Abstract
Apple (Malus domestica Borkh.) is an economically important fruit. The use of nitrate by plants plays a crucial role in their growth and development, and its absorption and dispersal are controlled by nitrate transport proteins (NRTs). In this study, we investigated the potential function of MdNRT2.4 under low-nitrogen (N) stress by overexpressing it in tobacco. Compared with plants treated with a normal nitrogen level (5 mM), the MdNRT2.4 overexpression lines under low-N stress (0.25 mM) exhibited significantly greater plant height and width, as well as larger leaves and a higher leaf density, than wild-type plants, suggesting that the overexpression of MdNRT2.4 enhances the low-N tolerance of tobacco. Enhanced antioxidant enzyme activities in the MdNRT2.4 overexpression plant lines promoted the scavenging of reactive oxygen species, which reduced damage to their cell membranes. GUS staining of pMdNRT2.4::GUS-transformed Arabidopsis thaliana lines showed that MdNRT2.4 was expressed in the roots, vascular bundles, seeds in fruit pods, and young anther sites, suggesting that MdNRT2.4 mediates the transport of nitrate to these tissues, indicating that MdNRT2.4 might promote nitrate utilization in apple and improve its tolerance to low-N stress. Experiments using yeast one-hybrid and dual-luciferase assays revealed that MdbHLH3 binds to the MdNRT2.4 promoter and activates its expression. MdbHLH3 belongs to the basic helix–loop–helix (bHLH) transcription factor (TF). It is speculated that MdbHLH3 may interact with the promoter of MdNRT2.4 to regulate N metabolism in plants and enhance their low-N tolerance. This study establishes a theoretical framework for investigating the regulatory mechanisms of low-N responsive molecules in apple, while simultaneously providing valuable genetic resources for molecular breeding programs targeting low-N tolerance.
1. Introduction
Nitrogen (N), an essential macronutrient for plant development, plays a significant role in the synthesis of vital substances, such as amino acids, nucleotides, chlorophyll, and vitamins [1,2]. Plants obtain inorganic nitrogen from the soil, mainly in the form of nitrate and ammonium [3]. Nitrate is the main form of nitrogen in most aerated soils, while ammonium nitrogen may be the main form in some acidic and/or anaerobic environments [4]. A lack of N can have a major effect on plant growth in both natural habitats and agricultural systems, given that soil erosion results in decreased soil quality [5,6]. However, the excessive use of chemical fertilizers can lead to soil eutrophication, which has a negative effect on the farmed land and surrounding ecosystems [7,8].
Plant nitrate transport proteins (NRTs) can be classified into several types based on their structure and function: the NO3− transporter 1 (NRT1/PTR), NO3− transporter 2 (NRT2), chloride channel (CLC), and slow anion channel homolog (SLAC1/SLAH) families [9]. Advanced plants benefit from the uptake and distribution of inorganic N (NH4+, NO3−) by transporter proteins, which include high-affinity transporters (HATs) and low-affinity transporters (LATs) with different binding strengths [10,11]. To ensure the entry of NO3− into roots, plants have developed NO3− transport systems with both high and low affinity [12]. Typically, NRT1 proteins function as LATs, and NRT2 proteins operate as HATs [13,14,15]. AtNRT1.2 encodes a low-affinity NRT that is predominantly found in root hairs and the epidermis and has the features of a typical transport protein [16]. Previous studies have shown that the overexpression of MdNRT1.1 can promote root development and N utilization in Arabidopsis thaliana (Arabidopsis) [17]; that MdNRT1.5 is involved in the transport of NO3− from roots to buds [18]; and that the overexpression of MdNRT1.7 enhances low-N tolerance in tobacco by modulating reactive oxygen species (ROS) clearance [19]. NRT2 genes comprise a major superfamily of transporters (MFS) [20]. Members of the NRT2 family have been well studied in recent years. Most studies on N utilization in apples have focused on MdNRT2 members, which have been reported to play an important role in enhancing NO3− absorption in low-N environments [21]. For example, increasing the concentration of sorbitol in the roots can induce the expression of MdNRT2.1 and MdNRT2.2 and promote the absorption of N by apple seedlings [22]. In Arabidopsis, AtNRT2.1 expression predominantly occurs near the mature roots and is influenced by NO3−. AtNRT2.1 expression was induced at low levels of NO3− and inhibited at higher NO3− concentrations [23]. In general, NRT2 proteins (NRT2s) are linked by cytoplasmic loops and include one MFS structural domain with a dual affinity for NO3− and twelve transmembrane structural domains, which are usually located in the cytoplasmic membrane [24,25,26]. The functional activity of NRT2 transporter proteins is usually dependent on auxiliary proteins or NRT3–NRT2 interactions [27]. Furthermore, AtNRT2.4 and AtNRT2.5 play a role in the movement of NO3− from its origin to its destination [28]. In wheat, TaNRT2.1 is involved in NO3− uptake at the post-flowering stage [29]. AtNRT2.7 expression is higher in branches than in roots [30].
Apple (Malus domestica Borkh.) belongs to the family Rosaceae and is mainly grown in temperate regions; apple fruits have high economic and nutritional value [31]. Enhancing N utilization is critically important for increasing crop production. Although N fertilizers enhance agricultural output, the inappropriate application of N fertilizers can lead to environmental contamination and negatively affect human health [32]. Consequently, research on apple NRT proteins is critically important for enhancing N-use efficiency. However, the physiological functions and regulatory mechanisms by which NRT2.4 regulates N utilization under low-N stress remain unclear. To elucidate the functional role of apple MdNRT2.4, we cloned its coding sequence from apple and induced its heterologous overexpression in tobacco. The regulatory interaction between MdbHLH3 and the MdNRT2.4 promoter was confirmed through yeast one-hybrid and dual-luciferase reporter systems. Additionally, to further verify the tissue-specific expression of MdNRT2.4, the GUS reporter gene driven by the MdNRT2.4 promoter was generated in Arabidopsis, and GUS staining analysis was performed. This study aimed to explore the molecular regulatory mechanism of MdNRT2.4 under low-N stress and, based on the results of its functional verification, provide a theoretical basis and technical strategies for nitrogen-efficient breeding of apples and other crops.
2. Materials and Methods
2.1. Plant Material
To determine the expression levels of MdNRT2.4 and MdbHLH3, two-month-old Malus hupehensis seedlings were used as materials. Genetic transformation for the overexpression of MdNRT2.4 and dual-luciferase assays were performed using two-month-old wild-type (WT) tobacco (Nicotiana benthamiana) grown in soil. The GUS expression activity driven by the MdNRT2.4 promoter was investigated in one-month-old WT Arabidopsis plants cultured in soil. They were cultivated in a plant growth chamber with a light intensity of 20,000 Lux under the conditions of 16 h/8 h (day/night) and 25 ± 2 °C.
2.2. Experimental Design
The T3-generation seeds of the MdNRT2.4 overexpression transformant lines (OE2, OE4, and OE6) with high, medium, and low expression levels and the seeds of the WT tobacco line were selected and sown in MS medium for subsequent root system observation. The seeds were cultured in a medium containing 11/12 N-free MS powder (PhytoTech, Kansas, USA) + 1/12 all-N MS powder (PhytoTech, Kansas, USA), while the control group seeds were cultured in the normal MS medium. Three seeds were sown in each bead line, including three biological replicates. The petri dishes containing the medium were oriented vertically, incubated for 7 days, and then transferred to a low-N medium for low-N stress treatment. The dimensions of the WT and OE lines’ roots were then measured. The plants were grown in a controlled environment growth chamber with a 16-h light/8-h dark photoperiod at a constant temperature of 25 ± 2 °C.
To further analyze the low-N tolerance of the MdNRT2.4 transformant lines, 4-week-old T3-generation MdNRT2.4 overexpression plant lines and WT plants were subjected to low-N stress treatment. In accordance with previous studies [19], a low-N nutrient solution (0.25 mM) was used for treatment, and a full-nutrient solution (5 mM) was used as the control. The control group was watered with Hoagland nutrient solution, and the low-N-treated group was treated with 0.25 mM Ca(NO3)2 and 0.25 mM KNO3 instead of 5 mM Ca(NO3)2 and 5 mM KNO3, respectively, in Hoagland nutrient solution. To ensure that the concentrations of Ca2+ and K+ remained unchanged, Ca(NO3)2 and potassium nitrate were replaced with CaCl2 and KCl [33], and samples were taken after 20 days of treatment. Treatments were performed on twenty tobacco plants in the experimental group and twenty in the control group. Twelve plants were used to determine dry and fresh weights and total N accumulation, and the remaining eight plants were used to determine other physiological indicators and the expression levels of related genes.
2.3. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The Polysaccharide Polyphenol Plant RNA Kit (Omega Bio-tek, Norcross, Georgia, USA) was employed to extract RNA from leaves, following the guidelines provided by the manufacturer. After RNA extraction and DNase treatment, RNA integrity was assessed using agarose gel electrophoresis, and the RNA dosage was controlled at 600 ng for the subsequent cDNA synthesis. The Revere Transcription Kit (Gene Star, Beijing, China) was employed for cDNA synthesis, followed by a ten-fold dilution for qRT-PCR. qRT-PCR which was performed using an analyzer (Qtower 3G, Jena, Germany). The 10 μL qRT-PCR reaction system mainly consisted of 5 μL PowerUp™SYBRTM Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), 0.5 μL of forward and reverse primers (Table S1), 2.0 μL of cDNA, and 2.0 μL of deionized water. The reaction procedure for qRT-PCR was as follows: pre-denaturation at 94 °C for 10 min and 40 cycles, with each cycle consisting of denaturation at 94 °C for 15 s, annealing at 60 °C for 30 s, and annealing at 72 °C for 1 min. The melting curve rose slowly from 65 °C to 95 °C, increasing by 0.5 °C every 15 s. The NtEF-1α gene was used as an internal reference in tobacco [19]. For every sample, three biological replicates were conducted, and the levels of expression were determined using the 2−ΔΔCt method. The primer sequences utilized in this study are presented in Table S1.
2.4. MdNRT2.4 Cloning and Sequence Analysis
Initially, total RNA was isolated from ‘Golden Crown’ apples, and then cDNA synthesis was performed using the Revere Transcription Kit (Gene Star, Beijing, China), which was used as a template for PCR amplification. PCR was performed with 35 cycles of 94 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min. Based on the MdNRT2.4 gene sequence (ID: XM_008359979.4) obtained from the apple transcriptome database (https://www.rosaceae.org/) (accessed on 5 September 2023), Oligo7 (Molecular Biology Insights, Inc., Cascade, ID, USA) was used to design specific primers (Table S1) to clone the gene [34], and they were verified by sequencing to obtain the full-length gene sequence. The target fragments were recovered and purified using a gel recovery kit (TaKaRa, Kusatsu, Japan). To clarify the molecular evolution of MdNRT2.4, the amino acid sequences of these 19 species were predicted using the NCBI online platform (https://www.ncbi.nlm.nih.gov/) (accessed on 2 October 2023). Phylogenetic trees were constructed for the NRT2.4 amino acid sequences of 19 plant species using MEGA 11 software. The neighbor-joining method was adopted, and 1000 bootstrap repetitions were used to evaluate the branch support. Additional parameters were set to their default values; the amino acid sequences of 19 species are shown in Table S2.
2.5. Expression Vector Construction and Tobacco Transformation
The Ready-to-Use Seamless Cloning Kit (Bioengineering, Shanghai, China) was used to ligate the target fragment to the pCAMBIA 1301-35s-EGFP vector. KpnI and XbaI were selected as the double digestion sites of this expression vector, and ligation was performed at 50 °C for 20 min. The recombinant plasmid was transformed into E. coli DH5α receptor cells (TransGen, Beijing, China). PCR was used to detect positive colonies, which were then sent to Shanghai Bioengineering Company Limited for sequencing (Sangon, Shanghai, China). The constructed overexpression vector was named pCAMBIA 1301-35s-MdNRT2.4-EGFP. After that, the recombinant plasmid was transformed into Agrobacterium tumefaciens GV3101 using the freeze–thaw method. The WT seeds were disinfected and sown into the common MS medium (Basebio, Chengdu, China). When the leaves grew larger, the leaf disk transformation method was used for genetic transformation of tobacco. The OD value of the transformed bacterial liquid was adjusted to 0.6 to 0.8. The entire transformation process should be carried out on a super-clean workbench (DiTu, Shanghai, China), and aseptic operation should be ensured throughout the process. The transformed strains were screened in the standard MS culture medium with 10 mg/L hygromycin (Hyg). The plants were maintained in a growth chamber under 16/8 h light/dark conditions at 25 ± 2 °C. The following cyclic program was used to conduct PCR identification on the screened transgenic tobacco: pre-denaturation at 94 °C for 5 min; denaturation at 94 °C for 30 s, annealing at 55–60 °C for 30 s, and extension at 72 °C for 2 min, 35 cycles, and extension at 72 °C for 5 min. The quantification of MdNRT2.4 expression in the examined transformant lines was conducted using qRT-PCR and semi-quantitative RT-PCR, using NtEF-1α as the internal reference. The primer sequences used are shown in Table S1.
2.6. MdNRT2.4 Expression Analysis of Genes Related to N Metabolism
Based on previous studies [19], the N-metabolism-related genes NtGDH1, NtGS1-3, NtGS2, and NtNR were used to further characterize the levels of NRT2.4 overexpression in tobacco under low-N stress. EF1-α was used as the reference gene for tobacco, and the expression levels of genes associated with N metabolism were examined using qRT-PCR. The qRT-PCR reaction procedure is described in 2.3. For each line, three biological replicates and then three technical replicates were conducted. The primers of the four N-metabolism-related genes are shown in Table S1.
2.7. Cis-Acting Element Analysis of the MdNRT2.4 Promoter and GUS Histochemical Staining
The promoter sequence of the first 2000 bp from the transcription start site of the MdNRT2.4 gene was obtained from the apple genome database. The cis-acting elements in the MdNRT2.4 promoter were predicted using the PlantCARE online software (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 9 December 2023). Using seamless cloning, the MdNRT2.4 promoter was integrated into the pBI121-GUS vector, and Hind III and BamH I were selected as the double digestion sites of this expression vector. Subsequently, the recombinant plasmid was transferred into A. tumefaciens after sequencing. The transformation of Arabidopsis was performed via the floral dip method. After seed collection, the transformant lines were screened in 1/2 MS medium containing 50 mg/L hygromycin (Hyg). According to the instructions provided in the kit, DNA was extracted from the positive plants by the low-throughput method and then verified by PCR. Different tissues and organs of the transformant Arabidopsis lines were sampled in different growth periods and stained using the staining method in the GUS staining kit (Coolaber CA1400, Beijing, China). Use 10 mL of staining solution for each reaction and the stained areas were observed using a light microscope. The primers used for cloning genes are shown in Table S1.
2.8. Subcellular Localization of MdNRT2.4
Primers were designed with the open reading frame (ORF) deletion stop codon of the MdNRT2.4 gene, and PCR amplification was performed using the cDNA of WT ‘Golden Crown’ apple leaves as the template (Primer reference Table S1), which was then integrated into the pCAMBIA 1301-35s-EGFP vector. KpnI and BamHI were selected as the double digestion sites of this expression vector. The empty vector pCAMBIA 1301-35s-EGFP (negative control) and the recombinant plasmid pCAMBIA 1301-35s-MdNRT2.4-EGFP were transferred into A. tumefaciens. A. tumefaciens carrying the recombinant plasmid and empty vector was transformed into Arabidopsis protoplasts using PEG-mediated transformation. Arabidopsis protoplasts were prepared based on the procedure described in a previous study [35]. After 48 h, confocal imaging was performed using a Leica TCSSP8 system (Leica Microsystems, Wetzlar, Germany) equipped with a 63×/1.40 NA oil-immersion objective and controlled by LAS X software (v3.5).
2.9. Yeast One-Hybrid and Dual-Luciferase Assays
The cloned MdNRT2.4 promoter sequence was inserted into the pHIS2 vector, and then the recombinant plasmid was transformed into Saccharomyces cerevisiae Y187; the inhibitory concentration of 3-amino-1,2,4-triazole (3-AT) was then determined [36]. Seven transcription factors (TFs) that responded to low-N stress were selected and cloned according to the transcriptome data, namely, MdbHLH2 (MD16G1274200), MdbHLH3 (MD06G1034300), MdNAC2 (MD06G1031700), MdAP2-1 (MD09G1183700), MdMYB2 (MD06G1211700), MdWRKY4 (MD15G1039500), and MdWRKY5 (MD12G1181000). Subsequently, to determine whether these transcription factors can bind to the MdNRT2.4 promoter, they were constructed separately in the pGADT7 vector and transferred into Saccharomyces cerevisiae Y187 along with the pHIS2-MdNRT2.4 recombinant plasmid. Spot-plating was performed in an SD-/THL medium using an established 3-AT inhibitory concentration, followed by incubation at 30 °C for 3–4 days.
The experimental results confirm that MdbHLH3 specifically binds to the promoter region of MdNRT2.4. Then, the CDS sequence of MdbHLH3 and the promoter sequence of MdNRT2.4 were inserted into the pGreenII 62-SK and pGreenII 0800-Luc vectors, respectively, and transferred into A. tumefaciens after sequencing. Finally, A. tumefaciens carrying the recombinant plasmids was injected into the abaxial surface of tobacco leaves and observed in a live imager. The primers associated with this experiment are shown in Table S1.
2.10. Measurement of Physiological Indicators
Three transformant lines and WT plants were selected for fresh weight measurements in the low-N and control treatments. Each seedling was divided into aboveground and belowground parts, rinsed with distilled water, and dried with blotting paper; the aboveground and belowground parts of the seedlings were weighed on an electronic scale and recorded. Subsequently, the samples were oven-dried at 90 °C until they reached a steady weight, followed by separate measurements of the dry weights of the seedlings’ aboveground and belowground segments. The chlorophyll content of the seedlings was measured using a SPAD instrument (Konica Minolta, Tokyo, Japan). The net photosynthetic rate of the seedlings was determined using a photosynthesis meter (LI-COR Biosciences, Lincoln, Nebraska, USA). A constant light intensity of 1000 μmol/m2/s (red and blue LEDs), a temperature of 25 °C, and a CO2 concentration of 400 μmol/mol were set, and the net photosynthetic rate in the steady state was automatically recorded. The total N content of the seedlings was measured using an AA3 continuous flow analyzer. According to the instructions provided in the kit (Solarbio, Beijing, China), the activities of superoxide dismutase (SOD), peroxidase (POD), proline (Pro) content, malondialdehyde (MDA) content, catalase (CAT) activity, plant soluble sugar content, and nitrate reductase (NR) activity were measured and analyzed using a UV spectrophotometer (Harde, Shandong, China).
2.11. Statistical Analysis
Three independent replications of each experiment were performed. One-way ANOVA of variance and Duncan’s test were performed using SPSS Software (SPSS Statistical Software 21.0), and the threshold for statistical significance was p-value < 0.05. Finally, the resulting data were plotted using Origin 2021.
3. Results
3.1. Molecular Characteristics of MdNRT2.4 and Analysis of Its Expression Under Low-N Stress
To clarify the molecular evolution of MdNRT2.4, we constructed phylogenetic trees for 19 species (Figure 1a). The amino acid sequences of these 19 species are shown in Table S2, and their scientific names are shown in Table S3. MdNRT2.4 was found to be more closely related to white pear PbNRT2.4 and Euonymus MsNRT2.4. They are both in the family Rosaceae, which is consistent with the observed evolutionary patterns. To clarify the sequence characteristics of MdNRT2.4, we analyzed the amino acid sequence of MdNRT2.4, which shed light on its evolutionary relationships. The results of the multiple sequence comparison revealed that the homologous amino acid sequences of MdNRT2.4 and 18 other species were highly conserved, which suggests that the genes that encode these proteins may play similar roles in plants (Figure S1). In the low-N treatment, the expression level of MdNRT2.4 began to increase on the fourth day and peaked on the eighth day. Moreover, the expression level in the low-N treatment was approximately five times higher than that in the normal-N treatment. After that, it slightly decreased, but the expression level in the low-N treatment was still higher than that in the normal-N treatment (Figure 1b).
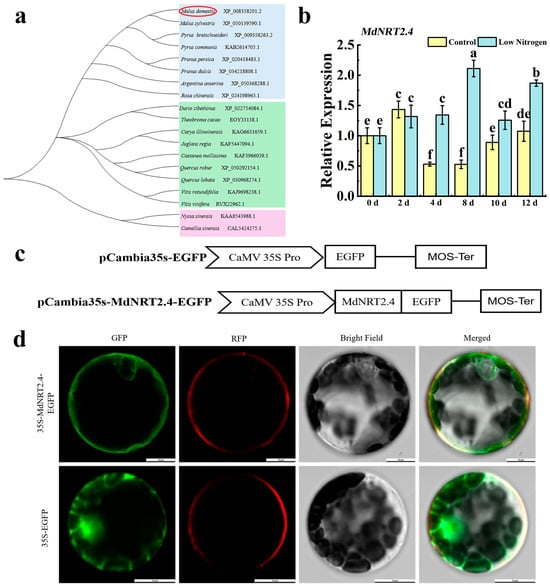
Figure 1.
Molecular characteristics of MdNRT2.4 and expression analysis under low-N stress. (a) Phylogenetic analysis of MdNRT2.4 and genes from 18 other species. The red circled part is ‘MdNRT2.4’. (b) Expression analysis of MdNRT2.4 under low-N stress. (c) The schematic structure of the recombinant vector. (d) The subcellular positioning of GFP-labeled MdNRT2.4 within the lower epidermal cells of Arabidopsis. Bright, visible field; GFP, green fluorescence; RFP, red fluorescence. The pCAMBIA 1301-35s-EGFP vector was used as a negative control; the scale of the measurement bar is 20 µm. The data are presented as mean ± SD (n = 3). Different letters on the bar graphs indicate significant differences at the p < 0.05 level.
The constructed pCAMBIA 1301-35s-MdNRT2.4-EGFP vector was transformed into Arabidopsis protoplasts using PEG-mediated transformation. pCAMBIA 1301-35s-EGFP was used as an empty control, and a plasma membrane marker (FM4-64 dyestuffs) was used to indicate where the cell membrane is located. The fluorescence of MdNRT2.4 was consistent with that of the plasma membrane marker, indicating that MdNRT2.4 was localized to the cell membrane (Figure 1d).
3.2. Phenotypic Analysis of Tobacco Overexpressing the MdNRT2.4 Transgene
Seven MdNRT2.4 overexpression plant lines (OE1-OE7) were obtained and characterized at the DNA and RNA levels (Figure 2a,b). After 20 days of treatment with Hoagland nutrient solution at a 5 mM N concentration, both the WT and OE lines showed regular growth, and there were no significant differences in the visual properties of tobacco. However, after 20 days of treatment with Hoagland nutrient solution at a 0.25 mM N concentration, the leaves of the WT and OE lines were significantly yellow, and the leaves of the OE tobacco lines were significantly larger than those of the WT line (Figure 2c). Early flowering occurred in tobacco under low-N conditions (Figure 2e). There were no significant differences in the roots of WT and OE lines under normal-N conditions (Figure 2d), but under low-N conditions, OE lines (especially OE6) had more primary and lateral roots than WT plants (Figure 2e). No notable differences in the dry weights of the aboveground and belowground parts were observed between the MdNRT2.4 transformant lines and the WT line in the normal-N (5 mM) treatment. However, in the low-N (0.25 mM) treatment, the dry weights of both the aboveground and belowground sections of the OE lines were considerably greater than those of the WT line (Figure 2f,g).
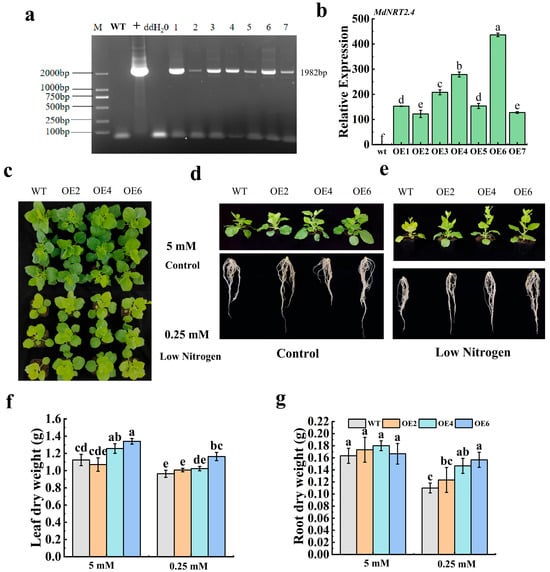
Figure 2.
Identification of overexpressed MdNRT2.4 in transformant lines and phenotypic analysis under low-N stress. (a) DNA verification of MdNRT2.4 overexpression tobacco plants. +, positive plasmid control; ddH2O, negative control; 1–7, OE1–OE7, MdNRT2.4 transformant lines. (b) Gene expression analysis of MdNRT2.4 overexpression plant lines. (c) Phenotypes of OE and WT lines under normal- and low-N treatments. (d) Surface and subterranean sections of OE and WT plants subjected to normal-N treatment. (e) Surface and subterranean segments of OE and WT when subjected to low-N treatment. (f,g) Leaf dry weight and root dry weight under normal-N and low-N treatments for OE and WT plants. The data are presented as mean ± SD (n = 3). Different letters on the bar graphs indicate significant differences at the p < 0.05 level.
3.3. Overexpression of MdNRT2.4 Enhances the Root Length of Tobacco
After sowing three MdNRT2.4 overexpression plant lines (OE2, OE4, and OE6) with high, medium, and low expression levels and WT tobacco seeds in normal MS medium and culturing for 7 days (Figure 3a), they were transferred to normal MS medium and low-N medium and cultured for 7 more days. The growth of the WT and three MdNRT2.4 overexpression plant lines was similar on MS medium (Figure 3b). However, a noticeable difference in the phenotypes of the WT and transformant lines was observed after seedlings were transferred to a low-N MS medium (Figure 3c). The three transformant lines exhibited significantly more lateral roots compared to the WT line, a finding that is consistent with the statistical analysis of root system quantification (Figure 3d). The root length of the transformant lines was 1.5-fold higher than that of the WT plants in the low-N treatment (Figure 3e).
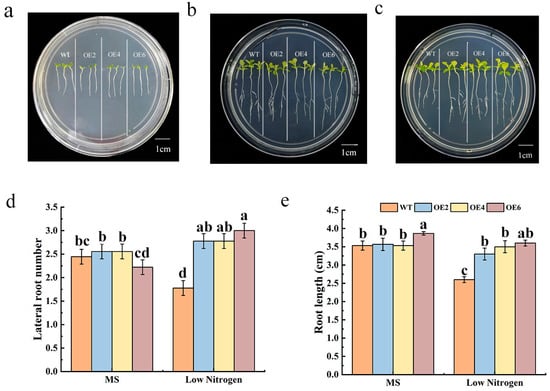
Figure 3.
A statistical comparison of the root lengths of OE and WT lines under normal-N and low-N conditions. (a) The growth of OE and WT plants 7 days after sowing in normal MS medium. (b) Seven days after sowing, the OE and WT plants were transferred to a normal MS medium and cultured for seven more days. (c) Following a 7-day seedling period, the OE and WT lines were relocated to a low-N medium for incubation for 7 days. (d) The root lengths of OE and WT on different media. The data are presented as mean ± SD (n = 3). (e) The number of lateral roots of OE and WT on different media. The data are presented as mean ± SD (n = 3). Different letters on the bar graphs indicate significant differences at the p < 0.05 level.
3.4. N-Metabolism-Related Enzymes and Growth Index Measurements in MdNRT2.4 Overexpression Plant Lines Under Low-N Stress
The leaves of WT and MdNRT2.4 overexpression transformant tobacco lines (OE2, OE4, and OE6) were collected after 15 days of low- and normal-N treatments, and relevant enzyme activities and physiological and biochemical indices were determined. We found that chlorophyll synthesis was impaired and leaves were yellow in plants subjected to low-N stress, which resulted in a decrease in the chlorophyll content. The WT was severely damaged, and the chlorophyll content was significantly lower in WT plants than in transformant lines (Figure 4a). Low-N stress also decreased the net photosynthetic rate in both transformant and WT tobacco. However, the transformant lines maintained a significantly higher photosynthetic rate compared to the WT, with the OE6 line exhibiting the most pronounced increase—approximately 1.3-fold that of the WT (Figure 4b). There was a significant difference in total N accumulation between the WT and transformant lines (Figure 4c). Under normal-N conditions, the total N accumulation in transgenic tobacco plants was significantly higher than in WT plants. When subjected to low-N treatment, the three overexpression lines still exhibited substantially greater N accumulation compared to the WT line, with levels approximately 1.5 times higher. The MDA content is often used as a measure of oxidative damage in plants [37]. The results indicated that the increase in MDA content under low-N stress exacerbated the damage to plants. The MDA content of the WT line was 19.78 nmol/g, while the highest MDA content of the transformant lines was 15.91 nmol/g in the OE2 line. The MDA content of the WT line was significantly higher than that of transformant lines. This indicates that genetically modified tobacco has a lower degree of leaf damage than the WT line and is in a better physiological state than the WT line (Figure 4d). Additionally, we assessed the antioxidant enzyme functions in the plants and observed no notable disparities in CAT, POD, or SOD activity between the WT and genetically modified lines in standard-N environments. Under low-N stress conditions (0.25 mM), the OE4 line exhibited the highest CAT activity, reaching 277.98 U/g—approximately 1.6-fold higher than that in the WT line. Similarly, the POD content in the OE4 line peaked at 5965.56 U/mg, representing a 1.7-fold increase over the WT. Meanwhile, SOD activity was maximized in the OE2 line at 199.64 U/g, roughly 1.5 times the level observed in the WT line (Figure 4f–h). Under low-N stress, the enzyme activities in the transformant lines surpassed those in WT plants. Under low-N stress, the proline contents of both transformant and WT lines increased significantly. The Pro content of the WT line was 46.90 μg/g. The Pro content of the OE6 line was the highest, 136.47 μg/g, which is about 2.9 times that of WT tobacco. The transformant lines had significantly higher Pro content than the WT line (Figure 4h). Under low-N conditions, soluble sugar levels were notably elevated across all lines (Figure 4i). The soluble sugar content in the WT line leaves was 69.11 mg/g, while that in the transformant lines could be as low as 87.29 mg/g, which was significantly higher than that in the WT line. This suggests that the accumulation of osmoregulatory agents was greater in MdNRT2.4 transformant lines than in the WT line. Under low-N conditions, the NR activity in the OE lines was about 1.5 times higher than that in the WT line, especially for OE6, in which it reached 71.20 NO2−h−1g−1 FW. Under normal-N conditions, the NR activity in the OE lines was also significantly higher than that in the WT line (Figure 4j).
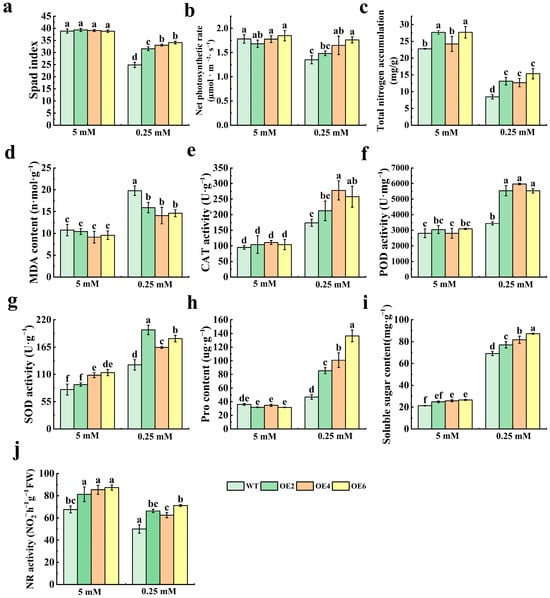
Figure 4.
Physiological indices measured in OE and WT lines under low-N and normal-N conditions. (a) Spad index. (b) Photosynthetic rate. (c) Total N content. (d) MDA content. (e) CAT activity. (f) POD activity. (g) SOD activity. (h) Pro content. (i) Soluble sugar content. (j) NR activity. The data are presented as mean ± SD (n = 3). Different letters on the bar graphs indicate significant differences at the p < 0.05 level.
3.5. Expression Analysis of N-Metabolism-Related Genes Under Low-N Stress Conditions in MdNRT2.4 Overexpression Plant Lines
To investigate the effects of MdNRT2.4 overexpression on other N-metabolism-related genes in tobacco under low-N stress, we analyzed the expression levels of four N-response-related genes: NtGDH1, NtGS1-3, NtGS2, and NtNR (Figure 5). All these genes showed increased expression in low-N environments compared with normal-N conditions. Notably, the expression of the NtGDH1 gene in the OE6 line was approximately four-fold higher than in the WT line (Figure 5a), while NtNR expression was nearly eight times greater in the OE6 plants compared to the WT controls (Figure 5d). These findings indicate that the overexpression of MdNRT2.4 can lead to the upregulation of the expression of N-metabolism-related genes, which can enhance the adaptation of plants to low-N stress.
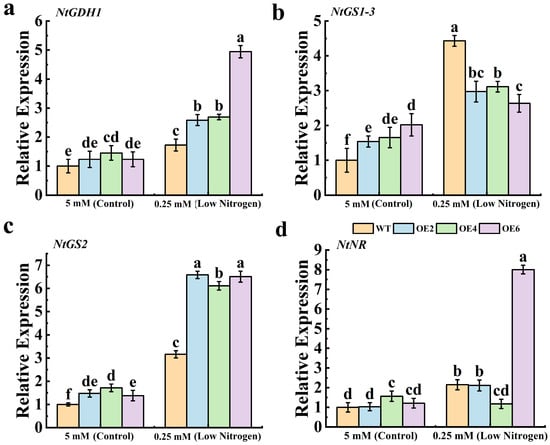
Figure 5.
Expression levels of genes related to N metabolism. (a) NtGDH1. (b) NtGS1-3. (c) NtGS2. (d) NtNR. The data are presented as mean ± SD (n = 3). Different letters on the bar graphs indicate significant differences at the p < 0.05 level.
3.6. Examination of Cis-Acting Elements Within the MdNRT2.4 Promoter
The cis-acting elements upstream of the gene coding region are key areas that regulate transcriptional initiation and gene expression. They perform the fine regulation of promoter activity by binding to specific proteins [38]. To clarify the role of cis-acting elements in the MdNRT2.4 promoter region in response to environmental stress, we used the PlantCARE online software to predict the cis-acting elements of the MdNRT2.4 promoter. There were common growth hormone-responsive elements (TGA-elements) and light-responsive elements (ACE) within the MdNRT2.4 promoter. Three salicylic acid-responsive elements (TCA elements) and two abscisic acid-responsive elements (ABREs) were detected, suggesting that hormonal signaling regulates MdNRT2.4 expression. In addition, a homeopathic regulatory element (CAT-box) associated with the expression of meristematic tissues, a homeopathic regulatory element (HD-Zip) involved in the differentiation of fenestrated cells, and a MYB binding site (MBS) involved in drought were identified (Figure 6a).
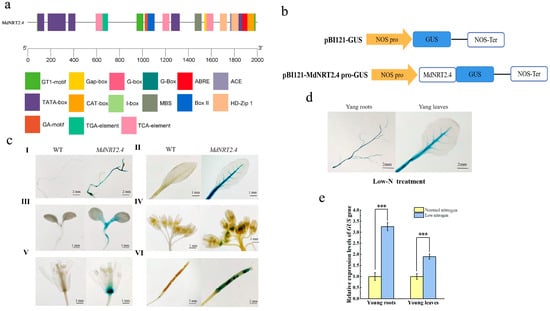
Figure 6.
Promoter analysis of MdNRT2.4. (a) Prediction of cis-acting elements in the MdNRT2.4 promoter. (b) The schematic structure of the recombinant vector. (c) Expression of MdNRT2.4 in different tissues of transformant Arabidopsis lines. I, root; II, vascular bundles; III, stem-tip meristem; IV, flower receptacle; V, anthers; VI, fruit pod. (d) Expression of MdNRT2.4 in young roots and young leaves of transformant Arabidopsis lines after low-N treatment. (e) GUS expression levels in young roots and leaves of transformant Arabidopsis lines after normal-N and low-N treatments. Asterisks indicate significant differences (***: p < 0.001).
3.7. Activity Analysis of pBI121-MdNRT2.4 Pro-GUS-Transformed Arabidopsis Lines at Various Growth Sites
To further investigate the expression pattern of MdNRT2.4, pBI121-MdNRT2.4 pro-GUS-transformed Arabidopsis lines were obtained (Figure 6c). Transformant Arabidopsis exhibited GUS staining activity at different growth sites, including the roots, vascular bundles, stem-tip meristematic tissues, young anthers, receptacles, and seeds in fruit pods. Observations of anthers isolated from flowers at different stages of maturity revealed strong GUS staining activity in the more tender parts, which gradually disappeared as the anthers matured (Figure 6c(IV)). In addition, we stained young roots and leaves after low-N treatment and found that the roots and vascular bundles also exhibited GUS staining activity (Figure 6d). The expression levels of the GUS reporter gene in young roots and young leaves under low-N conditions were analyzed. It was found that low-N treatment induced approximately three-fold higher GUS expression in young roots and two-fold higher expression in young leaves compared to normal nitrogen conditions (Figure 6e).
3.8. MdbHLH3 Binds to the MdNRT2.4 Promoter
To verify whether seven transcription factors responding to low-N can bind to the MdNRT2.4 promoter, we conducted yeast one-hybrid experiments. The structure of the recombinant vector is shown in Figure 7a. First, we found that the self-activating activity of the MdNRT2.4 promoter was inhibited by 45 mM 3-AT. We found that pHIS2-MdNRT2.4 and pGADT7-MdbHLH3 were co-transferred into Saccharomyces cerevisiae Y187 and could grow on SD-/THL + 45 mM 3-AT triple-defect medium. However, yeast cells combined with other transcription factors that bind to the pHIS2-MdNRT2.4 vector could not (Figure S2). MdbHLH3 could bind to the MdNRT2.4 promoter (Figure 7b). Additionally, we confirmed the relationship between MdbHLH3 and the MdNRT2.4 promoter through a dual-luciferase assay (Figure 7d), which showed that MdbHLH3 was able to bind to MdNRT2.4 and activate its expression. The expression of MdbHLH3 under low-N conditions increased after day 2, and it peaked on day 10. The expression levels were roughly 17 times greater in the low-N treatment than in the normal-N treatment.
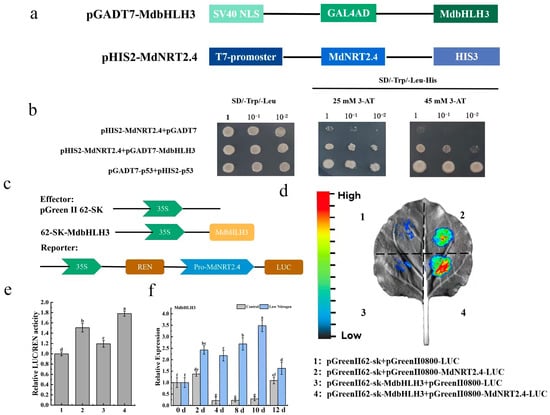
Figure 7.
MdbHLH3 directly binds to the MdNRT2.4 promoter. (a) The schematic structure of the recombinant vector. (b) The yeast one-hybrid assay assessing the binding of MdbHLH3 to the promoter of MdNRT2.4. Negative control: pHIS2-MdNRT2.4+pGADT7; positive control: pGADT7-p53+pHIS-p53. (c) The schematic structure of the recombinant vector. (d) Dual-luciferase imaging analysis, where 1 and 3 are negative controls, and 2 is the positive control. (e) Dual-luciferase activity analysis. (f) Expression analysis of MdbHLH3 under low-N stress. The data are presented as mean ± SD (n = 3). Different letters on the bar graphs indicate significant differences at the p < 0.05 level.
4. Discussion
The application of appropriate amounts of N fertilizer can effectively enhance the carbon sequestration potential of forest ecosystems. However, excessive N input may intensify the global greenhouse effect by stimulating soil emissions of nitrous oxide (N2O). Notably, plant NRTs play a core role in regulating the efficiency of NO3− assimilation within root systems [39,40]. The expression of NRT2.4 was moderately induced in both the shoots and roots of Arabidopsis [41]; NRT2.5 transcripts were predominantly found in the roots and, to a lesser extent, in the shoots [42]. We found that the expression of MdNRT2.4 was upregulated under low-N stress in apple (Figure 1b), suggesting that it might positively regulate the response to low-N stress. To study the role of MdNRT2.4, we transfected it into Nicotiana benthamiana and obtained MdNRT2.4 overexpression tobacco lines. We also found that the overexpression of MdNRT2.4 enhanced the biomass of tobacco under low-N stress (Figure 2f,g) and that the OE lines had more primary and lateral roots than the WT line (Figure 2c–e), suggesting that the overexpression of MdNRT2.4 led to increased levels in the roots, which is consistent with the results of previous studies. For example, in Arabidopsis, the expression of NRT2.4 in the roots increases 12-fold during N starvation on a low-N medium [43]. Under low-N stress, the expression of NtGDH1, NtGS1-3, NtGS2, and NtNR was significantly upregulated (Figure 5), suggesting that MdNRT2.4 may improve low-N tolerance by activating their expression [44].
Increasing the rate of photosynthesis can greatly enhance crop production and the N utilization efficiency [45]. The photosynthetic rate and total N accumulation of the transformant lines overexpressing MdNRT2.4 were significantly higher than those of the WT line (Figure 4b,c). This might be attributed to the synergistic enhancement mechanism of nitrogen absorption and photosynthesis. Under normal growth conditions, organelles generate intracellular reactive oxygen species (ROS) in minimal quantities; under abiotic stress, ROS accumulate in large amounts. In addition to organelles, the primary production of ROS occurs in the plasma membrane and plastids under stress [46]. SOD, CAT, and POD can act as ROS-scavenging enzymes in plants. These antioxidant enzymes are present in various parts of plant cells and efficiently neutralize ROS [47]. Under low-N stress, antioxidant enzyme activities were significantly higher in MdNRT2.4 overexpression transformant lines than in WT plants (Figure 4e–g). This indicates that enhanced antioxidant enzyme activity allows transformant lines to scavenge ROS faster and reduces damage to the cell membrane. Consequently, the elevated expression of MdNRT2.4 promoted the activity of antioxidant enzymes under low-N stress conditions and improved the resilience of plants to stress. The initial reaction of plants to abiotic stress, which also leads to the generation of ROS, results in the generation of MDA as a secondary product [48]. MDA is often used as an indicator of lipid peroxidation, which is a consequence of oxidative stress [49]. Under low-N stress, the MDA levels in the WT line notably exceeded those in MdNRT2.4 overexpression plant lines (Figure 4d), indicating that genetically modified tobacco had relatively little damage, as well as increased stress tolerance and improved resistance to adverse conditions. Under abiotic stress, plants can activate the ROS-scavenging system and increase the content of osmoregulatory substances to enhance stress resistance [50]. The levels of proline and soluble sugars in the transformant lines exceeded those in the WT line (Figure 4h,i), suggesting that damage to the transformant lines is reduced under low-N conditions. Under low-N stress, the NR activity in the transformant lines was significantly higher than that in the WT line (Figure 4j). The results indicated that MdNRT2.4 played an important role in regulating nitrate uptake, transport, and assimilation. Nitrate metabolism can significantly promote plant respiration [51]. When plants absorb nitrate (NO3⁻), they need to consume a large amount of energy (ATP) and reducing power (NADH) to convert it into ammonium (NH4⁺), which directly induces an increase in the mitochondrial respiratory rate. Accumulating evidence indicates that nitrate transporters often exhibit pleiotropy in the acquisition of other nutrients. For example, studies have shown that nitrate signaling can simultaneously achieve the synergistic activation of nitrate response genes and phosphorus response genes through NRT1.1B-SPX4, thereby achieving the nutritional balance of nitrogen and phosphorus in plants [52]. This indicates that the nitrate transport mediated by MdNRT2.4 may also affect the in vivo balance of other minerals. This study confirmed that the overexpression of MdNRT2.4 can enhance the low-N tolerance of tobacco. In apple, MdNRT2.4 or its homologous genes can be overexpressed through transgenic or gene-editing techniques to cultivate varieties that grow better under low-N conditions. In addition, this gene can also be applied to economic crops such as tomatoes and potatoes, enhancing their adaptability to low-N environments by improving the nitrogen absorption capacity of their root systems and providing new strategies for sustainable agriculture. However, it should be pointed out that this study is mainly based on the results obtained from the tobacco model system, and its direct applicability to perennial woody plants (such as apple) still needs further verification. In addition, the influence of complex environmental factors in the field on the function of MdNRT2.4 also awaits further research.
NRT2.1, NRT2.4, and NRT2.5 facilitate the absorption of NO3− by the roots [43,53]. In environments with low N, the expression of NRT2.4 increased rapidly to high levels, and the expression of NRT2.1 increased slowly. The expression of AtNRT2.5 was higher in the root hair areas of both the primary and lateral roots, and GUS activity was absent in the older root regions. NRT2.4 was expressed in the primary veins of source leaves and shoot vascular tissues, and it mediated the transport of NO3− to the phloem for remobilization under low-N conditions. We found that NRT2.4 was expressed in both the young roots and vascular tissues of Arabidopsis (Figure 6a,b). We also found that NRT2.4 was expressed in stem-tip meristems, receptacles, young anthers, and seeds (Figure 6c). This suggests that MdNRT2.4 plays a role in the transport of NO3− to these tissues, which may promote the utilization of NO3− in apple and improve its tolerance to low-N stress. In our study, the expression levels of MdNRT2.4 in the young roots and young leaves of pMdNRT2.4::GUS-transformed Arabidopsis lines were upregulated after exposure to low-N stress compared with that under normal N conditions, which confirms that MdNRT2.4 may be involved in the transport of N metabolites under low-N stress. Subcellular localization analyses revealed that MdNRT2.4 and NRT2.5 were both localized to the plasma membrane [42]. This plasma membrane localization is consistent with the role of NRT2.4 as a transporter of NO3−.
Numerous genes associated with N have been discovered in plants, including structural genes, TFs, and protein kinases (PKs) [54]. TFs and PKs are involved in the N signaling pathway in response to N stress [54,55]. Certain TFs have been shown to play a role in regulating the expression of NRT genes, such as MdATG18a, which positively regulates the expression of three genes encoding high-affinity NTRs: MdNRT2.1, MdNRT2.4, and MdNRT2.5; thus, MdATG18a might have a significant effect on N acquisition [33]. MdMYB88 and MdMYB124 can be combined with the MdNRT1.7 and MdNRT2.4 promoters to increase the uptake, distribution, and reuse of nitrate under N-limited conditions, thereby affecting nitrate remobilization in apple plants [21]. MdMYB10 binds to the MdNRT2.4-1 promoter and activates its expression to enhance nitrate uptake [56]. Similarly, a yeast one-hybrid assay verified the interaction between MdbHLH3 and MdNRT2.4 (Figure 7b); we also verified that MdbHLH3 could bind to the MdNRT2.4 promoter and activate its expression via LUC reporter gene analysis (Figure 7d). MdbHLH3 is an alkaline helix–loop–helix transcription factor (bHLH TF) that regulates anthocyanin biosynthesis and also regulates malic acid and carbohydrate accumulation [57,58]. Previous research indicates that MdbHLH3 attaches to the MdPFPβ promoter to induce its activation and thus promotes the accumulation of soluble sugars and accelerate photosynthesis in apple [59]. It has been confirmed that TabHLH489 is involved in nitrate signal transduction in wheat [60]. In apple rootstocks, MhbHLH130 can bind to the chalcone synthase gene (CHS) promoter to regulate flavonoid biosynthesis to improve plant N absorption efficiency [61]. We hypothesized that MdbHLH3 may interact with the promoter of MdNRT2.4 to regulate N metabolism in plants and enhance the low-N tolerance of plants. The expression of MdbHLH3 was upregulated under low-N conditions (Figure 7f), which further demonstrates that MdbHLH3 activates the expression of the MdNRT2.4 promoter under low-N conditions. Some factors that negatively regulate NRT gene family members have also been found. For example, in vivo and in vitro experiments have verified that the ABA-reactive transcription factor MdABI5 can bind to the MdNRT1.5 promoter and inhibit its expression, thus regulating the transport of nitrate from roots to buds [18]. NIGT1.1 was shown to be a negative regulator of NRT2.1 in Arabidopsis according to LUC reporter gene analysis [62]. In conclusion, the analysis of interactions between MdbHLH3 and the MdNRT2.4 promoter revealed that the MdbHLH3 TF plays an important role in the nitrate transport process.
5. Conclusions
This study demonstrated that MdNRT2.4 was upregulated under low-N stress, and its overexpression enhanced low-N tolerance in transgenic lines. This improvement is associated with elevated activities of antioxidant enzymes, including POD, CAT, and SOD, which facilitate faster ROS scavenging and mitigate oxidative damage to cell membranes. Tissue-specific expression analysis revealed that the MdNRT2.4 promoter drives expression in roots, vascular bundles, seeds in fruit pods, and young anther sites of transformant Arabidopsis plants. Additionally, yeast one-hybrid and dual-luciferase assays confirmed that MdbHLH3 binds to the MdNRT2.4 promoter and activates its transcription (Figure 8). These findings provide novel insights into the molecular mechanisms underlying apple’s adaptation to low-N stress.
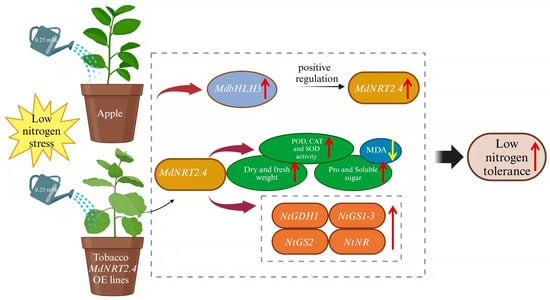
Figure 8.
A working model for the role of MdNRT2.4 in response to low-N stress. POD: peroxidase; 0.25 mM: 0.25 mM low-N nutrient solution; CAT: catalase; SOD: superoxide dismutase; Pro: proline; MDA: malondialdehyde. An upward arrow represents an increase and a downward arrow represents a decrease. The image was drawn using Biorender online software (https://www.biorender.com/).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11060662/s1, Figure S1: Multiple sequence alignment of MdNRT2.4; Figure S2: Yeast one-hybrid assay assessing the binding of MdbHLH2, MdNAC2, MdAP2-1, MdMYB2, MdWRKY4 and MdWRKY5 to the promoter of MdNRT2.4; Table S1: The primers used in this study; Table S2: Protein sequences of different species; Table S3: The Latin names of 19 species.
Author Contributions
J.L.: curating data, drafting original drafts, and visualizing; K.L.: gene cloning, vector construction; C.S.: data survey, data curation; Q.H.: visualization; X.N.: data curation; Q.D.: investigations, provision of programs; D.H.: supervision, provision of materials; Q.W.: supervision, review, provision of funds and materials. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number: 32460727); Guizhou Provincial Basic Research Program (Natural Science) (Grant number: Qiankehejichu ZD [2025] 069 and Qiankehe-ZK [2023] Yiban 103); the Open Project Fund of the Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education) (Grant number: Qianjiaoji [2022] 431).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zeng, Z.; Song, Z.; Wang, F.; Tian, D.; Mi, W.; Huang, X.; Wang, J.; Song, L.; Yang, Z.; et al. Vital roles of soil microbes in driving terrestrial nitrogen immobilization. Glob. Change Biol. 2021, 27, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, T.; Sakakibara, H. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 2016, 68, 2501–2512. [Google Scholar] [CrossRef]
- Miller, A.J.; Cramer, M.D. Root Nitrogen Acquisition and Assimilation. Plant Soil 2005, 274, 1–36. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, L.; Jin, S.; Guo, Q.; Hou, J. The influence of plants on the migration and transformation of nitrogen in plant-soil systems: A review. J. Soil Sci. Plant Nutr. 2022, 22, 4084–4102. [Google Scholar] [CrossRef]
- Wang, Q.; Li, S.; Li, J.; Huang, D. The utilization and roles of nitrogen in plants. Forests 2024, 15, 1191. [Google Scholar] [CrossRef]
- Yang, J.; Udvardi, M. Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. J. Exp. Bot. 2018, 69, 855–865. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, C.; Huang, D.; Dong, Q.; Li, P.; Ma, F. High-efficient utilization and uptake of N contribute to higher NUE of ‘Qinguan’ apple under drought and N-deficient conditions compared with ‘Honeycrisp’. Tree Physiol. 2019, 39, 1880–1895. [Google Scholar] [CrossRef] [PubMed]
- Dechorgnat, J.; Nguyen, C.T.; Armengaud, P.; Jossier, M.; Diatloff, E.; Filleur, S.; Daniel-Vedele, F. From the soil to the seeds: The long journey of nitrate in plants. J. Exp. Bot. 2011, 62, 1349–1359. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Hsu, P.-K.; Tsay, Y.-F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Hao, D.-L.; Zhou, J.-Y.; Yang, S.-Y.; Qi, W.; Yang, K.-J.; Su, Y.-H. Function and regulation of ammonium transporters in plants. Int. J. Mol. Sci. 2020, 21, 3557. [Google Scholar] [CrossRef]
- Forde, B.G. Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Nacry, P.; Bouguyon, E.; Gojon, A. Nitrogen acquisition by roots: Physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 2013, 370, 1–29. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef]
- Zou, X.; Liu, M.Y.; Wu, W.H.; Wang, Y. Phosphorylation at Ser28 stabilizes the Arabidopsis nitrate transporter NRT2.1 in response to nitrate limitation. J. Integr. Plant Biol. 2020, 62, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Yan, Y.; Liu, W.; Zhang, W.; Gao, L.; Tian, Y. Knock-down of CsNRT2.1, a cucumber nitrate transporter, reduces nitrate uptake, root length, and lateral root number at low external nitrate concentration. Front. Plant Sci. 2018, 9, 722. [Google Scholar] [CrossRef]
- Liu, G.; Rui, L.; Yang, Y.; Liu, R.; Li, H.; Ye, F.; You, C.; Zhang, S. Identification and functional characterization of MdNRT1.1 in nitrogen utilization and abiotic stress tolerance in Malus domestica. Int. J. Mol. Sci. 2023, 24, 9291. [Google Scholar] [CrossRef]
- Liu, Y.J.; Gao, N.; Ma, Q.J.; Zhang, J.C.; Wang, X.; Lu, J.; Hao, Y.J.; Wang, X.F.; You, C.X. The MdABI5 transcription factor interacts with the MdNRT1.5/MdNPF7.3 promoter to fine-tune nitrate transport from roots to shoots in apple. Hortic. Res. 2021, 8, 236. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, K.; Li, J.; Huang, D. Overexpression of apple MdNRT1.7 enhances low nitrogen tolerance via the regulation of ROS scavenging. J. Biol. Macromol. 2024, 293, 139358. [Google Scholar] [CrossRef]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.S.; Chaillou, S.; Ferrario-Méry, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, K.; Kan, Z.; Dang, H.; Feng, S.; Yang, Y.; Li, L.; Hou, N.; Xu, L.; Wang, X.; et al. The regulatory module MdBT2-MdMYB88/MdMYB124-MdNRTs regulates nitrogen usage in apple. Plant Physiol. 2021, 185, 1924–1942. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Liu, C.; Xu, X.; Xing, Y.; Liu, J.; Lyu, M.; Feng, Z.; Zhang, X.; Qin, H.; Jiang, H.; et al. Effects of magnesium on nitrate uptake and sorbitol synthesis and translocation in apple seedlings. Plant Physiol. Biochem. 2023, 196, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Noguero, M.; Lacombe, B. Transporters involved in root nitrate uptake and sensing by Arabidopsis. Front. Plant Sci. 2016, 7, 1391. [Google Scholar] [CrossRef] [PubMed]
- Galvan, A.; Fernández, E. Eukaryotic nitrate and nitrite transporters. Cell. Mol. Life Sci. 2001, 58, 225–233. [Google Scholar] [CrossRef]
- Yan, N. Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef]
- Tong, J.; Walk, T.C.; Han, P.; Chen, L.; Shen, X.; Li, Y.; Gu, C.; Xie, L.; Hu, X.; Liao, X.; et al. Genome-wide identification and analysis of high-affinity nitrate transporter 2 (NRT2) family genes in rapeseed (Brassica napus L.) and their responses to various stresses. BMC Plant Biol. 2020, 20, 464. [Google Scholar] [CrossRef]
- Kotur, Z.; Mackenzie, N.; Ramesh, S.; Tyerman, S.D.; Kaiser, B.N.; Glass, A.D.M. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012, 194, 724–731. [Google Scholar] [CrossRef]
- Jia, L.; Hu, D.; Wang, J.; Liang, Y.; Li, F.; Wang, Y.; Han, Y. Genome-wide identification and functional analysis of nitrate transporter genes (NPF, NRT2 and NRT3) in maize. Int. J. Mol. Sci. 2023, 24, 12941. [Google Scholar] [CrossRef]
- Taulemesse, F.; Le Gouis, J.; Gouache, D.; Gibon, Y.; Allard, V. Post-flowering nitrate uptake in wheat is controlled by N status at flowering, with a putative major role of root nitrate transporter NRT2.1. PLoS ONE 2015, 10, e0120291. [Google Scholar] [CrossRef]
- Okamoto, M.; Kumar, A.; Li, W.; Wang, Y.; Siddiqi, M.Y.; Crawford, N.M.; Glass, A.D. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006, 140, 1036–1046. [Google Scholar] [CrossRef]
- Peng, H.; Yi, Y.; Li, J.; Qing, Y.; Zhai, X.; Deng, Y.; Tian, J.; Zhang, J.; Hu, Y.; Qin, X.; et al. A haplotype-resolved genome assembly of Malus domestica ‘Red Fuji’. Sci. Data 2024, 11, 592. [Google Scholar] [CrossRef]
- Wu, Y.; Su, S.X.; Wang, T.; Peng, G.H.; He, L.; Long, C.; Li, W. Identification and expression characteristics of NLP (NIN-like protein) gene family in pepper (Capsicum annuum L.). Mol. Biol. Rep. 2023, 50, 6655–6668. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jia, X.; Huo, L.; Che, R.; Gong, X.; Wang, P.; Ma, F. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018, 41, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, F.; Farhadi, A.; Rahimi-Mianji, G.; Gholizadeh, M. Molecular genetics and bioinformatics analysis of EDG1 and AKIRIN2 genes in Iranian fat-tailed and nonfat-tailed sheep breeds. Small Rumin. Res. 2016, 144, 263–268. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Fu, D.; Chen, Y.; Gao, F. Yeast one-hybrid screening for transcription factors of IbbHLH2 in purple-fleshed sweet potato. Genes 2023, 14, 1042. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Kong, Y.; Zhao, W.; Wang, F. Measurement of cellular MDA content through MTBE-extraction based TBA assay by eliminating cellular interferences. J. Pharm. Biomed. 2024, 248, 116332. [Google Scholar] [CrossRef]
- Govardhana, M.; Kumudini, B.S. In-silico analysis of cucumber (Cucumis sativus L.) Genome for WRKY transcription factors and cis-acting elements. Comput. Biol. Chem. 2020, 85, 107212. [Google Scholar] [CrossRef]
- Liu, K.H.; Liu, M.; Lin, Z.; Wang, Z.F.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425. [Google Scholar] [CrossRef]
- Ma, Q.; Zhao, C.; Hu, S.; Zuo, K. Arabidopsis calcium-dependent protein kinase CPK6 regulates drought tolerance under high nitrogen by the phosphorylation of NRT1.1. J. Exp. Bot. 2023, 74, 5682–5693. [Google Scholar] [CrossRef]
- Svietlova, N.; Zhyr, L.; Reichelt, M.; Grabe, V.; Mithöfer, A. Glutamine as sole nitrogen source prevents induction of nitrate transporter gene NRT2.4 and affects amino acid metabolism in Arabidopsis. Front. Plant Sci. 2024, 15, 1369543. [Google Scholar] [CrossRef] [PubMed]
- Lezhneva, L.; Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Boutet-Mercey, S.; Zoufan, P.; Sakakibara, H.; Daniel-Vedele, F.; Krapp, A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014, 80, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Lezhneva, L.; Boutet-Mercey, S.; Orsel, M.; Bréhaut, V.; Miller, A.; Daniel-Vedele, F.; Sakakibara, H.; et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 2012, 24, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Hou, Q.; Deng, H.; Xiao, L.; Cai, X.; Shang, C.; Qiao, G. Overexpression of PavHIPP16 from Prunus avium enhances cold stress tolerance in transgenic tobacco. BMC Plant Biol. 2024, 24, 536. [Google Scholar] [CrossRef]
- Hu, W.; Zhao, M.; Zhang, S.; Li, Y.; Dai, J.; Gu, C.; Li, X.; Yang, L.; Qin, L.; Liao, X. Optimized leaf storage and photosynthetic nitrogen trade-off promote synergistic increases in photosynthetic rate and photosynthetic nitrogen use efficiency. Physiol. Plant. 2023, 175, e14013. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D.; et al. ROS generated from biotic stress: Effects on plants and alleviation by endophytic microbes. Front. Plant Sci. 2022, 13, 1042936. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Pospíšil, P. Malondialdehyde enhances PsbP protein release during heat stress in Arabidopsis. Plant Physiol. Bioch. 2023, 202, 107984. [Google Scholar] [CrossRef]
- Sofo, A.; Dichio, B.; Xiloyannis, C.; Masia, A. Effects of different irradiance levels on some antioxidant enzymes and on malondialdehyde content during rewatering in olive tree. Plant Sci. 2004, 166, 293–302. [Google Scholar] [CrossRef]
- Miura, K.; Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef]
- Bloom, A.J.; Sukrapanna, S.S.; Warner, R.L. Root Respiration Associated with Ammonium and Nitrate Absorption and Assimilation by Barley 1. Plant Physiol. 1992, 99, 1294–1301. [Google Scholar] [CrossRef]
- Hu, B.; Jiang, Z.; Wang, W.; Qiu, Y.; Zhang, Z.; Liu, Y.; Li, A.; Gao, X.; Liu, L.; Qian, Y.; et al. Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 2019, 5, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, B.; Chu, C. Towards understanding the hierarchical nitrogen signalling network in plants. Curr. Opin. Plant Biol. 2020, 55, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.F.; Li, H.L.; An, X.H.; Song, L.Q.; You, C.X.; Zhao, L.L.; Tian, Y.; Wang, X.F. MdMYB10 affects nitrogen uptake and reallocation by regulating the nitrate transporter MdNRT2.4-1 in the red flesh apple. Hortic. Res. 2022, 9, uhac016. [Google Scholar] [CrossRef]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Yu, J.Q.; Gu, K.D.; Sun, C.H.; Zhang, Q.Y.; Wang, J.H.; Ma, F.F.; You, C.X.; Hu, D.G.; Hao, Y.J. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 2021, 19, 285–299. [Google Scholar] [CrossRef]
- Yu, J.Q.; Gu, K.D.; Zhang, L.L.; Sun, C.H.; Zhang, Q.Y.; Wang, J.H.; Wang, C.K.; Wang, W.Y.; Du, M.C.; Hu, D.G. MdbHLH3 modulates apple soluble sugar content by activating phosphofructokinase gene expression. J. Integr. Plant Biol. 2022, 64, 884–900. [Google Scholar] [CrossRef]
- Yang, F.; Li, X.; Liu, S.; Lyu, J.; Ge, Z.; Bai, M.Y. TabHLH489 suppresses nitrate signaling by inhibiting the function of TaNLP7-3A in wheat. J. Integr. Plant Biol. 2024, 67, 1162–1178. [Google Scholar] [CrossRef]
- Wang, X.; Chai, X.; Gao, B.; Deng, C.; Günther, C.S.; Wu, T.; Zhang, X.; Xu, X.; Han, Z.; Wang, Y. Multi-omics analysis reveals the mechanism of bHLH130 responding to low-nitrogen stress of apple rootstock. Plant Physiol. 2023, 191, 1305–1323. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Konishi, M.; Kiba, T.; Sakuraba, Y.; Sawaki, N.; Kurai, T.; Ueda, Y.; Sakakibara, H.; Yanagisawa, S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 2018, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).