Abstract
α-Linolenic acid (ALA) is an important essential omega-3 (ω-3) polyunsaturated fatty acid for the maintenance of human health. Although ALA has traditionally been obtained from plant sources, microbial fermentation has emerged as a promising alternative for its sustainable and cost-effective production. However, most of the present approaches rely on genetically modified organisms, which present regulatory and consumer-acceptance concerns. In this study, we aimed to develop a high-ALA-producing strain of Aspergillus oryzae, a Generally Recognized As Safe (GRAS) microorganism widely used in food production in Japan, through self-cloning, a form of genetic engineering that utilizes only the host’s own DNA. To achieve this, an endogenous ω-3 desaturase gene (fad3), which catalyzes the conversion of linoleic acid to ALA, was identified via BLASTP analysis. Subsequently, a multicopy A. oryzae strain (Aofad3-MC) overexpressing fad3 was constructed. This strain increased ALA production, with ALA comprising 30.7% of the total lipids. Furthermore, k-mer analysis confirmed the absence of foreign vector sequences, verifying that Aofad3-MC was constructed through self-cloning. In addition to the identification of the A. oryzae ω-3 desaturase gene, this study provides a microbial platform for the sustainable production of ALA, with potential applications across the food, feed, and related industries.
1. Introduction
Polyunsaturated fatty acids (PUFAs) are lipid components that play important roles in the maintenance of human health [1]. PUFAs are mainly classified into ω-6 and ω-3 fatty acids, based on their structural configuration [2]. In recent years, an imbalance in human intake of these fatty acids has been reported, characterized by the excessive consumption of ω-6 fatty acids and the insufficient intake of ω-3 fatty acids. This imbalance increases the risks of cardiovascular, chronic, cancer, inflammatory, autoimmune, and obesity-related diseases [3,4,5,6]. α-Linolenic acid (ALA, C18:3 n-3), an ω-3 fatty acid, has been demonstrated to provide various health benefits, including blood pressure and triglyceride reduction, anti-inflammatory effects, and cognitive function improvement [7,8]. In addition, ALA is an essential precursor for the synthesis of long-chain ω-3 fatty acids, including eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). However, as an essential fatty acid, ALA cannot be synthesized in the human body and therefore must be obtained from the diet [9]. ALA-rich oils are limited to only a few plant sources, such as perilla and flaxseed, and their supply is constrained by low productivity often due to environmental factors, such as climate and soil conditions [10]. Consequently, commercially available oils containing high concentrations of ALA are traded at relatively high prices; therefore, there is a growing need for alternative, low-cost, and sustainable methods for the production of ALA-containing oils.
Microbial fermentation has been explored as alternative ALA production method. ALA is biosynthesized from stearic acid (C18:0) through sequential desaturation reactions at the Δ9, Δ12, and ω-3 (Δ15) positions (Figure 1). In the final step of this biosynthetic pathway, ω-3 (Δ15) desaturase is essential for the conversion of linoleic acid (C18:2, an ω -6 fatty acid) to ALA (an ω-3 fatty acid) [11]. Omega-3 desaturase genes have been identified in various organisms, including plants [12,13], nematodes [14], yeast [15,16], algae [17], cyanobacteria [18], and fungi [19,20]. Microbial fermentation ALA production methods have been attempted by introducing these ω-3 desaturase genes into heterologous microbial hosts [21]. For example, the heterologous expression of the Fusarium moniliforme ω-3 desaturase gene (Fm1) in the oleaginous yeast Yarrowia lipolytica was reported in a previous study to enable the accumulation of ALA to reach 28.1% of the total lipids [19]. Subsequent studies using recombinant Y. lipolytica have demonstrated that low-temperature cultivation conditions can enhance ALA productivity further [22]. Filamentous fungi have also been studied for their ALA production abilities. For example, ALA biosynthesis was enabled in Mucor circinelloides by introducing the ω-3 desaturase gene Δ15D from Mortierella alpina [20]. However, products derived from recombinant heterologous genes are subject to regulatory frameworks such as the Cartagena Protocol and often face limited social acceptance [23]. Therefore, sustainable industrial ALA production requires the selection of suitable host microorganisms without the use of genetically modified organisms.
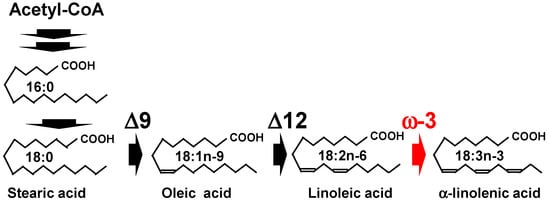
Figure 1.
The α-linolenic acid (ALA) biosynthesis pathway. Δ9, Δ12, and ω-3 indicate the respective desaturase enzymes. The red arrow indicates ω-3 desaturase, a key enzyme responsible for ALA production.
Aspergillus oryzae is a filamentous fungus that has long been used in the manufacture of traditional Japanese fermented foods such as miso, soy sauce, and sake. Based on many years of dietary use, the United States Food and Drug Administration has designated A. oryzae as a Generally Recognized As Safe (GRAS) organism [24,25]. This fungus has a high protein secretion capacity, including high levels of hydrolytic enzymes, such as amylases, proteases, and cellulases [26,27]. Various genetic engineering techniques have been established for A. oryzae [28,29,30]. Due to these characteristics and advancements, A. oryzae is widely used for the production of enzymes [31], and is useful as a platform for the biosynthesis and expression of various proteins and natural products [32].
A. oryzae is also known to be an oleaginous fungus, with its dry cell weight containing a relatively high lipid content [33]. A. oryzae mainly produces four fatty acids: palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), and linoleic acid (C18:2) [34]. Its fatty acid content and composition are suitable for applications in food, feed, and biomass and give it potential for future lipid production. Ongoing research aims to enhance its fatty acid production [35]. For example, one study reported the knockout of the acyl-CoA synthetase gene faaA (AO090011000642), which converts fatty acids into fatty acyl-CoA, resulting in an approximately 9.2-fold increase in fatty acid production [36]. In another study, fatty acid production was increased by the overexpression of a lipase gene (AO090701000644) and a diacylglycerol O-acyltransferase gene (AO090011000863) [37]. Another study reported that the introduction of heterologous lipid-related enzymes (e.g., Δ6-desaturase, Δ6-elongase, and DGAT2) derived from M. alpina enabled the accumulation of functional ω-6 fatty acids, including γ-linolenic acid (GLA) and dihomo-γ-linolenic acid (DGLA), which were not detected in the wild-type strains [38]. These metabolic engineering strategies demonstrate that A. oryzae is a suitable host for customizing fatty acid production processes.
However, the biosynthesis of ALA and other ω-3 fatty acids by A. oryzae has been relatively underexplored. Sakuradani et al. previously reported that A. oryzae natively produces small amounts of ALA [34]. Based on this result, we hypothesized that A. oryzae may possess ω-3 desaturase. However, the A. oryzae ω-3 desaturase gene has not yet been identified, and no attempts have yet been made to enhance ALA production in A. oryzae. In this study, we first aimed to identify the ω-3 desaturase gene in A. oryzae and then we attempted to enhance ALA production by overexpressing this endogenous gene. This approach enables ALA production based on self-cloning, a form of genetic engineering that utilizes only the host’s own DNA sequences, potentially facilitating industrially viable, socially acceptable, low-cost, and sustainable ALA production.
2. Materials and Methods
2.1. Media
Czapek-Dox (CD) medium (6.0 g/L NaNO3, 0.52 g/L KCl, 1.52 g/L KH2PO4, 10 g/L D-glucose, 0.49 g/L MgSO4·7H2O, 0.001 g/L FeSO4·7H2O, 0.0088 g/L ZnSO4·7H2O, 0.00004 g/L CuSO4·5H2O, 0.0001 g/L Na2B4O7·10H2O, and 0.00005 g/L {NH4}6Mo7O24·4H2O), glucose yeast extract NaCl (GYN) medium (20 g/L glucose, 10 g/L yeast extract, 30 g/L NaCl), and yeast extract peptone dextrose (YPD) medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L D-glucose) were used for A. oryzae culture. A. oryzae transformants were selected on CD-pyr medium supplemented with 0.1 mg/L pyrithiamine. For evaluating ALA production, a low nitrogen source concentration medium, ssCD medium (2 g/L NaNO3, 0.17 g/L KCl, 0.51 g/L KH2PO4, 30 g/L starch, 0.16 g/L MgSO4·7H2O, 0.0017 g/L FeSO4·7H2O, 0.017 g/L EDTA, 0.0073 g/L ZnSO4·7H2O, 0.0036 g/L H3BO3, 0.0017 g/L MnCl2·4H2O, 0.00053 g/L CoCl2·5H2O, 0.00053 g/L CuSO4·5H2O, 0.00036 g/L {NH4}6Mo7O24·4H2O) was used. All media were adjusted to a pH of 6.5 and sterilized. For the preparation of solid plates, agar was added to the medium at a concentration of 2%.
2.2. A. oryzae Strains and Genomic DNA
In this study, the A. oryzae strain RIB40 (NBRC 100959), a representative experimental strain of A. oryzae was obtained from the Biological Resource Center, National Institute of Technology and Evaluation (NBRC, Chiba, Japan) and used as the wild-type strain. To extract the genomic DNA, the RIB40 strain was cultured in YPD medium, the mycelia were collected using a microspatula and dehydrated with paper towels, and the genomic DNA was extracted from the mycelia using the ISOPLANT kit (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions.
2.3. Construction of Plasmid and DNA Fragments for A. oryzae Transformation
The A. oryzae gene registered as locus tag AO090010000714 in the GenBank genetic sequence database (https://www.ncbi.nlm.nih.gov/gene/5999798, accessed on 1 October 2025) was predicted to be a ω-3 desaturase gene (fad3). To obtain DNA fragments for the overexpression of fad3, two plasmids were constructed and cloned in Escherichia coli for plasmid propagation. The plasmid map and the schematic diagram of the constructed DNA fragments are shown in Figure S1, and the corresponding gene sequences are listed in Table S1.
2.3.1. Plasmid pPTRI [Ptef1-fad3-Tfad3]
To obtain a vector DNA fragment, the plasmid pPTRI (Takara Bio, Kusatsu, Japan), containing the A. oryzae pyrithiamine resistance marker gene, ptrA, and the E. coli ampicillin resistance marker gene, AmpR, was digested using the NdeI restriction enzyme (Nippon Gene, Tokyo, Japan) and purified using a QIAEX II gel extraction kit (Qiagen, Hilden, Germany). The tef1 promoter region (Ptef1) [39] and fad3 gene, including the terminator region (Tfad3), were then amplified via PCR using KAPA HiFi polymerase (KAPA Biosystems, Wilmington, MA, USA), using the genomic DNA of RIB40 as a template. The primer sets used in each PCR are listed in Table S2. The fragments were then purified using a QIAEX II gel extraction kit. The two purified DNA fragments were employed as insert DNA fragments. The pPTRI plasmid [Ptef1-fad3-Tfad3] was constructed via Gibson assembly using these three DNA fragments with NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA), according to the manufacturer’s protocol.
2.3.2. Plasmid pPTRI [PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3]
To obtain a vector DNA fragment, pPTRI [Ptef1-fad3-Tfad3] was digested with the restriction enzyme NotI and purified using a QIAEX II Gel Extraction Kit. The amyB promoter region (PamyB) [40] and the fad3 gene, including the terminator region, were amplified via PCR and then purified (Table S2). The two purified DNA fragments were employed as insert DNA fragments. The plasmid pPTRI [PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3] was constructed via Gibson assembly using these three DNA fragments with NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA).
2.3.3. Cloning and Preparation of DNA Fragments for A. oryzae Transformation
The constructed plasmids were cloned and amplified in E. coli strain NEB 5-alpha competent cells (New England Biolabs). The plasmids were extracted using a QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). Plasmids containing the target sequences were digested with the MfeI restriction enzyme (New England Biolabs, Ipswich, MA, USA) and purified. In this way, the DNA fragments ptrA-Ptef1-fad3-Tfad3 and ptrA-PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3 for the transformation of A. oryzae were prepared.
2.4. Transformation of A. oryzae
Prepared the DNA fragments were introduced into the protoplasts of the A. oryzae RIB40 strain. Protoplast preparation and transformation were conducted according to the protocol for the pPTRI transformation vector (Takara Bio Inc., Shiga, Ogasu City, Japan) [41]. Transformants were selected by spreading on CD-pyr medium and culture at 30 °C for 5–7 days. The successful introduction of the DNA fragment was confirmed by PCR using the genomic DNA of the obtained strains. The strain containing a single-copy DNA fragment (ptrA-Ptef1-fad3-Tfad3) was named Aofad3, and the strain containing a multi-copy DNA fragment (ptrA-PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3) was named Aofad3-MC.
2.5. Spore Suspension Preparation
To induce spore formation, the RIB40 strain was cultured on CD agar, while the Aofad3 and Aofad3-MC strains were cultured on CD-pyr agar at 30 °C for 4 days. The resulting spores were suspended in a solution containing 1 g/L Tween 80 and 4 g/L NaCl. The optical density of the prepared spore suspensions was measured at 600 nm (OD600) and used to adjust the inoculation volume at a final OD600 of 0.02.
2.6. Evaluation of ALA Production
To examine ALA production by the RIB40, Aofad3, and Aofad3-MC strains, spore suspensions of each strain were inoculated into 100 mL of ssCD medium in a 500 mL unbaffled Erlenmeyer flask at a final OD600 of 0.02. The spore suspensions were then cultured at 30 °C and 100 rpm for 2–4 days. The dry cell weight (DCW) and total intracellular lipid content of each culture sample were measured, and their fatty acid compositions were analyzed via gas chromatography (GC). The experimental methods are described in detail in the following sections.
2.7. Comparison of Dry Cell Weights
Mycelia were collected by filtering the culture medium through preweighed Miracloth. Excess moisture was removed, then the Miracloth pieces containing the mycelia were placed into preweighed 50 mL tubes and frozen at −80 °C. The frozen samples were then freeze-dried (FDU-810, EYELA, Tokyo, Japan). The 50 mL tubes containing the dried samples and Miracloth were weighed, and the dry cell weights (DCW) were calculated. The results were presented as the mean and SD of the DCW (g/L) obtained from three independent experiments.
2.8. Comparison of Total Intracellular Lipid Contents
Approximately 200 mg of the dried samples were placed into tubes containing stainless steel beads (133 mg) and pulverized using a bead-beating device (FastPrep-24 5G; MP Biomedicals, Irvine, CA, USA). The pulverized dry samples were then added to preweighed 15 mL tubes containing 2 mL of sterile water. A minimum of 100 mg of the pulverized dry sample was added. Then, 20 µL of acetic acid, 5 mL of methanol, and 2.5 mL of chloroform were added to the tubes, and the samples were vortexed for 10 min. Subsequently, 2.5 mL of chloroform and 1 mL of sterile water were added, and the mixture was inverted to homogenize the aqueous and chloroform layers, followed by vortexing for 10 min. Next, the mixture was centrifuged at approximately 583× g for 1 min, and the chloroform layer containing the dissolved lipids was separated from the aqueous layer. The chloroform layer was transferred to a preweighed tube using a Pasteur pipette and dried using a heating block. The dried tubes were weighed, and the total intracellular lipid content was measured. The results were presented as the mean and SD of the total content of intracellular lipids (g/L) obtained from three independent experiments per group. The dried lipids were dissolved in 100 µL of methanol and stored at −80 °C.
2.9. Analysis of Intracellular Fatty Acid Composition by Gas Chromatography
Total lipid samples were dissolved in 100 µL of methanol, then mixed with 500 µL of 0.5 N sodium methoxide. A 2 g/L methyl heptadecanoate solution in hexane (200 µL) was used as an internal control (IC). The mixture was vortexed at room temperature for 60 min to allow transesterification. Methanol without lipids was used as the negative control for the experiment. After transesterification, the pH of each mixture was neutralized by adding 40 µL of 98% sulfuric acid (H2SO4), followed by vortexing with 500 µL of hexane for 30 min for fatty acid methyl esters (FAMEs) extraction. The samples were then centrifuged at approximately 21,000× g for 40 s, after which the upper hexane layer was collected and transferred to vials for gas chromatography (GC) analysis. FAMEs were analyzed using a GC-2030A system (Shimadzu, Kyoto, Japan) equipped with a flame ionization detector (FID) and hydrogen generator (Precision, Peak Scientific, Inchinnan, UK). The samples were injected into an HP-INNOWAX capillary column (30 m × 0.25 mm, Agilent Technologies, Santa Clara, CA, USA) with a split ratio of 15 and an injector temperature of 260 °C. Hydrogen was used as the carrier gas, and a flow rate of 1.5 mL/min was applied. The oven temperature was maintained at 180 °C for 15 min. The temperature of the FID was set at 260 °C.
Prior to sample analysis, each commercially available single FAME standard was individually analyzed by GC to determine its retention time. The retention times obtained were then compared with those of the sample peaks to identify the peaks of C16:0, C17:0, C18:0, C18:1, C18:2, and C18:3. The peak areas were corrected with theoretical correction factors (TCFs) according to the AOCS official method Ce 1j-07 to calculate the weight ratios of each FAME [42]. To determine the intracellular fatty acid composition (%), the weight ratios of individual FAMEs were divided by the total weight ratio excluding the internal standard C17:0. The intracellular fatty acid composition of ALA (%) was then multiplied by the intracellular lipid content (g/L) to calculate the ALA titer (mg/L). Finally, the obtained ALA titer was divided by the DCW (g/L) to calculate the ALA yield (mg/g dry weight).
2.10. Evaluation of Growth and Fatty Acid Production in the Solid Culture
For the evaluation of growth and fatty acid production, 100 mg of pre-gelatinized rice (Alpha-Foods, Izumo, Japan), 250 µL of water, and 10 µL of spore suspension were placed in a 2 mL plastic tube and incubated at 30 °C for 5 days. After incubation, 500 μL of methanol and a stainless bead were added to the tube, and the mixture was pulverized using a bead-beating device. The resulting cell disruption solution was used for both growth and fatty acid composition analyses. To evaluate fungal growth, we used the nucleic acid-based method described by Ohuchi et al. [43]. In brief, 50 µL of the disrupted cell solution was transferred to a 1.5 mL microtube and mixed with 500 µL of 0.5 N perchloric acid. The mixture was then heated at 100 °C for 15 min to extract the nucleic acids, vortexed again, and centrifuged at approximately 21,000× g for 40 s. The nucleic acid concentration in the supernatant was then quantified spectrophotometrically at an optical density of 260 nm (OD260) using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). For fatty acid composition analysis, 100 μL of the cell solution was analyzed according to a described above method.
2.11. Quantitative Analysis of the Carbohydrate Concentration in the Culture Medium by HPLC
For HPLC analysis, 1 mL of the culture supernatant was collected from the medium without fungal inoculation (day 0) and from day 2 and day 4 cultures. Each supernatant was filtered through a 0.22-µm hydrophilic nylon filter and analyzed using an LC-VP system (Shimadzu) equipped with a refractive index detector (RID-10A; Shimadzu, Kyoto, Japan). The mobile phase consisted of 14 mM H2SO4, which was delivered through an HPX-87H column (300 × 7.8 mm; Bio-Rad, Hercules, CA, USA) at a flow rate of 0.7 mL/min. The column oven was maintained at 60 °C, and the refractive index detector was set at 40 °C. Each sample (200 µL) was injected using an SIL-10ADvp autosampler (Shimadzu, Kyoto, Japan), with the needle rinsed using 200 µL of rinse solution at a flow rate of 35 µL/s.
Data analysis was conducted in LabSolutions software (version 5.73, Shimadzu, Kyoto, Japan). The calibration curves were prepared using 1.0% (w/v) standard solutions of glucose, maltose, and starch. The concentrations of each sugar in the samples were individually quantified based on the prepared calibration curves. The total quantity of starch and its hydrolysates was expressed as ‘total glucose equivalents’ by converting the concentrations of maltose and starch using the correction factors of 1.05 and 1.11, respectively.
2.12. qPCR-Based Quantification of fad3 Gene Copy Number
To evaluate the relative copy number of the fad3 gene in strains RIB40, Aofad3, and Aofad3-MC, quantitative real-time PCR (qPCR) was performed using a TaqMan probe-based detection system. The endogenous single-copy gene pyrG was used as a reference gene (https://www.ncbi.nlm.nih.gov/gene/5998543, accessed on 1 October 2025). Primer-probe mixtures (PP Mix) were prepared by mixing 50 µM of forward primer, 50 µM of reverse primer (50 µM), and 100 µM of TaqMan probe and adjusting the final concentrations to 2.5 µM for each primer and 0.5 µM for the probe using sterile ultrapure water. The primer and probe sets used for each target gene are shown in Table S3.
The qPCR reaction mix was prepared by mixing 80 µL of the prepared PP Mix, 200 µL of 2× DirectAce qPCR Mix (Nippon gene, Tokyo, Japan), 0.8 µL of 50× ROX Passive Reference, and 79 µL of sterile ultrapure water. The solution was vortexed thoroughly and briefly centrifuged to collect all liquid at the bottom. Next, 18 µL of the qPCR mix was dispensed into each well of a 96-well PCR plate, followed by the addition of 2 µL of the template DNA (100 ng/µL) to a final volume of 20 µL per well. Genomic DNA extracted from the stains RIB40, Aofad3, and Aofad3-MC were used as the DNA templates. Each well was thoroughly mixed by pipetting at least ten times. The plate was then sealed with a MicroAmp Optical Adhesive cover and centrifuged at approximately 583× g for 1 min to eliminate air bubbles.
qPCR was conducted in a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The thermal cycling conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 45 cycles of denaturation at 95 °C for 15 s and annealing/extension at 65 °C for 1 min. The reactions were performed using the standard mode.
Data analysis was performed using the instrument software (version 1.2.4), with the threshold manually set at 0.1 and the baseline set automatically. The cycle threshold (Ct) values for both fad3 and pyrG were determined, and the average Ct values from three independent experiments were used for the further calculations. To evaluate the amplification efficiency (E) of each primer-probe set, a standard curve was generated using serial dilutions of RIB40 genomic DNA (100, 10, and 1 ng/µL). The amplification efficiencies for fad3 and pyrG detection (Efad3 and EpyrG) were separately calculated from the slope of the standard curve using the following equation:
E = 10^(–1/slope),
The calculated amplification efficiencies were Efad3 = [1.86] and EpyrG = [1.86], respectively. The relative copy number of fad3s in the transformants was estimated based on the differences in Ct values compared to the wild type. First, the average Ct values for the target (fad3) and reference (pyrG) genes were determined for each strain. The following ΔCt values were calculated:
ΔCt_fad3 = Ct_fad3 (transformant) − Ct_fad3 (RIB40),
ΔCt_pyrG = Ct_pyrG (transformant) − Ct_pyrG (RIB40),
The relative fold change in the fad3 gene copy number was calculated using the following equation:
Fold change = (Efad3)^ΔCt_fad3/(EpyrG)^ΔCt_pyrG,
These fold change values were used as estimates of the fad3 gene copy number in each transformant relative to that in the wild type. This calculation approach was based on the modified ΔΔCt method described previously [44].
2.13. Whole-Genome Sequencing (WGS) Analysis
Genomic DNA of the Aofad3-MC strain was used for WGS analysis. Library preparation was performed using a TruSeq DNA PCR-Free Kit (Illumina, San Diego, CA, USA) and sequencing was conducted by Macrogen Japan Corp. using the NovaSeq X system with 150 bp paired ends. The obtained WGS data of Aofad3-MC have been deposited in the DDBJ Sequence Read Archive (SRA) under accession number DRR710002.
2.14. Prediction of the Copy Numbers of fad3 and Evaluation of Self-Cloning Using WGS Data
The Illumina raw data were preprocessed using trimmomatic-0.39 to discard low-quality reads and adapter sequences (-phred33 ILLUMINACLIP: TruSeq3-PE.fa:2:30:10 LEADING:15 TRAILING:15 SLIDINGWINDOW:4:15 MINLEN:32) [45]. The processed reads were mapped to the A. oryzae RIB40 genome sequence (GCF_000184455.2) using BWA-mem (ver. 0.7.18-r1234-dirty) with the default settings [46]. Processing of the mapped reads was performed using SAMtools (version 1.12) [47,48], and the read depth was calculated using the SAMtools depth. The whole genome sequence data of RIB40 and Aofad3-MC were subjected to k-mer analysis using the open-source bioinformatics analytical program, GeneEditScan (version 1.0.0) [49].
2.15. RNA-seq and Bioinformatics Analysis
To investigate the transcriptomic changes associated with the overexpression of fad3, RNA-seq was performed. The RIB40 and Aofad3-MC strains were cultured in 100 mL of ssCD medium in 500 mL Erlenmeyer flasks at 30 °C with shaking at 100 rpm for 4 days. Two spatulas of mycelium, a stainless-steel bead, 350 µL of RA1 buffer (NucleoSpin RNA kit, Macherey-Nagel, Düren, Germany), and 3.5 µL of 1 M DTT were mixed in a bead-beating tube, and the samples were disrupted using a bead-beating device. The total RNA was then extracted according to the manufacturer’s protocol for the NucleoSpin RNA kit. The RNA concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and RNA quality was assessed using an Agilent Bioanalyzer. Library preparation was performed using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA), and sequencing was performed using an Illumina NovaSeq 6000 system with 150 bp paired-end reads. RNA-seq data were analyzed using the nf-core RNA-seq pipeline (nf-core/rnaseq, version 3.13.0) [50], with the genome sequence and gene annotation of A. oryzae RIB40 (GCF_000184455.2) as reference files. The workflow was executed with default parameters, except for specifying CDS as the feature type and product as the group type for featureCounts. Gene expression differences between the two strains were estimated based on transcripts per million (TPM) values.
3. Results
3.1. Identification and Characterization of the ω-3 Desaturase Gene in A. oryzae
To identify candidate A. oryzae ω-3 desaturase genes, a BLASTP search was performed using the amino acid sequence of the ω-3 desaturase gene (Fm1) from F. moniliforme (Accession No. ABB88516.1) [19]. Among the genes analyzed, AO090010000714 showed the highest similarity to Fm1, with a sequence identity of 49.7% (Figure S2). This gene contains three conserved histidine-rich motifs (H-boxes 1–3) at positions 93–97, 129–133, and 321–325, which are conserved in the ω-3 desaturase genes of various organisms and are essential for desaturase activity [11]. Therefore, AO090010000714 was designated ‘fad3’, and considered a candidate A. oryzae ω-3 desaturase gene.
To characterize the function of fad3, an overexpression strain (Aofad3) was constructed by randomly introducing a single-copy DNA fragment (ptrA-Ptef1-fad3-Tfad3), in which fad3 was placed under the control of the high-expression promoter (Ptef1) (Figure S1C). The RIB40 and Aofad3 strains were cultured in ssCD medium at 30 °C with shaking at 100 rpm for 96 h, and the intracellular fatty acid composition was subsequently analyzed via GC. In the resulting spectra, the peak corresponding to ALA was faint in the RIB40 strain under the experimental conditions used (Figure 2A, left). In contrast, a distinct ALA peak was observed in the Aofad3 strain (Figure 2A, right). In Aofad3, the peak area of linoleic acid (C18:2) was found to be decreased, while those of palmitic acid (C16:0), stearic acid (C18:0), and oleic acid (C18:1) appeared largely unchanged. In the negative control sample, only the IC peak was observed, and no other background peaks were detected. Growth comparison on nutrient-rich GYN agar plates revealed that the RIB40 strain exhibited slightly but significantly faster mycelial growth than the Aofad3 strain at all measured time points (Figure 2B). The high expression of fad3 and ALA production may have some impact on mycelial growth.
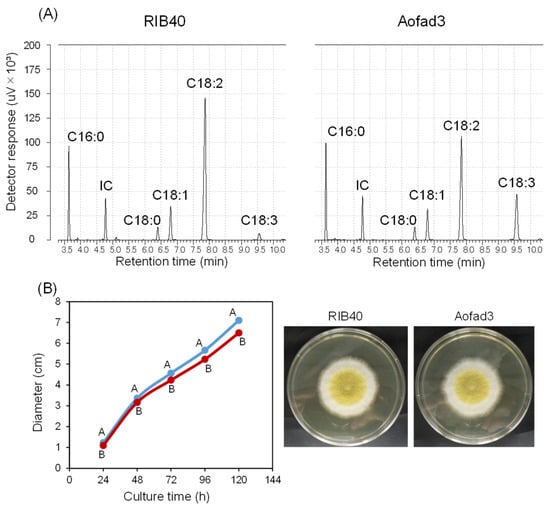
Figure 2.
Functional characterization of fad3 via single-copy integration. (A) Gas chromatography analysis results of the fatty acids obtained from Aspergillus oryzae strains RIB40 and Aofad3. Peaks corresponding to C16:0 (palmitic acid), C18:0 (stearic acid), C18:1 (oleic acid), C18:2 (linoleic acid), and C18:3 (α-linolenic acid) were detected. The IC peak represents methyl heptadecanoate, which was added to each sample at a final concentration of 0.5 g/L. The y-axis (detector response, μV × 103) is scaled to a range of 0–200 for both strains. (B) Comparison of the mycelial growth of the RIB40 and Aofad3 strains on GYN agar. Spore suspensions (5 µL) were inoculated onto GYN plates, and mycelial growth diameters (cm) were measured at 24, 48, 72, 96, and 120 h [51]. The blue line represents the RIB40 strain, and the red line represents the Aofad3 strain. Values represent the mean ± SD of three independent experiments (n = 3). Statistical analysis was performed using one-way ANOVA, followed by Tukey’s HSD test when significant differences were detected. Groups with significant differences (p < 0.05) are indicated by different letters. Representative images of the growth at 96 h are shown on the right.
3.2. Increasing the fad3 Copy Number to Enhance ALA Production
To further increase fad3 expression, another strain, named Aofad3-MC, was constructed by randomly introducing a multicopy DNA fragment (ptrA-PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3), in which multiple copies of fad3 were placed under the control of two high expression promoters, PamyB and Ptef1 (Figure S1D). The wild-type strain, RIB40, and the two transformants, Aofad3 and Aofad3-MC, which contained introduced fad3 expression fragments in single and multiple copies, respectively, were cultured in ssCD medium at 30 °C and 100 rpm. The dry cell weight (DCW, g/L) was not significantly different among the three strains at any time point (Figure 3A), indicating that the fad3 expression level does not affect growth. Although the total lipid content (g/L) at 48 h was comparable among the strains, Aofad3-MC showed a slight but statistically significant increased lipid content at 96 h compared with the other strains (Figure 3B). In addition, HPLC analysis of the culture supernatants revealed that higher copy numbers of the introduced fad3 gene were associated with increased concentrations of residual starch and its hydrolysis products (maltose and glucose) (Figure 3C). At 48h, the concentrations of residual starch and its hydrolysis products were 25 g/L for strain RIB40, 28.2 g/L for Aofad3, and 31.2 g/L for Aofad3-MC. By 96 h, these levels had decreased to 9.6 g/L for RIB40, 11.5 g/L for Aofad3, and 12.5 g/L for Aofad3-MC. These results indicate that the overexpression of fad3 may affect lipid production and carbon source utilization.
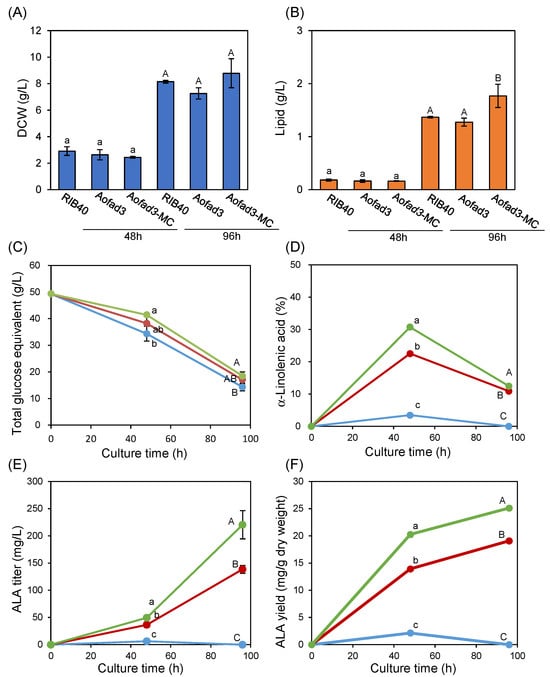
Figure 3.
Comparative analysis of fad3 single- and multicopy strains. A. oryzae strains RIB40, Aofad3, and Aofad3-MC were compared after 48 and 96 h of cultivation. (A) Dry cell weight (DWC, g/L) and (B) total lipid content (lipids, g/L) are shown. (C) The concentrations of residual starch and its hydrolysis products (maltose and glucose) in the medium are shown as glucose equivalents (g/L). (D) The intracellular ALA composition (%). (E) ALA titer (mg/L). (F) ALA yield (mg/g dry weight) of the mycelia. In panels (C–F), RIB40, Aofad3, and Aofad3-MC are represented in blue, red, and green lines, respectively. Data are shown as mean ± SD (n = 3), and statistical analysis was conducted as in Figure 2B. Lowercase letters indicate group of 48 h cultivation, and uppercase letters indicate group of 96 h cultivation.
In contrast, large differences in intracellular fatty acid composition were noted between the strains (Figure 3D). At 48 h, ALA accounted for 3.5% of the total fatty acids in strain RIB40, whereas this proportion was 22.5% in strain Aofad3, and 30.7% in strain Aofad3-MC. By 96 h, the ALA composition had decreased to 10.9% in strain Aofad3 and 12.5% in Aofad3-MC, while ALA was undetectable in strain RIB40 (Figure 3D, Table 1). In addition, the highest ALA titer (220 mg/L) and yield (25 mg/g dry weight) were observed in strain Aofad3-MC at 96 h (Figure 3E,F). These results show that ALA production increases in line with the introduced fad3 copy number. Regarding other fatty acids, only linoleic acid (C18:2) decreased with increasing fad3 copy number, while the compositions of other fatty acids remained mostly unchanged (Table 1). This finding suggesting that fad3 specifically catalyzes the conversion of linoleic acid to ALA. Notably, the ALA content decreased between 48 h and 96 h in all strains (Figure 3D), while the stearic acid (C18:0) and oleic acid (C18:1) contents increased approximately 1.5-fold and 2-fold, respectively, during the same period, including in the control strain RIB40 (Table 1).

Table 1.
Intracellular fatty acid composition (%) of A. oryzae strains.
3.3. Relationship Between fad3 Copy Number, Expression Level, and ALA Production
During the transformation of A. oryzae, DNA fragments can integrate randomly into multiple genomic locations. Therefore, the inserted DNA fragments (ptrA-Ptef1-fad3-Tfad3 and ptrA-PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3) may be placed throughout the genome rather than being arranged in tandem, and simple PCR cannot accurately determine their copy number. To accurately determine the number of fad3 copies in the transformants, we performed qPCR and whole-genome sequencing (WGS). The qPCR analysis of genomic DNA from strains RIB40, Aofad3, and Aofad3-MC revealed that Aofad3 and Aofad3-MC contained approximately 1.9-fold and 6.4-fold more copies of fad3, respectively, compared to RIB40 (Table 2). In addition to qPCR, WGS was performed on the Aofad3-MC strain to confirm the fad3 copy number. The newly generated raw sequence reads are available at the DDBJ DRA database under run accession DRR710002. The obtained reads were mapped to the A. oryzae RIB40 reference genome, and the read depth across the genome was calculated using the ‘depth’ tool in SAMtools software. The genome-wide median read depth was found to be 31×, which represents the typical depth for single-copy regions. In comparison, the fad3 gene and its terminator region (1,540,592–1,542,299) showed a read depth of 218×, indicating an approximately 7-fold higher coverage than the genome-wide median (Figure S3). Meanwhile, the 5 kb regions upstream (1,535,591–1,540,591) and downstream (1,542,300–1,547,300) of fad3 showed average read depths of 32× and 35×, respectively (Figure S3), which were comparable to the genome-wide median.

Table 2.
Copy number evaluation of the introduced fad3 gene.
This consistency supports the notion that the elevated read depth was localized specifically to the inserted fad3 gene region. Collectively, these results suggest that the fad3 copy number is approximately 6–7 times higher than that of the single-copy genomic regions. These findings were generally consistent with the copy number estimated by qPCR analysis (Table 2, Figure S3). The unexpectedly high copy number in the Aofad3-MC strain may be due to the integration of the introduced DNA fragment into multiple genomic loci during transformation.
The copy number analysis clearly demonstrated that higher fad3 copy numbers were correlated with increased ALA production. However, the increase in ALA production was not proportional to the increase in copy number (Figure 3C, Table 1 and Table 2). Therefore, to investigate the gene expression levels, RNA-seq was performed using the total RNA extracted from the RIB40 and Aofad3-MC strains after 96 h of cultivation. The newly generated RNA-seq reads are available at the DDBJ DRA database under run accessions DRR702295 (RIB40 strain) and DRR702296 (Aofad3-MC strain). Among the approximately 12,000 genes encoded in the A. oryzae genome, 6068 genes showed less than 35% variation in expression, whereas 126 exhibited over 10-fold differences. The fad3 gene exhibited 1400-fold higher expression in Aofad3-MC than in RIB40 (Figure S4). In contrast, previously reported elongase and other desaturase genes, which are involved in the ALA biosynthesis pathway [37], demonstrated expression changes of less than 35% (Figure S4). These results suggest that fad3 expression was upregulated while the expression of elongase and other desaturase genes was unaffected. As this analysis was based on a single replicate (n = 1), these findings should be interpreted as approximate expression trends rather than definitive conclusions.
3.4. K-mer Analysis to Confirm the Absence of Foreign Sequences
The Aofad3-MC strain was constructed by the homologous recombination of a DNA fragment composed solely of A. oryzae-derived sequences (ptrA-PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3) (Figure S1D). However, because the plasmid vector backbone was also used during transformation, the unintended integration of E. coli-derived sequences (e.g., ampicillin resistance gene and origin of replication) into the genome was possible. Confirming the presence or absence of such foreign sequences is important not only to exclude their potential effects on gene expression but also to validate that the strain qualifies as a self-cloned strain. To confirm the absence of non-A. oryzae-derived sequences, k-mer analysis was conducted in GenEditScan software (version 1.0.0) using the WGS data from the wild-type RIB40 strain (Genbank accession: SRR1835311) and the Aofad3-MC strain (DDBJ accession: DRR710002). Sequencing reads were split into 20-mers and mapped to the plasmid sequence (pPTRI [PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3]) (Figure 4A and Figure S1B). In both strains, many reads aligned to the inserted gene region (2465–9176 bp) (Figure 4B, upper panel). Reads mapping to E. coli-derived vector backbone sequences were detected faintly in RIB40, but were absolutely absent in Aofad3-MC (Figure 4B, orange vs. blue plot). The read depth and Bonferroni-adjusted p-values (–log10 Bonferroni) indicating the statistical significance of the differences between the RIB40 and Aofad3-MC were calculated at each nucleotide position. A threshold of –log10 Bonferroni ≥ 2 (p < 0.01) was used to indicate statistical significance [49]. No significant differences were found in the vector region between the two strains (Figure 4B, lower panel). These results indicate that no unintended foreign sequences were detected in the genome of Aofad3-MC.
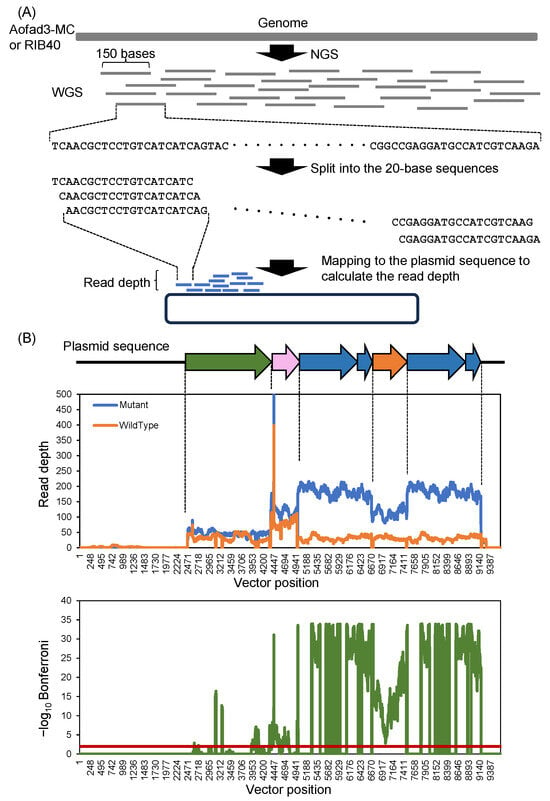
Figure 4.
K-mer analysis for the detection of foreign DNA sequences in Aofad3-MC. (A) Schematic overview of the K-mer-based analytical strategy. (B) The plasmid sequence pPTRI [PamyB-fad3-Tfad3-Ptef1-fad3-Tfad3] is shown linearly, with the read depth plotted at each base position (B, upper panel). The data from RIB40 and Aofad3-MC are shown in orange and blue, respectively. The statistical significance of the differences between the RIB40 and Aofad3-MC strains was shown using Bonferroni-adjusted p-values, which are represented in green as –log10 Bonferroni at each base position (B, lower panel). Base positions with a Bonferroni-adjusted p-values below 0.01 (−log10 Bonferroni > 2) were considered to be statistically different. The red horizontal line indicates the criterion (−log10 Bonferroni = 2), and base positions with values above this line are considered statistically significant.
In the Aofad3-MC strain, the highest read depth was observed at the fad3 gene and its terminator region (Figure 4B, blue plot). In contrast, the amyB and tef1 promoter regions showed approximately half the read depth compared to fad3, consistent with the construct design in which two copies of fad3 were expressed under different promoters. These observations support the validity of the construct and the accuracy of the mapping results. At the ptrA region, the mapping depth was relatively low in both strains, and only a few significant differences were observed, suggesting that the introduced DNA fragment may have been cleaved during transformation, leading to the partial deletion of the ptrA sequence upon chromosomal integration. However, it should be noted that the total quantity of WGS data differed between the two strains, which may have affected the number of mapped reads.
3.5. Production of ALA on Solid Media
In addition to liquid culture, the potential for ALA production via solid-state cultivation was investigated. The strains RIB40, Aofad3, and Aofad3-MC were inoculated on precooked dried rice and cultured statically at 30 °C for 5 days. After cultivation, lipids were extracted from the mycelia, and their fatty acid composition was analyzed using GC. The analysis revealed ALA contents of 3.6% and 7.7% in Aofad3 and Aofad3-MC strains, respectively (Figure 5A), while no ALA was detected in RIB40 strain. These results clarify that ALA production in A. oryzae via fad3 overexpression is feasible under both solid-state and liquid cultivation conditions. In addition, the comparison of mycelial growth based on nucleic acid concentration (OD260) revealed no significant differences in growth among the three strains (Figure 5B), indicating that the introduced genes did not affect the organisms’ growth.
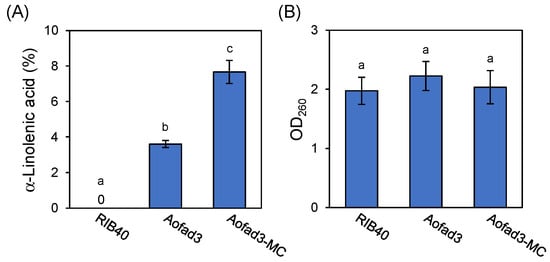
Figure 5.
Evaluation of ALA production and mycelial growth in the precooked dried rice culture. (A) Comparison of the intracellular ALA composition and (B) mycelial growth among the RIB40, Aofad3, and Aofad3-MC strains. Mycelial growth was assessed based on the nucleic acid content. Data are shown as mean ± SD (n = 3), and statistical analysis was conducted as in Figure 2B. Groups with significant differences (p < 0.05) are indicated by different lowercase letters.
4. Discussion
In this study, we identified AO090010000714 as an ω-3 desaturase gene (fad3) in A. oryzae (Figure S2 and Figure 2A), and found that overexpression of fad3 by increasing the copy number significantly enhanced ALA production (Figure 3C, Table 1). Collectively, the results suggested that fad3 encodes a functional ω-3 desaturase and that low expression of fad3 leads to low production of ALA. Moreover, fad3 overexpression facilitated the conversion of linoleic acid to ALA without affecting the production of other fatty acids (Table 1), indicating high substrate specificity. In some organisms, ω-3 desaturase has been reported to exhibit bifunctional Δ12 activity, converting stearic acid to linoleic acid [19], and to have broad substrate specificity [11]. In contrast, the A. oryzae fad3 gene does not appear to exhibit bifunctional activity. In addition, the absence of other ω-6 fatty acids, such as γ-linolenic acid, in A. oryzae is a beneficial feature for the accumulation of ALA [34,35]. Such high substrate specificity and simple fatty acid composition offer significant advantages for applications that require the precise regulation of intracellular lipids. A. oryzae not only shows promising potential as a sustainable and safe platform for ALA production but also holds promise for future expansion to the production of other functional lipids.
The high-copy Aofad3-MC strain reached an ALA content of 30.7% (Table 1). This level is higher than those reported in previous microbial fermentation systems, such as Yarrowia lipolytica (28–30%) [19,22], Lipomyces starkeyi (24.9%) [15], Saccharomyces cerevisiae (6.3%) [52], cyanobacteria (23.6%) [18], Tetraselmis sp. (18.7%) [53], and Rhodosporidium kratochvilovae (10.3%) [54]. However, the major existing sources of ALA, such as flaxseed and perilla oils, contain approximately 55–60% ALA content [55]. In addition, although the highest ALA titer reported in microbial systems to date was 1.4 g/L in Yarrowia lipolytica [22], the Aofad3-MC strain produced only 0.22 g/L (Figure 3E). Therefore, further enhancement of ALA productivity is required for practical industrial applications. Our results demonstrated that increasing the copy number of fad3 enhanced ALA production (Figure 3D, Table 1). While the Aofad3 strain contained approximately two copies of fad3, Aofad3-MC contained six to seven copies (Table 2, Figure S3), producing maximum ALA contents of 25.0% and 30.7% (Figure 3D), respectively. This result suggests that ALA production is not solely determined by the gene copy number. Given that RNA-seq analysis suggested sufficient increase in fad3 transcription levels in the Aofad3-MC strain, ALA production may be limited by enzyme activity and other metabolic factors. The conversion of linoleic acid (C18:2) to α-linolenic acid (C18:3) by ω-3 desaturase is known to vary significantly among fungal species. For example, large differences in desaturase activity have been reported between the genera Fusarium and Magnaporthe [19]. In addition, ω-3 desaturases have been shown to exhibit high activity and stability under low-temperature conditions [15,22]. In L. starkeyi, ALA production was demonstrated to increase during low-temperature cultivation [15]. Based on these observations, improving ALA productivity in A. oryzae may be possible through the comparative analysis of ω-3 desaturase, enzyme engineering, and optimization of the cultivation conditions, particularly temperature.
The observed lipid metabolic shifts suggest that metabolic engineering approaches could further enhance ALA production. In all strains, the ALA content peaked at 48 h but decreased significantly by 96 h (Figure 3D). In contrast, the stearic acid (C18:0) and oleic acid (C18:1) levels increased by approximately 1.5- and 2-fold, respectively, during the same time period (Table 1). These results indicate that A. oryzae accumulates highly unsaturated fatty acids, such as ALA and linoleic acid (C18:2), during the early cultivation phase, later shifting to accumulating saturated and monounsaturated fatty acids during the later stages. This metabolic shift likely reflects the need for highly unsaturated fatty acids to maintain membrane fluidity during early growth [56], and the promotion of the accumulation of storage lipids, such as triacylglycerols (TAGs), which are typically composed of less unsaturated fatty acids, during nutrient limitation in the later phase [57,58]. In fact, the DCW increased only 2- to 3-fold between 48 h and 96 h (Figure 3A), whereas the total lipid content increased 7- to 10-fold (Figure 3B), supporting the idea of active lipid accumulation. In addition, desaturase expression is feedback-regulated according to membrane fatty acid unsaturation levels [59,60,61]. For example, in S. cerevisiae, the Δ9 desaturase gene OLE1 is regulated by the transcription factors Mga2 and Spt23 [60,61]. Although direct evidence in filamentous fungi is lacking, a similar mechanism may contribute to the observed decrease in ALA content in A. oryzae. The decrease in ALA from 48 h to 96 h may be due to multiple metabolic pathways, including incorporation into membrane phospholipids and TAGs, feedback inhibition of desaturase, and ALA degradation. Therefore, metabolic engineering strategies may be effective for maintaining and improving ALA accumulation.
The industrial use of genetically modified organisms (GMOs) often requires strict containment measures to prevent their release into the natural environment, which can lead to higher operational costs compared to using naturally occurring microbes. In addition, due to consumer preferences for non-genetically modified food ingredients, the use of GMOs poses challenges in gaining acceptance for industrial or commercial use [62]. In contrast, microorganisms engineered by introducing DNA sequences derived from the same species are considered “self-cloning” and are not classified as GMOs for consumer labeling purposes in countries such as Japan, the United States, and Australia [63]. This classification allows for easier application in food and feed compared to conventional GMOs. For example, in Japan, self-cloning microorganisms are exempt from the regulations that govern recombinant organisms under Cartagena domestic law [23]. In the United States, while microorganisms created using DNA from different taxonomic genera must be reported to the Environmental Protection Agency, self-cloning microorganisms are not subject to such reporting requirements [64]. The Aofad3-MC strain developed in this study using a self-cloning approach can be classified as a non-GMO (Figure 4), which offers a regulatory advantage for industrial applications [21].
ALA production technology using A. oryzae is expected to expand into diverse fields, such as food, feed, and cosmetics, and contribute to the construction of a sustainable society. ALA is widely used for its health benefits in the food and supplement industries and also has industrial applications due to its properties as a drying oil, such as in inks. The self-cloned strain developed in this study enables high-level ALA production, making the practical supply of ALA via liquid cultivation and extraction from fungal biomass possible in the future. Currently, the use of A. oryzae biomass as mycoprotein is rapidly expanding, and products such as alternative meats and vegan cheeses made from koji mold have recently appeared on the market [65,66,67]. Combined with the ALA-rich A. oryzae strain obtained in this study, this approach could add health functionality to mycoproteins.
In addition, the new strain developed in this study could be useful in the livestock feed industry [68,69]. Solid-state fermentation of starch-rich grains or food waste using A. oryzae can effectively increase the ALA content of feed. Feeding livestock ALA-enriched diets leads to the accumulation of ALA in their bodies and its partial conversion into DHA and EPA. These accumulated ω-3 fatty acids are thought to contribute to the maintenance and improvement of human health. Currently, high-value-added eggs and pork produced using flaxseed oil–enriched feed are available [70]. Similar effects are expected from ALA-enriched feed produced using A. oryzae (Figure S5). In addition, ALA-rich oil has been reported to reduce methane production in the rumen [71,72]. Methane emitted from the rumen—the first stomach compartment in ruminants such as cattle—during belching is a significant contributor to greenhouse gas emissions. ALA-rich oil derived from A. oryzae may therefore help reduce the environmental impact of livestock farming.
As the global population grows and protein shortages intensify (the “protein crisis”), ALA production using A. oryzae holds great social and industrial significance as a sustainable and low environmental impact technique. A. oryzae possesses multiple advantages, such as safety, strong degradative ability, and resource recyclability, making it a promising fermentation platform for simultaneous food loss and waste upcycling and ALA production. The ALA-rich strains developed in this study are applicable not only to fermented foods but also to the low-cost production of ALA from food residues and agricultural by-products. Potential applications span food, feed, supplements, cosmetics, and beyond, positioning this technology as a key contributor to a sustainable society.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation11100585/s1, Figure S1: Schematic diagrams of plasmid maps and excised DNA fragments; Figure S2: Amino acid sequence comparison between Fm1 and AO090010000714 (fad3); Figure S3: The profile of reed depth in RIB40 genome around fad3 locus; Figure S4: Comparison of expression levels of fad3 and other fatty acid biosynthesis-related genes; Figure S5: Future applications of ALA production by A. oryzae in food and feed; Table S1: Nucleotide sequences used for plasmid construction and gene introduction into A. oryzae; Table S2: List of primer sequences and template used for PCR in plasmid construction; Table S3. List of primer and probe sequences used for qPCR.
Author Contributions
Conceptualization, J.M.; methodology, H.K., H.S., T.T., E.K., W.T., R.H., S.S., K.-I.K. and J.M.; validation, H.K., T.T. and J.M.; formal analysis, H.K., T.T. and J.M.; investigation, H.K. and J.M.; resources, H.K. and J.M.; data curation, H.K. and J.M.; writing—original draft preparation, H.K.; writing—review and editing, H.K. and J.M.; visualization, H.K.; supervision, J.M.; project administration, J.M.; funding acquisition, J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study was conducted with funding from the NARO Innovation Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. Whole-genome sequencing and RNA-seq data supporting the results of this study have been deposited in the DDBJ Sequence Read Archive under accession numbers DRR702295, DRR702296, and DRR710002.
Acknowledgments
This article is an extended version of our paper [73] presented at 2022 Annual Meeting of the Japan Society for Bioscience, Biotechnology and Agrochemistry, Kyoto, Japan, 15–18 March 2022. Abstract in the Program Book of the Japan Society for Bioscience, Biotechnology, and Agrochemistry, March 2022. Only the abstract was disclosed for the purpose of conference presentation; no detailed data were presented, and therefore this does not constitute duplicate publication.
Conflicts of Interest
J.M., E.K., W.T., S.S. and K.-I.K. are inventors on Japanese patent JP2023123456 which is related to this report. The patent is assigned to the National Agriculture and Food Research Organization.
References
- Mititelu, M.; Lupuliasa, D.; Neacșu, S.M.; Olteanu, G.; Busnatu, Ș.S.; Mihai, A.; Popovici, V.; Măru, N.; Boroghină, S.C.; Mihai, S.; et al. Polyunsaturated fatty acids and human health: A key to modern nutritional balance in association with polyphenolic compounds from food sources. Foods 2024, 14, 46. [Google Scholar] [CrossRef]
- Wall, R.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. Fatty acids from fish: The anti-inflammatory potential of long-chain omega-3 fatty acids. Nutr. Rev. 2010, 68, 280–289. [Google Scholar] [CrossRef]
- Bodur, M.; Yilmaz, B.; Ağagündüz, D.; Ozogul, Y. Immunomodulatory effects of omega-3 fatty acids: Mechanistic insights and health implications. Mol. Nutr. Food Res. 2025, 69, e202400752. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, S.; Motti, M.L.; Meccariello, R. ω-3 and ω-6 polyunsaturated fatty acids, obesity and cancer. Nutrients 2020, 12, 2751. [Google Scholar] [CrossRef]
- Simopoulos, A. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef]
- Crawford, M.A.; Sinclair, A.J.; Hall, B.; Ogundipe, E.; Wang, Y.; Bitsanis, D.; Djahanbakhch, O.B.; Harbige, L.; Ghebremeskel, K.; Golfetto, I.; et al. The imperative of arachidonic acid in early human development. Prog. Lipid. Res. 2023, 91, 101222. [Google Scholar] [CrossRef]
- Akbari, M.; Ostadmohammadi, V.; Moosazadeh, M.; Asemi, Z.; Heydari, S.T.; Tabrizi, R.; Chamani, M.; Lankarani, K.B.; Mobini, M.; Kolahdooz, F. The effects of alpha-lipoic acid supplementation on inflammatory markers among patients with metabolic syndrome and related disorders: A systematic review and meta-analysis of randomized controlled trials. Nutr. Metab. 2018, 15, 39. [Google Scholar] [CrossRef]
- Wendland, E.; Farmer, A.; Glasziou, P.; Neil, A. Effect of α linolenic acid on cardiovascular risk markers: A systematic review. Heart 2006, 92, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Hussein, N.; Ah-Sing, E.; Wilkinson, P.; Leach, C.; Griffin, B.A.; Millward, D.J. Long-chain conversion of [13C]Linoleic acid and α-linolenic acid in response to marked changes in their dietary intake in men. J. Lipid. Res. 2005, 46, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Harris, W.S. Alpha-Linolenic Acid: A Gift from the Land? Circulation 2005, 111, 2872–2874. [Google Scholar] [CrossRef][Green Version]
- Wang, M.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. ω3 Fatty acid desaturases from microorganisms: Structure, function, evolution, and biotechnological use. Appl. Microbiol. Biotechnol. 2013, 97, 10255–10262. [Google Scholar] [CrossRef]
- Yeom, W.W.; Kim, H.J.; Lee, K.-R.; Cho, H.S.; Kim, J.-Y.; Jung, H.W.; Oh, S.-W.; Jun, S.E.; Kim, H.U.; Chung, Y.-S. Increased production of α-linolenic acid in soybean seeds by overexpression of lesquerella FAD3-1. Front. Plant Sci. 2020, 10, 1812. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Yang, S.-M.; Shang, Z.-W.; Xu, J.; Zhao, D.-G.; Wang, H.-B.; Shen, Q. Genome-wide analysis of the fatty acid desaturase gene family reveals the key role of PfFAD3 in α-linolenic acid biosynthesis in perilla seeds. Front. Genet. 2021, 12, 735862. [Google Scholar] [CrossRef]
- Spychalla, J.P.; Kinney, A.J.; Browse, J. Identification of an animal ω-3 fatty acid desaturase by heterologous expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 1142–1147. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, T.; Maehara, T.; Kamisaka, Y.; Ara, S.; Takaku, H.; Yaoi, K. Identification and characterization of Δ12 and Δ12/Δ15 bifunctional fatty acid desaturases in the oleaginous yeast Lipomyces starkeyi. Appl. Microbiol. Biotechnol. 2018, 102, 8817–8826. [Google Scholar] [CrossRef]
- Buček, A.; Matoušková, P.; Sychrová, H.; Pichová, I.; Hrušková-Heidingsfeldová, O. Δ12-fatty acid desaturase from Candida parapsilosis is a multifunctional desaturase producing a range of polyunsaturated and hydroxylated fatty acids. PLoS ONE 2014, 9, e93322. [Google Scholar] [CrossRef][Green Version]
- Sun, K.; Meesapyodsuk, D.; Qiu, X. Molecular cloning and functional analysis of a plastidial ω3 desaturase from Emiliania huxleyi. Front. Microbiol. 2024, 15, 1381097. [Google Scholar] [CrossRef]
- Chen, G.; Qu, S.; Wang, Q.; Bian, F.; Peng, Z.; Zhang, Y.; Ge, H.; Yu, J.; Xuan, N.; Bi, Y.; et al. Transgenic expression of delta-6 and delta-15 fatty acid desaturases enhances omega-3 polyunsaturated fatty acid accumulation in Synechocystis sp. PCC6803. Biotechnol. Biofuels 2014, 7, 32. [Google Scholar] [CrossRef]
- Damude, H.G.; Zhang, H.; Farrall, L.; Ripp, K.G.; Tomb, J.-F.; Hollerbach, D.; Yadav, N.S. Identification of bifunctional Δ12/ω3 fatty acid desaturases for improving the ratio of ω3 to ω6 fatty acids in microbes and plants. Proc. Natl. Acad. Sci. USA 2006, 103, 9446–9451. [Google Scholar] [CrossRef]
- Kabir Khan, M.A.; Yang, J.; Hussain, S.A.; Zhang, H.; Garre, V.; Song, Y. Genetic modification of Mucor circinelloides to construct stearidonic acid producing cell factory. Int. J. Mol. Sci. 2019, 20, 1683. [Google Scholar] [CrossRef]
- Hassane, A.M.A.; Eldiehy, K.S.H.; Saha, D.; Mohamed, H.; Mosa, M.A.; Abouelela, M.E.; Abo-Dahab, N.F.; El-Shanawany, A.-R.A. Oleaginous fungi: A promising source of biofuels and nutraceuticals with enhanced lipid production strategies. Arch. Microbiol. 2024, 206, 338. [Google Scholar] [CrossRef]
- Cordova, L.T.; Alper, H.S. Production of α-linolenic acid in Yarrowia lipolytica using low-temperature fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 8809–8816. [Google Scholar] [CrossRef]
- Senoo, S.; Shintani, T.; Nieda, S.; Shintani, T.; Kariyama, M.; Gomi, K. Construction of self-cloning Aspergillus oryzae strains with high production of multiple biomass-degrading enzymes on solid-state culture. J. Biosci. Bioeng. 2024, 137, 204–210. [Google Scholar] [CrossRef]
- Taylor, M.J.; Richardson, T. Applications of microbial enzymes in food systems and in biotechnology. Adv. Appl. Microbiol. 1979, 25, 7–35. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Microorganisms & Microbial-Derived Ingredients Used in Food (Partial List). Available online: https://www.fda.gov/food/generally-recognized-safe-gras/microorganisms-microbial-derived-ingredients-used-food-partial-list (accessed on 25 August 2025).
- Naeem, M.; Manzoor, S.; Abid, M.-U.-H.; Tareen, M.B.K.; Asad, M.; Mushtaq, S.; Ehsan, N.; Amna, D.; Xu, B.; Hazafa, A. Fungal proteases as emerging biocatalysts to meet the current challenges and recent developments in biomedical therapies: An updated review. J. Fungi 2022, 8, 109. [Google Scholar] [CrossRef]
- Tanaka, M.; Gomi, K. Induction and repression of hydrolase genes in Aspergillus oryzae. Front. Microbiol. 2021, 12, 677603. [Google Scholar] [CrossRef]
- Mizutani, O.; Kudo, Y.; Saito, A.; Matsuura, T.; Inoue, H.; Abe, K.; Gomi, K. A Defect of LigD (Human Lig4 Homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 2008, 45, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.-J.; Hu, S.; Wang, B.-T.; Jin, L. Advances in genetic engineering technology and its application in the industrial fungus Aspergillus oryzae. Front. Microbiol. 2021, 12, 644404. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.-J.; Wang, B.-T.; Wang, Z.-D.; Jin, L.; Han, P. CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wu, Y.; Long, S.; Feng, S.; Jia, X.; Hu, Y.; Ma, M.; Liu, J.; Zeng, B. Aspergillus oryzae as a cell factory: Research and applications in industrial production. J. Fungi 2024, 10, 248. [Google Scholar] [CrossRef]
- Sheng, Y.; Qiu, S.; Deng, Y.; Zeng, B. Recent Advances in Heterologous Protein Expression and Natural Product Synthesis by Aspergillus. J. Fungi 2025, 11, 534. [Google Scholar] [CrossRef]
- Meng, X.; Yang, J.; Xu, X.; Zhang, L.; Nie, Q.; Xian, M. Biodiesel production from oleaginous microorganisms. Renew. Energy 2009, 34, 1–5. [Google Scholar] [CrossRef]
- Sakuradani, E.; Kobayashi, M.; Shimizu, S. Δ9-fatty acid desaturase from arachidonic acid-producing fungus. unique gene sequence and its heterologous expression in a fungus, Aspergillus. Eur. J. Biochem. 1999, 260, 208–216. [Google Scholar] [CrossRef]
- Tamano, K. Advancements in lipid production research using the koji-mold Aspergillus oryzae and future outlook. Front. Fungal Biol. 2024, 5, 1526568. [Google Scholar] [CrossRef] [PubMed]
- Tamano, K.; Bruno, K.S.; Koike, H.; Ishii, T.; Miura, A.; Umemura, M.; Culley, D.E.; Baker, S.E.; Machida, M. Increased production of free fatty acids in Aspergillus oryzae by disruption of a predicted Acyl-CoA synthetase gene. Appl. Microbiol. Biotechnol. 2015, 99, 3103–3113. [Google Scholar] [CrossRef]
- Wong, P.S.; Tamano, K.; Aburatani, S. Improvement of free fatty acid secretory productivity in Aspergillus oryzae by comprehensive analysis on time-series gene expression. Front. Microbiol. 2021, 12, 605095. [Google Scholar] [CrossRef]
- Tamano, K.; Cox, R.S.; Tsuge, K.; Miura, A.; Itoh, A.; Ishii, J.; Tamura, T.; Kondo, A.; Machida, M. Heterologous production of free dihomo-γ-linolenic acid by Aspergillus oryzae and its extracellular release via surfactant supplementation. J. Biosci. Bioeng. 2019, 127, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, N.; Matsui, J.; Kawai, Y.; Kato, A.; Yoshino, S.; Ohmiya, K.; Tsukagoshi, N. Utilization of the TEF1-α Gene (TEF1) promoter for expression of polygalacturonase genes, pgaA and pgaB, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 1998, 50, 85–92. [Google Scholar] [CrossRef]
- Kanemori, Y.; Gomi, K.; Kitamoto, K.; Kumagai, C.; Tamura, G. Insertion analysis of putative functional elements in the promoter region of the Aspergillus oryzae Taka-amylase A gene (amyB) using a heterologous Aspergillus Nidulans amdS-lacZ fusion gene system. Biosci. Biotechnol. Biochem. 1999, 63, 180–183. [Google Scholar] [CrossRef]
- Takara Bio Inc. Transformation of A. oryzae by the Protoplast-PEG Method Using pPTR I DNA. Available online: https://catalog.takara-bio.co.jp/com/tech_info_detail.php?mode=2&masterid=M100003051&unitid=U100004022 (accessed on 24 September 2025).
- American Oil Chemists’ Society. AOCS Official Method Ce 1j-07: Determination of Cis-, Trans-, Saturated, Monounsaturated, and Polyunsaturated Fatty Acids in Extracted Fats by Capillary GLC; American Oil Chemists’ Society: Urbana, IL, USA, 2013. [Google Scholar]
- Ouchi, K.; Ishido, T.; Sugama, S.; Nojiro, K. Studies on the ecology of yeast in koji. III. Estimation of the growth of Aspergillus oryzae during koji making. J. Brew. Soc. Jpn. 1967, 62, 1029–1033. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids. Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Itoh, T.; Onuki, R.; Tsuda, M.; Oshima, M.; Endo, M.; Sakai, H.; Tanaka, T.; Ohsawa, R.; Tabei, Y. Foreign DNA detection by high-throughput sequencing to regulate genome-edited agricultural products. Sci. Rep. 2020, 10, 4914. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-Core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Heredero, M.; Garrigues, S.; Gandía, M.; Marcos, J.F.; Manzanares, P. Rational Design and Biotechnological Production of Novel AfpB-PAF26 Chimeric Antifungal Proteins. Microorganisms 2018, 6, 106. [Google Scholar] [CrossRef]
- Sayanova, O.; Haslam, R.; Guschina, I.; Lloyd, D.; Christie, W.W.; Harwood, J.L.; Napier, J.A. A bifunctional Δ12,Δ15-desaturase from Acanthamoeba castellanii directs the synthesis of highly unusual n-1 series unsaturated fatty acids. J. Biol. Chem. 2006, 281, 36533–36541. [Google Scholar] [CrossRef]
- Adarme-Vega, T.C.; Thomas-Hall, S.R.; Lim, D.K.Y.; Schenk, P.M. Effects of long chain fatty acid synthesis and associated gene expression in microalga Tetraselmis sp. Mar. Drugs 2014, 12, 3381–3398. [Google Scholar] [CrossRef]
- Cui, J.; He, S.; Ji, X.; Lin, L.; Wei, Y.; Zhang, Q. Identification and characterization of a novel bifunctional Δ12/Δ15-fatty acid desaturase gene from Rhodosporidium kratochvilovae. Biotechnol. Lett. 2016, 38, 1155–1164. [Google Scholar] [CrossRef]
- Ciftci, O.N.; Przybylski, R.; Rudzińska, M. Lipid components of flax, perilla, and chia seeds. Eur. J. Lipid Sci. Technol. 2012, 114, 794–800. [Google Scholar] [CrossRef]
- Falcone, D.L.; Ogas, J.P.; Somerville, C.R. Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol. 2004, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef]
- Ratledge, C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 2004, 86, 807–815. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant unsaturated fatty acids: Biosynthesis and regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Martin, C.E.; Oh, C.-S.; Jiang, Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 271–285. [Google Scholar] [CrossRef]
- Chellappa, R.; Kandasamy, P.; Oh, C.-S.; Jiang, Y.; Vemula, M.; Martin, C.E. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 2001, 276, 43548–43556. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; De Vries, R.P.; Garrigues, S. Strategies for the development of industrial fungal producing strains. J. Fungi 2023, 9, 834. [Google Scholar] [CrossRef]
- Hanlon, P.; Sewalt, V. GEMs: Genetically engineered microorganisms and the regulatory oversight of their uses in modern food production. Crit. Rev. Food Sci. Nutr. 2021, 61, 959–970. [Google Scholar] [CrossRef]
- Leenes, R.E.; Kosta, E. (Eds.) Bridging Distances in Technology and Regulation; Wolf Legal Publishers (WLP): Oisterwijk, The Netherlands, 2013. [Google Scholar]
- Rousta, N.; Hellwig, C.; Wainaina, S.; Lukitawesa, L.; Agnihotri, S.; Rousta, K.; Taherzadeh, M.J. Filamentous fungus Aspergillus oryzae for food: From submerged cultivation to fungal burgers and their sensory evaluation-a pilot study. Foods 2021, 10, 2774. [Google Scholar] [CrossRef]
- Prime Roots. Available online: https://www.primeroots.com/ (accessed on 5 August 2025).
- Formo Bio GmbH. Available online: https://www.eatformo.com/process/ (accessed on 5 August 2025).
- Ghiyasi, M.; Rezaei, M.; Sayyahzade, H. Effect of prebiotic (Fermacto) in low protein diet on performance and carcass characteristics of broiler chicks. Int. J. Poult. Sci. 2007, 6, 661–665. [Google Scholar] [CrossRef][Green Version]
- Zahirian, M.; Seidavi, A.; Solka, M.; Nosrati, M.; Corazzin, M. Dietary supplementation of Aspergillus oryzae meal and its effect on performance, carcass characteristics, blood variables, and immunity of broiler chickens. Trop. Anim. Health Prod. 2019, 51, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Good Balance Meat. Available online: http://www.goodbalancemeat.jp/data/ (accessed on 5 August 2025).
- Sato, Y. Food by-products and methane-inhibiting feeds for ruminants. Nihon Chikusan Gakkaiho 2023, 94, 161–168. [Google Scholar] [CrossRef]
- Martin, C.; Rouel, J.; Jouany, J.P.; Doreau, M.; Chilliard, Y. Methane output and diet digestibility in response to feeding dairy cows crude linseed, extruded linseed, or linseed Oil1. J. Anim. Sci. 2008, 86, 2642–2650. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.; Kotake, E.; Tsuzuki, W.; Suzuki, S.; Kusumoto, K.; Hattori, R. Omega-3 Fatty Acid Production by Self-Cloning Koji Mold toward Food Waste Utilization. In Proceedings of the 2022 Annual Meeting of Japan Society for Bioscience, Biotechnology, and Agrochemistry, Kyoto, Japan, 15–18 March 2022. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).