastes for enzyme production exemplifies a closed-loop process within the bioeconomy, where microbes transform organic residues into high-value enzymes. These enzymes are then utilized across various industrial sectors—such as biofuel production, food processing, pharmaceuticals, and bioremediation [12]—contributing to resource efficiency, waste minimization, and circularity by reintegrating biological materials into the production cycle.
Different sets of enzymes can be obtained successfully through microbial submerged fermentation, which is well suited to the liquid nature of wastewater. Among the most important industrial enzyme producers, members of the genus Bacillus are microorganisms of choice for their simple, scalable cultivation, supported by well-studied biochemistry, physiology, and genome [13]. Their ability to efficiently export metabolites across the cytoplasmic membrane, including proteases, α-amylase, lipase, cellulase, etc. [11,14,15,16], simplifies downstream processing compared to bacteria that produce proteins intracellularly [17]. Notably, as their fermentative metabolism enables the rapid utilization of a broad range of substrates [18], Bacillus spp. emerge as key players in zero-waste initiatives and green manufacturing.
Given the increasing interest in harnessing diverse industrial wastewater streams for biotechnological applications, selecting the most suitable substrate for a specific purpose requires a carefully designed and systematic strategy. This study introduces a pragmatic approach to evaluating the enzymatic potential of industrial wastewaters by employing well-characterized commercial bacterial strains during the initial screening phase. Utilizing these strains accelerates the screening process, ensures reproducible enzyme production, and provides a standardized platform for comparing diverse waste streams. This approach is particularly valuable in cases where the native microbial community is limited or where the physicochemical properties of the wastewater hinder effective microbial isolation. Although a potential drawback of using defined commercial strains is the risk of underestimating the full bioconversion capacity of certain wastes, this trade-off is a deliberate and calculated aspect of the research design. Crucially, it is addressed in subsequent stages, where the most promising waste streams can be revisited using native isolates or strains more suitable for targeted optimization. Importantly, the scalability of this strategy has been demonstrated in 5 L bioreactors—even when using selected commercial strains—as effective waste–strain combinations identified during screening can be readily adapted for larger-scale bioprocess applications. This initial phase of the stepwise framework enables systematic substrate evaluation, setting the stage for future strain optimization and scalable enzyme production.
2. Materials and Methods
2.1. Industrial Wastewater Sources
Drawing from literature highlighting the capacity of Bacillus sp. to produce a wide range of enzymes using agro-industrial, brewery, and starch-based waste substrates [19,20,21], four distinct industrial wastewater samples were collected from local facilities in Québec, Canada, to evaluate their potential as fermentation media: beverage wastewater (BW) from Brasserie Rural (Cookshire-Eaton, QC, Canada), pulp and paper mill activated sludge (PPMS) from Kruger Inc. (Trois-Rivières, QC, Canada), food industry wastewater (FIW) from Diana Food Canada Inc. (Champlain, QC, Canada) (fruit processing from cranberries, blueberries, and strawberries) (Champlain), and starch industry wastewater (SIW) from ADM-Ogilvie (Candiac, QC, Canada). These samples were collected directly from the effluent output points prior to any tertiary treatment. In all cases, a basic pretreatment stage was performed, consisting of coarse screening to remove large particulates and debris. Immediately after collection, the samples were stored in sterile containers, kept at 4 °C, and transported to the laboratory within 24 h. Upon arrival, their pH was measured (ranging from 2.45 to 6.65), and samples were either processed immediately or stored at 4 °C for no more than 72 h prior to use in fermentation experiments.
To characterize their suitability for microbial growth and enzyme production, a suite of analytical techniques was employed. Total organic carbon (TOC) and total nitrogen (TN) were quantified using a Shimadzu VCPH analyzer, calibrated with standard solutions to ensure precision within ±2% error. This provided insight into the carbon and nitrogen pools available for bacterial metabolism. Mineral composition—including phosphorus, sodium, iron, potassium, calcium, and sulfur—was determined via inductively coupled plasma optical emission spectrometry (ICP-OES) using an ICP-5110 Dual View spectrometer (Agilent Technologies, Santa Clara, CA, USA), with calibration against multi-element standards to achieve detection limits below 0.01 mg/L. Sugar profiles (glucose, fructose, lactose, sucrose, galactose, xylose, and trehalose) were analyzed using high-performance liquid chromatography (HPLC) equipped with a refractive index detector, employing an Aminex HPX-87H column and a mobile phase of 5 mM H2SO4 at 0.6 mL/min. These measurements elucidated the nutrient diversity and concentration, critical for fermentation conditions to specific enzyme production goals.
2.2. Microorganisms
Three industrially relevant Bacillus strains were selected for their enzymatic versatility: Bacillus licheniformis ATCC 14580, known for producing amylase, protease, and lipase; Bacillus amyloliquefaciens ATCC 23842, recognized for high amylase and protease yields; and Bacillus megaterium (laboratory strain), capable of protease and lipase synthesis. These strains were chosen for their ability to secrete enzymes extracellularly, facilitating recovery and reducing downstream processing costs [13].
2.3. Inoculum Preparation
The pre-seed followed a two-stage protocol to ensure optimal bacterial adaptation and vigor. Initially, a loopful of bacterial culture was transferred from an agar plate to 10 mL of nutrient broth (10 g/L peptone, 3 g/L beef extract, 5 g/L NaCl, pH 7) in a 50 mL Erlenmeyer shake flask. The inoculated flask was incubated at 30 °C with shaking at 180 rpm for 16–18 h, reaching the late exponential phase (approximately 108 CFU/mL for B. licheniformis and B. megaterium, 107 CFU/mL for B. amyloliquefaciens). Five (5) mL of this culture was subsequently transferred to 250 mL of the respective sterilized wastewater in a 1 L flask and incubated under identical conditions to acclimate the bacteria to the wastewater’s nutrient profile, enhancing their metabolic readiness for fermentation.
In this study, all wastewaters were sterilized in an autoclave for 15 min at 121 °C before any experiments.
2.4. Screening of Wastewater Sources for Enzyme Production
Screening experiments assessed the wastewaters’ capacity to support enzyme production by culturing each Bacillus strain in 200 mL of BW, PPMS, FIW, or SIW, alongside a nutrient broth control. Cultures were maintained at 30 °C, pH 7, and 180 rpm for 48 h in an orbital shaker. Post-incubation, samples were analyzed for bacterial growth (CFU/mL) and enzyme activities (protease, amylase, lipase), providing a basis for selecting the most promising strain–wastewater combinations.
2.5. Growth Conditions
Bacterial growth was quantified by serial dilution in 0.8% (w/v) saline, followed by spread plating 100 μL aliquots onto nutrient agar plates. After incubation at 30 °C for 24 h, colonies were enumerated to calculate CFU/mL, with all measurements conducted in duplicate to ensure reproducibility (standard deviation <5%).
2.6. Total Solids (TSs) and Total Volatile Solids (TVSs)
A well-homogenized aliquot of 25 to 50 mL of the sample was transferred into a pre-weighed porcelain crucible. The sample was dried at 105 °C for 24 h in a drying oven to remove moisture. The crucible was then cooled in a desiccator and weighed to determine total solids (TSs).
To determine total volatile solids (TVSs), the dried sample was further combusted at 550 °C for 2 h in a muffle furnace. The crucible was again cooled in a desiccator and weighed. TVSs were calculated as the weight loss between 105 °C and 550 °C.
TSs (g/L) = (weight after 105 °C drying − tare weight)/volume of sample (L)
TVSs (g/L) = (weight after 105 °C drying − weight after 550 °C ashing)/volume of sample (L)
2.7. Suspended Solids (SSs) and Suspended Volatile Solids (SVSs)
To quantify the suspended solids (SSs), a known volume (typically 25 to 100 mL) of the sample was filtered through a pre-weighed glass fiber filter (Whatman GF/C or equivalent). The filter containing the retained solids was dried at 105 °C for 24 h, cooled in a desiccator, and weighed.
For suspended volatile solids (SVSs), the dried filter was ashed at 550 °C for 2 h in a muffle furnace, cooled in a desiccator, and reweighed.
SSs (g/L) = (weight after 105 °C drying − tare weight of filter)/volume of sample (L)
SVSs (g/L) = (weight after 105 °C drying − weight after 550 °C ashing)/volume of sample (L)
All measurements were performed in triplicate, and results are expressed as mean ± standard deviation.
2.8. Protease Assay
Protease activity was measured using a casein hydrolysis method. A reaction was set up by mixing 1 mL of diluted enzyme with 1 mL of 1% casein in 50 mM Tris-HCl buffer (pH 7) and incubating it at 50 °C for 10 min. To stop the reaction, 2 mL of 15% trichloroacetic acid was added, and the mixture was centrifuged for 10 min at 10,000 rpm at 4 °C. After centrifugation, 0.5 mL of the supernatant was taken and mixed with 2.5 mL of 2% sodium carbonate and 0.25 mL of 1 N Folin’s reagent. The mixture was incubated at room temperature for 30 min. Absorbance was measured at 660 nm using a spectrophotometer. One unit of protease activity is the amount of enzyme that releases 1 μg of tyrosine under these conditions [22].
2.9. Lipase Assay
Lipase activity was determined using p-nitrophenyl palmitate (pNPP) as the substrate. The assay buffer was prepared by mixing 25 mL of 100 mM Tris-HCl (pH X), 5 mL of 100 mM CaCl2·2H2O, 0.15 mL of Triton X-100, and 19.85 mL of distilled water. To prepare the substrate solution, 1 mL of 20 mM pNPP was dissolved in 19 mL of the assay buffer. For the assay, 2.76 mL of the substrate solution was mixed with 0.24 mL of enzyme solution. The reaction was carried out at 30 °C for 30 min, and the release of p-nitrophenol was monitored by measuring the absorbance at 410 nm. One unit (U) of lipase activity was defined as the amount of enzyme that releases 1 μmol of p-nitrophenol per minute under the assay conditions [23].
2.10. Amylase Assay
Amylase activity was determined via the DNS method. The reaction mixture, containing 0.1 mL of appropriately diluted enzyme and 0.9 mL of 1.0% (w/v) corn starch in 50 mM Tris-HCl buffer (pH 7.0), was incubated at 50 °C for 10 min. The amount of reducing sugars liberated was determined using the dinitrosalicylic (DNS) acid method. After the reaction, 1.5 mL of DNS reagent was added, and the mixture was boiled for 10 min before being diluted with 5 mL of distilled water. The absorbance was then measured at 540 nm. One unit of α-amylase activity was defined as the amount of enzyme that released 1 µmol of reducing end groups per minute. A standard curve was plotted using D-glucose. The α-amylase activity is represented by the mean value of two determinations, each performed in duplicate [24].
2.11. Optimization of Fermentation Parameters Using Box–Behnken Design
The study of optimal conditions for enzyme production in 1 L shaking flasks was conducted using a set of 17 runs designed according to the Box–Behnken design. Three factors—total solids (TSs), inoculum size, and temperature—were investigated, with the design points positioned at the middle of the edges of the experimental domain, typically coded as −1, 0, and +1. This experimental approach efficiently models the response surface, requiring fewer experimental runs compared to traditional factorial techniques. Enzyme activities were measured as responses to the design, and a model for optimal conditions was developed. The optimal conditions determined from previous work at the flask level serve as the baseline for further optimization in a 5 L bioreactor.
The relationship between the studied variables and the resulting activities of enzymes will be considered via the following second-order polynomial equation:
where Y is the predicted enzyme yield (U/mL), β0 is the intercept (a constant), and βi, βii, and βij are regression coefficients for linear, squared, and interaction effects, respectively. Xi and Xj are the parameters under consideration. Data regression analysis was performed using Design Expert® (Version 7.0.0) software.
2.12. Bench Scale—5 Liter Bioreactors Fermentation
Scaling up protease production in a 5 L Sartorius Biostat B plus bioreactor represents a critical transition step in the research process, aimed at maintaining enzyme yield and activity for large-scale production.
The fermentation studies were conducted using a 5 L glass bioreactor with a working volume of 3 L, controlled via a programmable logic control (PLC) system to monitor key parameters such as dissolved oxygen (DO), pH, temperature, agitation, aeration rate, and antifoam (Figure 1). Nutrient concentrations of wastewater were optimized based on prior shake flask studies, and the wastewater media were autoclaved at 121 °C and 15 lbs/in2 pressure for 30 min to ensure sterility. Aseptic conditions were maintained during the transfer of the inoculum, which was incubated at 30 °C for 15 h. The selected microorganisms and wastewater media, based on earlier experiments, were cultured under specific conditions for each microorganism in the bioreactor. Dissolved oxygen levels were maintained at 30–50% by adjusting aeration and agitation. Samples were collected every 12 h to monitor progress, with the goal of achieving cell concentrations of 108 CFU/mL for B. megaterium, 107 CFU/mL for B. amyloliquefaciens, and optimizing enzyme production.
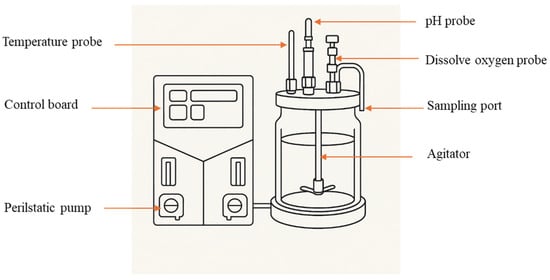
Figure 1.
Schematic diagram of the 5 L Sartorius bioreactor.
3. Results and Discussion
3.1. Characterization of Wastewaters
The nutrient profiles of the four wastewaters were pivotal in determining their suitability for enzyme production. SIW’s high sugar content (e.g., 31.9 g/L glucose, 11.4 g/L fructose, 9.1 g/L lactose) and balanced TOC (207 g C/L) and TON (17.5 g N/L) positioned it as an ideal substrate for amylase production, as Bacillus species efficiently metabolize sugars into amylolytic enzymes [25]. Its acidic pH (3.99) required neutralization to 7, aligning with the optimal range for Bacillus growth (pH 6.5–7.5). BW offered a nutrient-rich profile (TOC 345 g C/L, TON 11 g N/L), but its lower sugar levels suggested versatility rather than specificity for amylase synthesis. PPMS, with modest TOC (14 g C/L) but elevated calcium (22 g/L) and iron (0.4 g/L), supported protease production, as these minerals enhance enzyme stability and microbial metabolism [26,27]. FIW’s high TOC (387 g C/L) was offset by negligible nitrogen (0.339 g N/L) and phosphorus (0.039 g/L), resulting in a C/N ratio unfavorable for bacterial growth, rendering it impractical without supplementation (Table 1).

Table 1.
Wastewater characteristics.
3.2. Screening of Wastewater Sources for Potential Enzyme Production
Based on the characteristics of these wastewaters, only minimal pretreatment was required, primarily consisting of pH adjustment to 7 with sodium hydroxide following sterilization at 121 °C for 15 min. Additionally, 1% Tween 20 was supplemented in the media to enhance lipase production. This approach enables the evaluation of enzyme production potential while closely maintaining the original nature of the wastewater, offering insights into its feasibility as a fermentation medium with minimal modification.
3.2.1. Bacterial Growth on Wastewater
On nutrient broth, the number of B. licheniformis and B. megaterium cells reached approximately 108 CFU/mL after 24 h and maintained to 48 h, and that of B. amyloliquefaciens was slightly lower, but still around 106–107 CFU/mL (Figure 2).
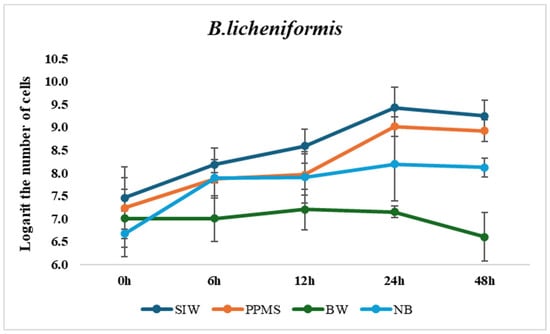
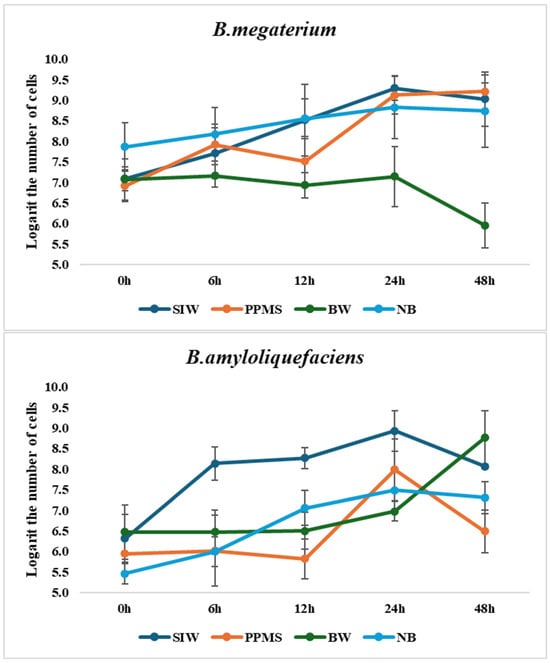
Figure 2.
Comparison of bacterial growth on various wastewaters and synthetic medium (NB).
FIW failed to support the growth of all strains, likely due to an imbalance in the carbon/nitrogen ratio leading to nitrogen starvation, which may inhibit bacterial growth while favoring yeast. Wastewaters with extremely unfavorable conditions require more complex treatment strategies to support effective microbial growth and enzyme production, such as mixing different wastewaters to adjust pH, compensate for nutrient deficiencies, and achieve a more balanced composition [28]. Therefore, the growth data for this wastewater were not included in the figure.
Although all three strains could grow on BW, their cell densities were lower. Notably, B. amyloliquefaciens exhibited an increased growth rate between 24 h and 48 h, suggesting its ability to utilize nutrients from this wastewater over a prolonged period. In contrast, the other strains showed minimal growth and a significant decline at 48 h.
The bacterial growth on SIW and PPMS was significantly comparable to that in synthetic medium, which means that these wastewater sources contain sufficient digestible nutrients for the division of cells, as nutrient availability and growth rate have a positive proportional relationship [29]. On both media, the peak cell densities of all three tested bacteria were observed at 24 h, with the exponential phase occurring between 12 and 24 h.
In conclusion, compared to the pure synthetic media, the growth of the studied strains on SIW and PPMS was basically similar, confirming that these types of wastewaters can provide enough digestible nutrients to support bacterial proliferation.
3.2.2. Enzyme Production Potential
After 48 h of incubation, Bacillus strains exhibited varying enzyme yields when cultivated on different types of wastewaters (Table 2). B. amyloliquefaciens was capable of producing both amylase and protease across all tested wastewaters, with amylase production particularly pronounced on SIW, reaching 4.26 U/mL alongside 11.64 U/mL of protease. This suggests its ability to synthesize amylase even under low-sugar conditions. In contrast, amylase activity from B. licheniformis and B. megaterium was detected mainly on SIW, indicating a more limited capacity for amylase production. While B. amyloliquefaciens did not exhibit any lipase activity in the tested media, B. licheniformis and B. megaterium showed minimal lipase production (0.01 U/mL) on PPMS. On the same medium, both strains also produced substantial amounts of protease, reaching 49.59 U/mL and 53.12 U/mL, respectively. On BW, only protease activity was detected in all three strains, while amylase production was observed exclusively in B. amyloliquefaciens. However, the enzyme levels were lower compared to those obtained on the other wastewater substrates, indicating a limited compatibility between this wastewater and the selected strains.

Table 2.
Enzyme activities on studied wastewaters.
In this study, B. licheniformis was selected for its potential in wastewater treatment and value-added applications, as its ability to produce novel enzymes and valuable bioactive compounds has attracted significant attention since the first report in 1945. To date, its unique genetic background and safety profile have made it highly relevant for biological applications in the food industry, pharmaceuticals, and bioremediation, resulting in the patenting of various B. licheniformis strains, along with associated methods and applications [12,30]. Evaluating its performance alongside other strains in wastewater treatment demonstrated the potential of SIW as an amylase- and protease-inducing medium, while PPMS specifically promotes protease production. Further studies with modifications may enhance the efficiency of B. licheniformis even further.
In conclusion, B. megaterium and B. licheniformis appear to be more efficient protease producers across various wastewater types. However, all strains had limited lipase production, suggesting that these studied wastewater sources are not suitable for selected Bacillus strains synthesizing lipase. Although BW shows high nutrient potential for enzyme production, SIW and PPMS appear to be more conducive to the desirable enzyme production, with higher enzyme yields observed in these media by the selected bacteria.
Based on the combined results of bacterial growth and enzyme activities, Bacillus amyloliquefaciens was selected to cultivate on SIW for amylase production, while Bacillus megaterium was chosen to ferment on PPMS for protease and lipase production. These strains were selected for their optimal enzyme production capabilities under the respective wastewater conditions.
3.3. Optimization of Enzyme Production
In the previous study, Bacillus megaterium demonstrated the ability to produce protease and minimal lipase when cultivated on PPMS and PPMS supplemented with Tween 20, respectively. To comprehensively assess this potential, a design-of-experiments approach was employed to identify key factors influencing protease and lipase production. A similar objective was pursued for Bacillus amyloliquefaciens on SIW. Among various response surface methodologies, the Box–Behnken design was chosen, considering three factors: temperature, total solids, and inoculum size. The Box–Behnken design was particularly advantageous due to its ability to reduce the number of experimental runs for a known bioprocess while maintaining three balanced levels, thereby avoiding extreme conditions unsuitable for bacterial growth and enzyme production. This statistical approach ensured the generation of a reliable dataset for model development, with a total of 17 experimental runs conducted across three enzyme production conditions.
Enzyme production is highly sensitive to cultural parameters, with various factors influencing yields to different extents depending on the bacterial strain and the specific enzyme produced. In this study, the primary goal is to develop a cost-effective enzyme production process by valorizing wastewater. Therefore, key variables and their relevant levels were selected based on their impact on both yield efficiency and economic feasibility, as well as their simplicity and practicality.
Temperature is one of the most critical physiological factors affecting microbial growth and enzyme production. It directly influences metabolic activity, enzymatic reaction rates, and microbial adaptability. Excessive temperatures can denature proteins and reduce bacterial viability, whereas temperatures that are too low slow down metabolism and enzyme synthesis. Furthermore, temperature regulation requires energy—higher temperatures demand more power, while lower temperatures may compromise microbial efficiency. Given these considerations, a moderate range of 30–37 °C suitable was chosen, as it supports optimal microbial activity while minimizing excessive energy costs. This range is commonly used in bioprocesses with Bacillus species due to its balance between efficiency and economic sustainability.
Nutritional factors, particularly media composition, play a key role in microbial fermentation. Instead of relying on pure chemical substrates, this study utilized low-cost industrial wastewaters as nutrient sources. However, careful selection was necessary to maximize nutrient availability while minimizing the impact of undesirable components. To support this, TSs were employed as an adjustment parameter, helping to ensure an efficient fermentation process without the need for additional chemical supplementation. As the wastewaters investigated in this study originated from established industrial operations, their quality could vary between batches, but typically within a manageable range. TSs thus provided a relatively stable and practical parameter for preliminary assessment. TSs reflect the sum of suspended and dissolved substances, encompassing both essential nutrients for microbial growth (such as organic carbon, nitrogen, and phosphorus) and potentially inhibitory compounds (such as phenolics and long-chain fatty acids). Therefore, the relationship between TSs, nutrient availability, and the presence of inhibitory substances was considered in selecting and optimizing wastewaters for enzyme production. In this study, optimization of TSs did not aim to eliminate undesirable substances, but rather to adjust the solids concentration to balance nutrient availability and toxicity risk. A TSs level that is too low may fail to support sufficient microbial growth due to nutrient limitation, whereas an excessively high TSs could introduce inhibitory effects that suppress enzyme production. Thus, adjusting TSs to an optimal range was critical for maximizing microbial performance and enzyme productivity while avoiding the need for further pretreatment, thereby enabling a more cost-effective and efficient valorization strategy.
Finally, inoculum size is another crucial factor, as it determines microbial adaptation to the fermentation environment and ultimately influences enzyme yields. This is particularly important in unconventional conditions such as wastewater-based fermentation, where microbial resilience plays a key role in process efficiency.
3.3.1. Optimization of Protease Production
Protease activity peaked at 42.56 U/mL (33.5 °C, 25 g/L TS, 5% inoculum) (Table 3). The comparison between predicted and experimental values showed no significant difference, indicating a good model fit and statistical significance (p ≤ 0.05) (Table 4).

Table 3.
Optimization of protease production by Bacillus megaterium on PPMS using Box–Behnken design.

Table 4.
Statistical analysis of Box–Benkhen design for protease production in Bacillus megaterium.
The first-order polynomial Equation (1) was fitted to the experimental protease activity, resulting in the following regression equation, which describes the relationship between the tested factors and enzyme production.
where A represents temperature, B represents total solids, and C represents inoculum size.
The results of the analysis indicate that the regression model is statistically significant, with a model F-value of 16.67 and only a 0.02% chance that this result could occur due to random noise. The lack-of-fit F-value of 3.11 suggests that the model fits the data well. Although the predicted R2 (0.5336) and adjusted R2 (0.6620) values are in reasonable agreement—indicating a consistent model fit—the moderate R2 values suggest that the model explains a fair portion of the variability in the response. This may limit its predictive accuracy to some extent and implies that additional influential factors may not have been captured in the current experimental design. Nonetheless, the Adeq precision value of 11.277 supports the model’s adequacy and reliability for guiding optimization within the design space.
The final linear equation highlights the relative positive impact of factors B and C on protease activity, with C having a stronger effect and factor A excluded due to its insignificance. The model is effective for predicting protease activity based on two out of three studied factors, and its linear nature suggests that no higher-order terms are needed for this analysis.
Since temperature had no significant effect on protease production, the lowest tested temperature was chosen to minimize energy consumption. Total solids and inoculum size positively affected the model; therefore, the maximum total solid concentration was maintained. However, the highest inoculum level was impractical for industrial applications, and the inoculum size was adjusted to 2% to ensure feasibility in large-scale operations.
The suggested conditions (30 °C, 25 g/L total solids, and 2% inoculum size) were tested in triplicate to validate the model. The experimental results closely aligned with the predicted values, confirming the model’s reliability despite the moderate predicted R2 value of 38.1 U/mL. Under these optimized conditions, Bacillus megaterium produced 39.6 ± 3.53 U/mL of protease, demonstrating the model’s effectiveness in predicting enzyme production.
3.3.2. Optimization of Lipase Production
As B. megaterium exhibited low lipase activity on PPMS alone, and non-ionic surfactants such as Tween are commonly used as low-cost inducers to stimulate bacterial lipase production [31], an additional step was included. Tween 20, containing medium-chain fatty acids, was selected for inclusion in the single-run optimization experiment, as the long-chain structure of Tween 80 had previously demonstrated a lower induction effect under the specific conditions of PPMS. In the presence of Tween 20 and Tween 80, the observed lipase activities were 0.07 ± 0.0012 U/mL and 0.02 ± 0.07 U/mL, respectively.
The Box–Behnken design (BBD) results revealed that maximum lipase production of 0.097 U/mL was achieved at 30 °C with 25 g/L total solids and 3,5% inoculum size in sludge. The correlation is expressed by the following equation:
where A represents temperature, B represents total solids, and C represents inoculum size.
The results indicate that, although A and B were reported to play a role in the model, the coefficients for both factors are relatively small, indicating that their individual effects on lipase production are modest. Additionally, factor C had no significant influence on the model, confirming that its exclusion does not affect predictive accuracy.
The model F-value of 47.16 indicates statistical significance, with only a 0.01% chance that such a high F-value occurred due to noise. The lack-of-fit F-value of 1.96 implies that the lack of fit is not significant relative to pure error, with a 26.93% chance that this large F-value could arise due to noise. A non-significant lack of fit is desirable, as it indicates the model fits the data well.
The predicted R2 of 0.7554 is in reasonable agreement with the adjusted R2 of 0.8964, with the difference being less than 0.2, which suggests a good model fit. Furthermore, the Adeq precision ratio of 23.323 indicates a high signal-to-noise ratio, confirming that the model can be reliably used to navigate the design space (Figure 3).
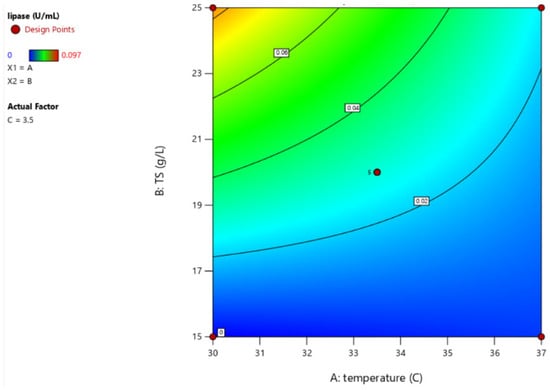
Figure 3.
Response surface of the optimal cultivation conditions for lipase production.
To confirm the statistical tests, while the significant factors—temperature (30 °C) and total solid concentration (25 g/L)—were maintained as per the model, the inoculum size was reduced to 2% from the initial 3.5%, as this factor had no significant impact on lipase production. This adjustment was made to improve the feasibility of large-scale optimization. Under these specific conditions, a lipase activity of 0.091 U/mL was recorded, showing an approximate 10% difference from the predicted value of 0.083 U/mL, confirming the model’s validity. The results from this study with PPMS, supplemented with 1% Tween 80, are quite similar to the findings for Bacillus megaterium AKG-1, which produced 0.116 U/mL of lipase in nutrient broth with added soybean oil (data converted for comparison based on the definition of one enzyme unit) [32]. Although lipase activity increased significantly from 0.01 U/mL before optimization, the final yield of approximately 0.1 U/mL remains too low for practical industrial applications, as recognized in other studies. For example, Bacillus megaterium F25 was reported to secrete lipase at a concentration of 0.583 U/mL in the presence of Tween 80 in shaking flask cultures [33]. As the experiment did not meet the necessary criteria for the project, further trials will not proceed.
3.3.3. Optimization of Amylase Production
The response variances of α-amylase (U/mL) are described in Table 5. The quadratic model was obtained from experimental amylases to explain the statistical interactions between each parameter and their effect on the process of α-amylase production. The correlation between the enzyme production and the optimization factors A, B, and C through the Box–Behnken design has presented in expressions of coded factors according to the following equation:
where A, B, and C are the coded values of temperature, total solids, and inoculum size, respectively.

Table 5.
Optimization of amylase production by Bacillus amyloliquefaciens on SIW using Box–Behnken design.
The given quadratic response surface model describes the influence of temperature (A), total solids (B), and inoculum size (C) on α-amylase activity (Y), incorporating linear, interaction, and quadratic terms (Table 6). The negative coefficient for temperature (−0.425) indicates that increasing temperature reduces α-amylase activity, with a strong quadratic effect (−0.566) suggesting an optimal temperature exists beyond which activity declines. In contrast, total solids has a positive linear effect (+0.575), enhancing activity, though the quadratic term (−0.141) implies diminishing returns at higher concentrations. The negative linear effect of inoculum size (−0.2325) and its quadratic term (−0.426) indicate a non-linear impact, where excessive inoculum may reduce enzyme activity. Interaction terms, such as the positive AB (+0.09) and negative AC (−0.155) and BC (−0.31), reveal that combined factors can enhance or inhibit α-amylase production, highlighting the need to optimize multiple parameters rather than individual factors in isolation. Overall, the model suggests that α-amylase activity is highly sensitive to these factors, and response surface methodology can be applied to determine optimal conditions for maximizing enzyme production.

Table 6.
Statistical analysis of Box–Behnken design for amylase production in Bacillus amyloliquefaciens.
The statistical analysis confirms that the model is highly significant, as indicated by the model F-value of 31.77, with only a 0.01% chance that such a high value could result from noise. The p-values reveal that temperature (A), total solids (B), inoculum size (C), the interaction term BC, and the quadratic terms A2 and C2 significantly influence α-amylase activity, while other terms may have a negligible effect. If multiple insignificant terms are present, model reduction could enhance efficiency without disrupting the hierarchical structure. The lack-of-fit F-value of 0.27 suggests that the model fits the data well, as the lack of fit is statistically insignificant. Furthermore, the predicted R2 (0.9037) and adjusted R2 (0.9454) are very close to 1, indicating that the model explains over 90% of the variation in α-amylase activity (Figure 4). The small difference between these values (less than 0.2) confirms the model’s predictive reliability and suggests that it generalizes well without overfitting. The high adjusted R2 also indicates that the model effectively captures the relevant predictors while minimizing the impact of unnecessary terms. Additionally, the Adeq precision ratio of 19.057, which far exceeds the minimum desirable threshold of 4, reflects a strong signal-to-noise ratio, reinforcing the model’s suitability for exploring the design space. These statistical indicators suggest that the model provides a reliable basis for optimizing and predicting α-amylase activity under different experimental conditions.
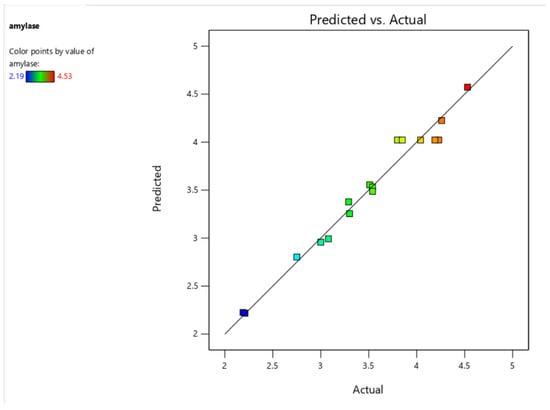
Figure 4.
Model predictions as a function of the experimental amylase activity (U/mL).
The experimental validation of the model’s optimal conditions confirmed that the maximum α-amylase production of 4.05 ± 0.05 U/mL can be achieved by incubating Bacillus amyloliquefaciens at 30 °C, using a medium containing 25 g/L total solids from starch wastewater and a 2% inoculum size (Figure 5). This result is highly consistent with the model’s predicted data (4.19 U/mL), further supporting the accuracy and reliability of the designed model.
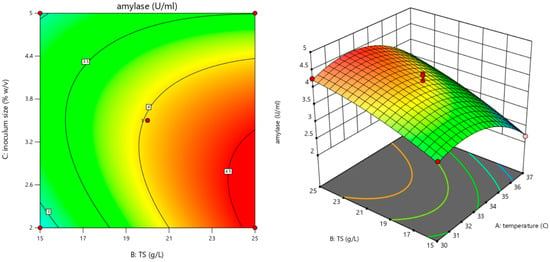
Figure 5.
Response surface of optimal conditions for amylase production.
3.4. Enzyme Production in 5 L Bioreactor
3.4.1. Production of Protease Using Bacillus megaterium
Protease production was assessed at 24 h intervals, showing significantly higher yields in 5 L bioreactors compared to shake flasks, highlighting the advantages of controlled fermentation conditions. In the first batch, Bacillus megaterium produced 52.91 U/mL after 24 h, decreasing to 43.73 U/mL at 48 h. In a second batch, protease activity peaked at 57.91 U/mL at 24 h—coinciding with maximum cell density—before declining to 40.18 U/mL at 48 h. The observed decline in activity after 24 h may be attributed to protease instability, product degradation, or nutrient depletion in the medium. These results are consistent with previous findings, although the maximum yields reported here are slightly lower than those in some literature. Differences can largely be explained by variations in medium composition, inoculum characteristics, and strain origin. For instance, Rajkumar et al. [34] reported a significantly higher yield of 78.5 U/mL from a B. megaterium strain isolated from red seaweed, using a complex medium enriched with glucose, yeast extract, gelatin, lactose, and seawater, and adjusted to an initial pH of 9. The incubation was carried out at 40 °C for 42 h. These optimized conditions, particularly the alkaline pH and complex nitrogen sources, may have favored protease expression.
In contrast, a marine-derived B. megaterium strain from the Gulf of Thailand showed a much lower activity (6.57 ± 0.25 U/mL) after 15 h at pH 5.0 and 30 °C [35]. This emphasizes the strong influence of environmental and nutritional parameters on protease productivity. An important observation from this study is the shorter fermentation time and higher yield in bioreactor cultures compared to shake flasks (see Figure 6). This improvement is likely due to better aeration, mixing efficiency, and pH/temperature control in bioreactors, which collectively enhance microbial growth and enzyme production, as also noted by Priya et al. [36].
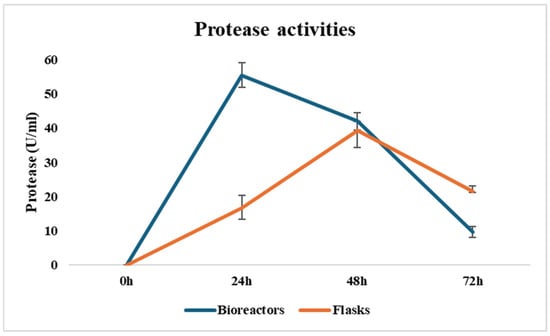
Figure 6.
Kinetics of protease production in flasks and bioreactors.
Similarly, B. megaterium AU02, an organic solvent-tolerant strain isolated from dairy effluents, produced 43.6 U/mL in shake flasks after 49 h using a medium containing skim milk and calcium chloride. Under optimized conditions in a 7 L fermenter, this value increased to 53 U/mL at approximately 32 h, aligning closely with the current findings. Overall, the current study demonstrates the potential of B. megaterium for efficient protease production under relatively simple culture conditions, with bioreactor cultivation significantly enhancing productivity. Further optimization of media components, particularly nitrogen and carbon sources, along with fed-batch or continuous fermentation strategies, could improve yields even further and facilitate industrial-scale applications.
3.4.2. Bioreactor Production of Amylase Using Bacillus amyloliquefaciens
The optimization model identified 25 g/L of total solids as the optimal concentration for protease production. However, it also suggested that increasing total solids could enhance amylase production, with a positive trend observed up to higher concentrations, despite diminishing returns. To validate this, an additional experiment was conducted using 40 g/L of total solids. Results from 1 L shake flasks confirmed that this concentration provided an optimal balance between maximizing amylase yield and maintaining manageable medium viscosity. Exceeding this concentration resulted in increased viscosity, which negatively impacted oxygen diffusion and mixing, critical parameters for microbial growth and enzyme production, especially in scaled-up fermentation systems. Based on this optimization, two 5 L bioreactor experiments were conducted using 40 g/L total solids, 30 °C incubation temperature, and a 2% inoculum size to evaluate amylase production under controlled and scalable conditions. In the first bioreactor trial, samples were collected every 12 h over a 72 h period. Bacillus amyloliquefaciens began secreting amylase during the exponential phase, with activity levels rising from 0.05 U/mL at 12 h to 4.15 U/mL at 36 h and peaking at 5.74 U/mL at 48 h. By 72 h, activity had declined to 4.34 U/mL, suggesting that maximum enzyme production occurred during the transition from the late exponential to early stationary phase, with a decline likely due to proteolytic degradation or depletion of key nutrients. The peak amylase activity in the second batch reached 5.72 U/mL, nearly identical to the first trial, indicating that extending the fermentation beyond 48 h offered no additional benefit and could even reduce enzyme yield due to stability issues. These findings confirm that bioreactor cultivation, with precise control over temperature and dissolved oxygen, significantly enhances amylase production compared to flask-scale fermentations, even under the same incubation duration (Figure 7). This underscores the scalability and industrial potential of the process.
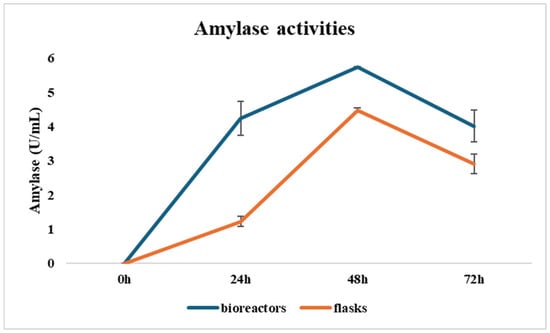
Figure 7.
Kinetics of amylase production in flasks and bioreactors.
B. amyloliquefaciens is widely recognized for its industrial utility, with applications spanning food processing, pharmaceuticals, agriculture, biofuels, and environmental biotechnology [37]. Its ability to convert diverse agro-industrial residues into high-value products like amylases aligns with current trends in circular bioeconomy and waste valorization. Numerous substrates such as kitchen waste [38], bread waste [39], wheat bran, rice husk, maize starch, and starchy tuber residues have been explored as low-cost feedstocks for enzyme production. However, reported amylase yields vary considerably across studies, influenced by multiple factors including strain genetics, substrate composition, pretreatment methods, fermentation strategies (submerged vs. solid-state), and differences in enzyme assay protocols.
4. Conclusions
This study investigated four local wastewater types in Quebec as alternative media for enzyme production, focusing on protease, amylase, and lipase using Bacillus species as part of a pragmatic screening strategy for potential waste substrates. Among them, SIW effectively supported amylase synthesis by B. amyloliquefaciens, achieving 5.73 ± 0.01 U/mL at 48 h under 40 g/L total solids, 30 °C, and a 2% inoculum size in a 5 L bioreactor. Meanwhile, B. megaterium produced both protease and lipase in PPMS, with significant protease activity warranting further scale-up experiments. The highest protease yield, 55.41 ± 3.54 U/mL, was recorded at 24 h under 25 g/L total solids, 30 °C, and a 2% inoculum in duplicated bioreactors. The simple preparation of wastewater—considering total solids, temperature, and inoculum size—and the adaptability of these enzymes for high yields in 5 L bioreactors, underscores the industrial relevance of this approach. The reproducibility observed at this scale implies a strong potential for larger-scale development. However, physical constraints encountered during scale-up, including limitations in oxygen transfer, mixing efficiency, temperature control, and foaming are anticipated to manifest more severely in complex media such as wastewater compared to synthetic substrates, owing to their heterogeneous and variable nature. Despite these challenges, repurposing wastewater not only mitigates environmental impact but also transforms waste into valuable bioproducts, aligning with the circular bioeconomy and enhancing sustainability by improving resource efficiency and lowering production costs.
This study is conducted from a foundational stage; therefore, selecting a subset of wastewater sources from several options serves as an initial step in identifying those with potential for enzyme production by designated Bacillus species. However, this approach inherently carries the risk of overlooking novel isolates that could enhance enzyme productivity due to their adaptation to specific environmental conditions. Consequently, the findings of this study should be regarded as preliminary and may serve as a reference for future experiments incorporating newly identified isolates.
Author Contributions
Writing original draft: V.N.; review and editing: A.N.; Supervision: J.-F.B.; supervision and funding acquisition: K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Institut National de la Recherche Scientifique (INRS).
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors would like to acknowledge the Institut National de la Recherche Scientifique (INRS) for their financial support.
Conflicts of Interest
There are no conflicts of interest to declare.
References
- Vea, E.B.; Romeo, D.; Thomsen, M. Biowaste valorisation in a future circular bioeconomy. Procedia CIRP 2018, 69, 591–596. [Google Scholar] [CrossRef]
- Maina, S.; Kachrimanidou, V.; Koutinas, A. A roadmap towards a circular and sustainable bioeconomy through waste valorization. Curr. Opin. Green Sustain. Chem. 2017, 8, 18–23. [Google Scholar] [CrossRef]
- Klitkou, A.; Fevolden, A.M.; Capasso, M. From Waste to Value: Valorisation Pathways for Organic Waste Streams in Circular Bioeconomies; Taylor & Francis: Abingdon, UK, 2019. [Google Scholar]
- Mahjoub, B.; Domscheit, E. Chances and challenges of an organic waste–based bioeconomy. Curr. Opin. Green Sustain. Chem. 2020, 25, 100388. [Google Scholar] [CrossRef]
- Lee, D.-J.; Kim, J.Y.; Park, J.; Choi, Y.-B.; Kim, J.K.; Choi, H.; Tsang, Y.F.; Kwon, E.E. Sustainable organic waste valorisation: A zero-waste approach. Chemosphere 2024, 365, 143365. [Google Scholar] [CrossRef]
- Ahmed, I.; Zia, M.A.; Afzal, H.; Ahmed, S.; Ahmad, M.; Akram, Z.; Sher, F.; Iqbal, H.M. Socio-economic and environmental impacts of biomass valorisation: A strategic drive for sustainable bioeconomy. Sustainability 2021, 13, 4200. [Google Scholar] [CrossRef]
- Godfrey, L.; Görgens, J.F.; Roman, H. Opportunities for Biomass and Organic Waste Valorisation: Finding Alternative Solutions to Disposal in South Africa; Routledge: Abingdon, UK, 2020. [Google Scholar]
- Khan, A. Sustainable Bioconversion of Waste to Value Added Products; Springer Nature: Berlin, Germany, 2021. [Google Scholar]
- Scarlat, N.; Dallemand, J.-F.; Monforti-Ferrario, F.; Nita, V. The role of biomass and bioenergy in a future bioeconomy: Policies and facts. Environ. Dev. 2015, 15, 3–34. [Google Scholar] [CrossRef]
- Kathi, S.; Singh, S.; Yadav, R.; Singh, A.N.; Mahmoud, A.E.D. Wastewater and sludge valorisation: A novel approach for treatment and resource recovery to achieve circular economy concept. Front. Chem. Eng. 2023, 5, 1129783. [Google Scholar] [CrossRef]
- Ravindran, R.; Hassan, S.S.; Williams, G.A.; Jaiswal, A.K. A review on bioconversion of agro-industrial wastes to industrially important enzymes. Bioengineering 2018, 5, 93. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 2016, 6, 174. [Google Scholar] [CrossRef]
- Schallmey, M.; Singh, A.; Ward, O.P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 2004, 50, 1–17. [Google Scholar] [CrossRef]
- Singh, S.; Mangla, J.; Singh, S. Evaluation of Aspergillus fumigatus NTCC1222 as a source of enzymes for detergent industry. Resour. Environ. Sustain. 2021, 5, 100030. [Google Scholar] [CrossRef]
- Özbek Yazıcı, S.; Özmen, I. Optimization for coproduction of protease and cellulase from Bacillus subtilis M-11 by the Box–Behnken design and their detergent compatibility. Braz. J. Chem. Eng. 2020, 37, 49–59. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 169. [Google Scholar] [CrossRef] [PubMed]
- Contesini, F.J.; Melo, R.R.d.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, D. Current development in genetic engineering strategies of Bacillus species. Microb. Cell Factories 2014, 13, 63. [Google Scholar] [CrossRef]
- Choubane, S.; Khelil, O.; Cheba, B.A. Bacillus sp. R2 α-amylase production optimization: Pasta cooking water as medium of amylase production. Afr. J. Biotechnol. 2015, 14, 3184–3189. [Google Scholar]
- Mazhar, H.; Abbas, N.; Hussain, Z.; Sohail, A.; Ali, S. Extracellular lipase production from Bacillus subtilis using agro-industrial waste and fruit peels. Punjab Univ. J. Zool. 2016, 31, 261–267. [Google Scholar]
- Sánchez Blanco, A.; Palacios Durive, O.; Batista Pérez, S.; Díaz Montes, Z.; Pérez Guerra, N. Simultaneous production of amylases and proteases by Bacillus subtilis in brewery wastes. Braz. J. Microbiol. 2016, 47, 665–674. [Google Scholar] [CrossRef]
- Ramkumar, A.; Sivakumar, N.; Gujarathi, A.M.; Victor, R. Production of thermotolerant, detergent stable alkaline protease using the gut waste of Sardinella longiceps as a substrate: Optimization and characterization. Sci. Rep. 2018, 8, 12442. [Google Scholar] [CrossRef]
- Shart, A.O.; Elkhalil, E.A. Biochemical characterization of lipase produced by Bacillus spp. isolated from soil and oil effluent. Adv. Enzym. Res. 2020, 8, 39–48. [Google Scholar] [CrossRef]
- Kherouf, M.; Habbeche, A.; Benamia, F.; Saoudi, B.; Kerouaz, B.; Ladjama, A. Statistical optimization of a novel extracellular alkaline and thermostable amylase production from thermophilic Actinomadura keratinilytica sp. Cpt29 and its potential application in detergent industry. Biocatal. Agric. Biotechnol. 2021, 35, 102068. [Google Scholar] [CrossRef]
- Abd-Elhalem, B.T.; El-Sawy, M.; Gamal, R.F.; Abou-Taleb, K.A. Production of amylases from Bacillus amyloliquefaciens under submerged fermentation using some agro-industrial by-products. Ann. Agric. Sci. 2015, 60, 193–202. [Google Scholar] [CrossRef]
- O’Hara, M.B.; Hageman, J.H. Energy and calcium ion dependence of proteolysis during sporulation of Bacillus subtilis cells. J. Bacteriol. 1990, 172, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Eijsink, V.; Matthews, B.; Vriend, G. The role of calcium ions in the stability and instability of a thermolysin-like protease. Protein Sci. 2011, 20, 1346–1355. [Google Scholar] [CrossRef]
- Lad, B.C.; Coleman, S.M.; Alper, H.S. Microbial valorization of underutilized and nonconventional waste streams. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab056. [Google Scholar] [CrossRef]
- Atlas, R. Handbook of Microbiological Media, 4th Edition ed.; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Muras, A.; Romero, M.; Mayer, C.; Otero, A. Biotechnological applications of Bacillus licheniformis. Crit. Rev. Biotechnol. 2021, 41, 609–627. [Google Scholar] [CrossRef]
- Boekema, B.K.H.L.; Beselin, A.; Breuer, M.; Hauer, B.; Koster, M.; Rosenau, F.; Jaeger, K.-E.; Tommassen, J. Hexadecane and Tween 80 Stimulate Lipase Production in Burkholderia glumae by Different Mechanisms. Appl. Environ. Microbiol. 2007, 73, 3838–3844. [Google Scholar] [CrossRef]
- Sekhon, A.; Dahiya, N.; Tewari, R.P.; Hoondal, G.S. Production of extracellular lipase by Bacillus megaterium AKG-1 in submerged fermentation. Indian J. Biotechnol. 2006, 5, 179–183. [Google Scholar]
- Karaman, F.; Incekara, U.; Arslan, N.P.; Albayrak, S.; Ortucu, S.; Taskin, M. A New Enzyme for Biodiesel Production and Food Applications: Lipase of Bacillus megaterium F25 Isolated From an Aquatic Insect Rhantus suturalis. GCB Bioenergy 2024, 16, e70009. [Google Scholar] [CrossRef]
- Rajkumar, R.; Jayappriyan, K.R.; Rengasamy, R. Purification and characterization of a protease produced by Bacillus megaterium RRM2: Application in detergent and dehairing industries. J. Basic Microbiol. 2011, 51, 614–624. [Google Scholar] [CrossRef]
- Uttatree, S.; Kobtrakool, K.; Ketsuk, A.; Kaenngam, W.; Thakolprajak, P.; Charoenpanich, J. A novel metal-tolerant, solvent and surfactant stable protease from a new strain of Bacillus megaterium. Biocatal. Agric. Biotechnol. 2017, 12, 228–235. [Google Scholar] [CrossRef]
- Priya, J.D.A.; Divakar, K.; Prabha, M.S.; Selvam, G.P.; Gautam, P. Isolation, purification and characterisation of an organic solvent-tolerant Ca2+-dependent protease from Bacillus megaterium AU02. Appl. Biochem. Biotechnol. 2014, 172, 910–932. [Google Scholar] [CrossRef] [PubMed]
- WoldemariamYohannes, K.; Wan, Z.; Yu, Q.; Li, H.; Wei, X.; Liu, Y.; Wang, J.; Sun, B. Prebiotic, probiotic, antimicrobial, and functional food applications of Bacillus amyloliquefaciens. J. Agric. Food Chem. 2020, 68, 14709–14727. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, B.; Prajapati, V.; Patel, K.; Trivedi, U. Kitchen waste for economical amylase production using Bacillus amyloliquefaciens KCP2. Biocatal. Agric. Biotechnol. 2020, 26, 101654. [Google Scholar] [CrossRef]
- Abd-Elhalim, B.T.; Gamal, R.F.; El-Sayed, S.M.; Abu-Hussien, S.H. Optimizing alpha-amylase from Bacillus amyloliquefaciens on bread waste for effective industrial wastewater treatment and textile desizing through response surface methodology. Sci. Rep. 2023, 13, 19216. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).