Bridging Ethnobotanical Knowledge and Multi-Omics Approaches for Plant-Derived Natural Product Discovery
Abstract
1. Introduction
2. Plant-Derived Natural Products
2.1. Traditional Medicine Based on Plant-Derived Natural Products
| Plant Name | Tissue Used | Administration | Medicinal Use/Treatment | Reference |
|---|---|---|---|---|
| Africa | ||||
| Acacia senegal (gum arabic) | Whole plant | Oral, topical | Bleeding, bronchitis, diarrhoea, gonorrhoea, leprosy, typhoid fever, upper respiratory tract infections | [32] |
| Aloe ferox (bitter aloe or Cape aloe) | Leaves | Oral | Anti-inflammatory, analgesic, calming, antiseptic, germicidal, antiviral, antiparasitic, anticancer | [33] |
| Artemisia herba-alba (wormwood) | Leaves, stems | Oral | Arterial hypertension, diabetes, bronchitis, diarrhoea, hypertension, neuralgias | [34] |
| Catharanthus roseus (Madagascar periwinkle) | Leaves, seeds, stems, petals | Oral | Anticancer, rheumatism, skin disorders, venereal diseases | [35] |
| China | ||||
| Aconitum napellus (monkshood) | Roots | Oral, topical, inhalation | Hypertension, haemorrhoids, colic, upper urinary tract cancer, kidney failure | [36] |
| Trichosanthes kirilowii (Chinese cucumber) | Seeds, fruits, pericarps, roots | Oral | Tumours, reduces fevers, swelling and coughing, abscesses, amenorrhea, jaundice, polyuria | [37] |
| Chrysanthemum spp. (mums) | Flowers, seeds | Oral, topical | Chest pain, high blood pressure, type 2 diabetes, fever, cold, headache, dizziness, and swelling | [38] |
| Panax ginseng (ginseng) | Leaves, stems, root | Oral | Fatigue, stress, asthma, cancer, diarrhoea, anxiety, mental health | [39] |
| Artemisia annua (sweet wormwood) | Leaves | Oral | Malaria, fever reduction, inflammation | [40] |
| America | ||||
| Allium sativum (garlic) | Cloves | Oral | Hypercholesterolemia, claudication, common cold, osteoarthritis | [41] |
| Hypericum perforatum (St. John’s wort) | Leaves, flowers, seeds | Oral | Depression, menopausal symptoms, attention-deficit hyperactivity disorder (ADHD) | [42] |
| Berberis vulgarisn (barberry) | Fruit, bark, root, stem | Oral | Fever, cough, liver disease, depression, hyperlipidaemia, hyperglycaemia and bleeding | [43] |
| Echinacea purpurea (purple coneflower) | Leaves, flower petals | Topical, oral | Infections, wounds, urinary tract infections, cold and flu | [44] |
| Taxus brevifolia (Pacific yew) | Bark | Oral | Breast and ovarian cancer | [45] |
| Digitalis lanata (woolly foxglove) | Leaves | Oral | Heart failure, arrhythmia, atrial fibrillation | [46] |
| India | ||||
| Terminalia arjuna (Arjuna) | Stem, bark, fruits leaves | Oral | Fractures, ulcers, antibacterial, antimicrobial, antioxidant, antiallergic, antifertility, anti-HIV | [47] |
| Andrographis paniculate (Kalmegh) | Whole plant | Oral | Cold, diarrhoea, fever, jaundice, as a health tonic for the liver and cardiovascular health, antioxidant | [48] |
| Mucuna Pruriens (Kauch) | Fruits, seed | Oral | Parkinson disease, sexual disorders | [49] |
| Acacia catechu (Khair) | Leaves, bark, wood | Oral | Mouth ulcer, anaemia, high blood pressure, dysentery, colitis, gastric problems, bronchial asthma, cough, leucorrhoea and leprosy | [50] |
| Catharanthus roseus (periwinkle) | Whole plant | Oral | Cancer, diabetes, vaginal discharge, tonsillitis, chest pain, high blood pressure, sore throat, intestinal pain, inflammation, toothache | [51] |
| Papaver somniferum (opium poppy) | Seeds | Oral, injection | Pain relief, cough suppressant, diarrhoea | [30] |
2.2. Chemical Characteristics of Plant-Derived Natural Products
| Plant Source | Bioactive Compound | Compound Class | Effects/Bioactivity | Mechanism of Action | Reference |
|---|---|---|---|---|---|
| Artemisia glabella | Arglabin | Terpene | Antitumor | Inhibits farnesyl transferase | [75] |
| Cannabis sativa | Cannabidiol | Cannabinoid | Anti-epileptic, anxiolytic, antipsychotic, and anticancer | Modulates CB1, CB2, 5HT1A receptors in the central nervous system | [76] |
| Capsicum annum | Capsaicin | Alkaloid | Chronic pain syndromes such as postherpetic neuralgia and musculoskeletal pain | Activates transient receptor potential vanilloid 1 (TRPV1) in sensory nerves | [77] |
| Colchicum spp. | Colchicine | Alkaloid | Gout | Scavenges reactive oxygen and nitrogen species, inhibits NF-kB, modulates activities of glutathione, catalase, and superoxide dismutase | [78] |
| Genista tinctoria | Genistein | Flavonoid | Anticancer, Alzheimer’s disease | Inhibits protein-tyrosine kinase, induces apoptosis, antimetastatic and antiangiogenic activity, antioxidant | [79] |
| Gossypium hirsutum | Gossypol | Terpene | Anti-infertility/male contraceptive, anticancer, antiviral, antimicrobial, antioxidant activities | Bcl-2 and sperm production inhibition, induces apoptosis, inhibits DNA polymerase and topoisomerase II | [80] |
| Tabebuia avellanedae | β-Lapachone | Quinone | Variety of cancers, especially solid tumours, anti-trypanosoma, antimicrobial, and antimalarial activities | Anticancer activity through formation of ROS in NQO1-positive cells, inhibits topoisomerase, modulates the mTOR pathway | [81] |
| Larrea tridentate | Masoprocol | Phenolic compound | Antineoplastic agent used in cancer chemotherapy | Inhibits 5-Lipoxygenase | [6] |
| Podophyllum emodi | Podophyllotoxin | Phenolic compound | Antitumour | Suppresses formation of mitotic spindles microtubules, cycle arrest via polymerisation of tubulin | [82] |
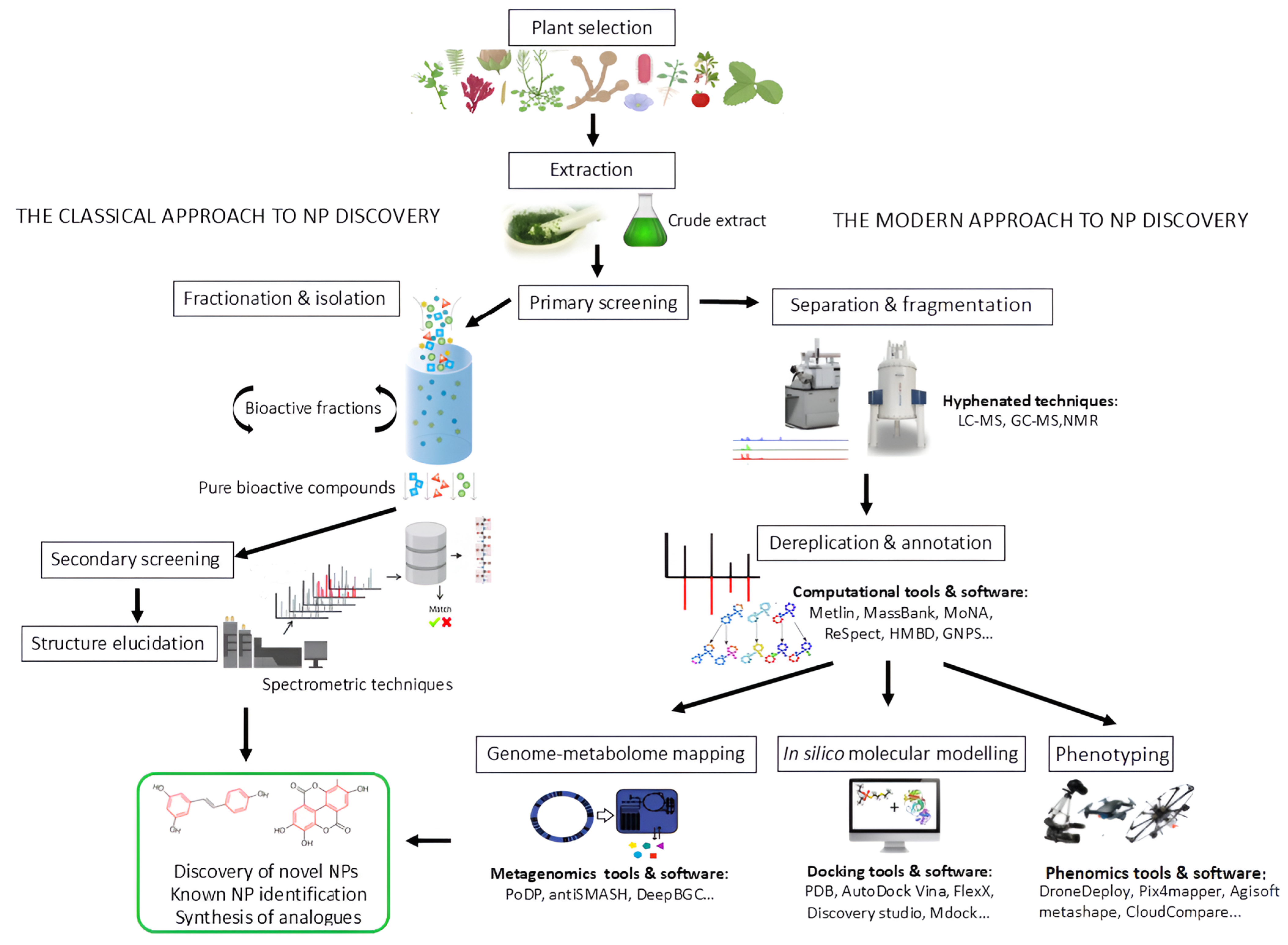
3. Classical Approaches in NP Research
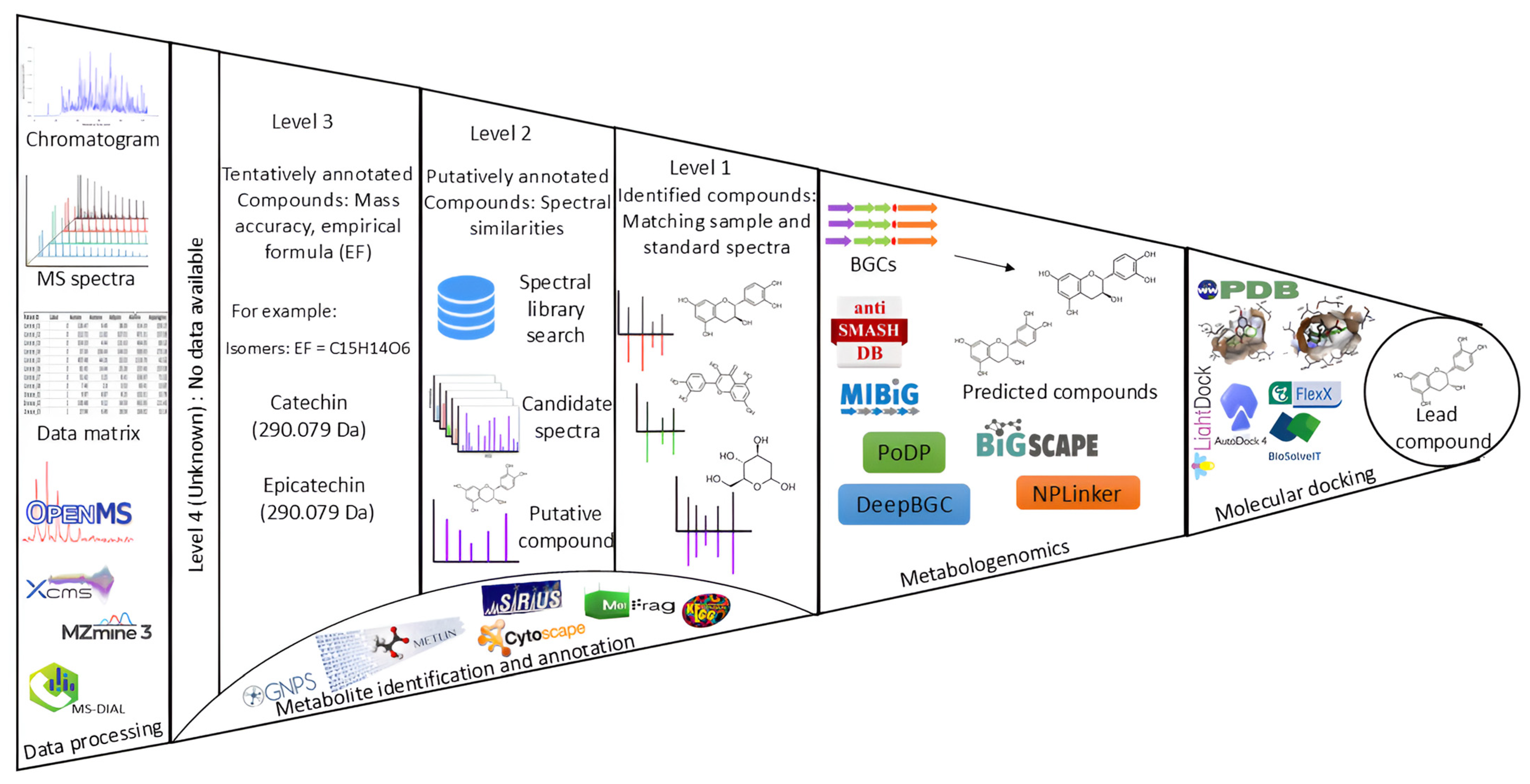
4. Computational Metabolomics in Plant-Derived NP Research
Technological Advancements in Metabolite Annotation
| Tool | Website | Description/Function/Role | References |
|---|---|---|---|
| Data processing and analysis | |||
| XCMS online | https://xcmsonline.scripps.edu | Nonlinear retention time alignment | [119] |
| MZmine | https://github.com/mzmine/mzmine | Mass detection, peak deconvolution, retention time alignment | [120] |
| OpenMS | https://www.openms.de/ | Peak picking, retention time alignment, baseline and noise filtering, metabolite quantification and identification | [121] |
| MS-DIAL | https://systemsomicslab.github.io/compms/msdial/main.html | Spectral deconvolution, peak identification, statistical analysis | [122] |
| HomologueDiscoverer | https://github.com/kevinmildau/homologueDiscoverer | Detection and omission of noise features and redundant features | [123] |
| Metabolite annotation libraries | |||
| METLIN | https://metlin.scripps.edu/ | Repository for searchable MS2 data (positive and negative modes) and neutral loss libraries acquired from standards | [152] |
| MassBank | https://massbank.eu/MassBank/ | Spectral data repository | [153,154] |
| ReSpect | https://github.com/shahab-sarmashghi/RESPECT | Spectra and taxonomy information repository | [147] |
| GNPS | https://gnps.ucsd.edu/ | Repository for spectral libraries, molecular network construction | [125] |
| Mass Spectral Networking, Embedding and Annotation | |||
| Classical Molecular Networking (MN) | https://gnps.ucsd.edu/ | Groups metabolites based on MS/MS spectra similarity, forming molecular networks | [125] |
| Feature-Based Molecular Networking (FBMN) | https://gnps.ucsd.edu/ | Enhances MN by using MS1 feature data to align nodes more precisely in molecular networks | [124] |
| msFeaST | https://github.com/kevinmildau/msFeaST | Integrates MS1 and MS2 information for improved feature selection in molecular networking | [140] |
| MS2LDA | https://ms2lda.org/ | Decomposition of molecular fragmentation, annotation and discovery of Mass2Motifs | [128] |
| MS2Query | https://github.com/iomega/ms2query | Integrates Spec2Vec and MS2Deepscore, ranks potential analogues and exact matches | [155] |
| Spec2Vec | https://github.com/iomega/spec2vec | Assess and rank spectral similarities | [133] |
| MS2Deepscore | https://github.com/matchms/ms2deepscore | Predicts structural similarity between MS/MS spectra | [134] |
| NAP | https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp | Improves in silico fragmentation candidate structure ranking | [135] |
| DEREPLICATOR | https://gnps.ucsd.edu/ | In silico identification of both peptidic and non-peptidic natural products | [136] |
| SIRIUS+CSI:FingerID | https://github.com/computational-metabolomics/sirius-csifingerid-galaxy | Analysis of isotope patterns, compound class prediction. | [156,157] |
| ClassyFire | https://bio.tools/ClassyFire | Large-scale automated chemical/metabolite classification | [138] |
| MolNetEnhancer | https://gnps.ucsd.edu/ | Enhances molecular networks by integrating chemical classification data to assign chemical ontologies to metabolites. Helps identify molecular families and structural variations | [132] |
| Bioactivity-Based Molecular Networking (BBMN) | https://gnps.ucsd.edu | Links molecular networks to bioactivity data to discover bioactive compounds | [139] |
| FERMO | https://fermo.bioinformatics.nl | Links bioactivity information to metabolite features in natural product discovery | [141] |
| Cytoscape | https://cytoscape.org/ | Network data integration, analysis, and visualisation | [127] |
| Metabologenomics | |||
| antiSMASH | https://github.com/antismash/antismash | Identify, annotate, and compare gene clusters that encode the biosynthesis of NPs | [142] |
| Paired Omics Data Platform (PoDP) | https://pairedomicsdata.bioinformatics.nl | Standardise links between genomic and metabolomics data in a computer readable format to further the field of natural products discovery | [146] |
| plantiSMASH | http://plantismash.secondarymetabolites.org/ | A specialised version of antiSMASH designed for the identification, annotation, and analysis of BGCs in plant genomes | [144] |
| NPLinker | https://github.com/nplinker/nplinker | Links BGCs with metabolomics data to facilitate the discovery of natural products by integrating and analysing paired omics datasets | [158] |
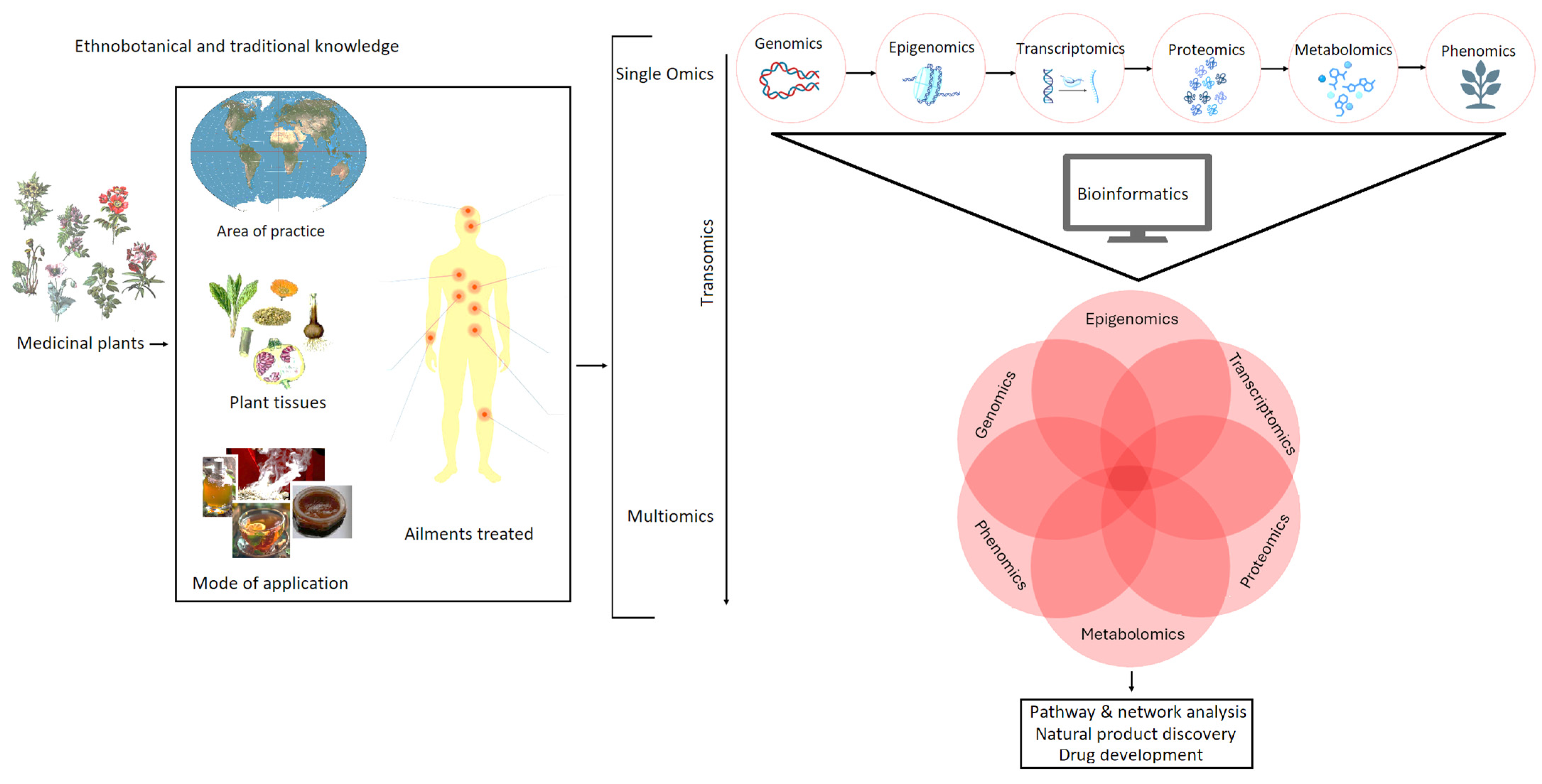
5. Integration of Omics Technologies for NP Discovery
5.1. Genomics: Uncovering Biosynthetic Potential
5.2. Transcriptomics: Understanding Gene Expression Dynamics
5.3. Proteomics: Linking Genotype to Phenotype
5.4. Applications of Integrated Omics in NP Discovery
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koehn, F.E.; Carter, G.T. The Evolving Role of Natural Products in Drug Discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Ramabulana, A.T.; Petras, D.; Madala, N.E.; Tugizimana, F. Metabolomics and Molecular Networking to Characterize the Chemical Space of Four Momordica Plant Species. Metabolites 2021, 11, 763. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Schwachtje, J.; Fischer, A.; Erban, A.; Kopka, J. Primed Primary Metabolism in Systemic Leaves: A Functional Systems Analysis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Shen, H.; Zamboni, N.; Heinonen, M.; Rousu, J. Metabolite Identification through Machine Learning—Tackling CASMI Challenge Using FingerID. Metabolites 2013, 3, 484–505. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef]
- Chemat, F.; Rombaut, N.; Sicaire, A.G.; Meullemiestre, A.; Fabiano-Tixier, A.S.; Abert-Vian, M. Ultrasound Assisted Extraction of Food and Natural Products. Mechanisms, Techniques, Combinations, Protocols and Applications. A Review. Ultrason. Sonochem. 2017, 34, 540–560. [Google Scholar] [CrossRef]
- Fabricant, D.S.; Farnsworth, N.R. The value of Plants Used in Traditional Medicine for Drug Discovery. Environ. Health Perspect. 2001, 109, 69–75. [Google Scholar] [CrossRef]
- Calixto, J.B. The Role of Natural Products in Modern Drug Discovery. Acad. Bras. Cienc. 2019, 91, e20190105. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Yang, X.; Feng, P.; Yin, Y.; Bushley, K.; Spatafora, J.W.; Wang, C. Cyclosporine Biosynthesis in Tolypocladium Inflatum Benefits Fungal Adaptation to the Environment. MBio 2018, 9, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Murray, V.; Chen, J.K.; Chung, L.H. The Interaction of the Metallo-Glycopeptide Anti-Tumour Drug Bleomycin with DNA. Int. J. Mol. Sci. 2018, 19, 1372. [Google Scholar] [CrossRef] [PubMed]
- Babashpour-Asl, M.; Kaboudi, P.S.; Barez, S.R. Therapeutic and Medicinal Effects of Snowdrop (Galanthus spp.) in Alzheimer’s Disease: A Review. J. Educ. Health Promot. 2023, 12, 128. [Google Scholar] [CrossRef]
- Medema, M.H. Computational Genomics of Specialized Metabolism: From Natural Product Discovery to Microbiome Ecology. mSystems 2018, 3, 10-1128. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Nuzillard, J.M.; Van Der Hooft, J.J.J.; Renault, J.H.; Bertrand, S. Accelerating Metabolite Identification in Natural Product Research: Toward an Ideal Combination of Liquid Chromatography-High-Resolution Tandem Mass Spectrometry and NMR Profiling, in Silico Databases, and Chemometrics. Anal. Chem. 2019, 91, 704–742. [Google Scholar] [CrossRef]
- Traxler, M.F.; Kolter, R. Natural Products in Soil Microbe Interactions and Evolution. Nat. Prod. Rep. 2015, 32, 956–970. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Alanjary, M.; Weber, T. The Evolution of Genome Mining in Microbes-a Review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Lima, R.D.C.L.; Gramsbergen, S.M.; Van Staden, J.; Jäger, A.K.; Kongstad, K.T.; Staerk, D. Advancing HPLC-PDA-HRMS-SPE-NMR Analysis of Coumarins in Coleonema Album by Use of Orthogonal Reversed-Phase C18 and Pentafluorophenyl Separations. J. Nat. Prod. 2017, 80, 1020–1027. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Sayed, A.M.; Elmaidomy, A.H. Natural Products’ Extraction and Isolation-Between Conventional and Modern Techniques. Front. Nat. Prod. 2022, 1, 873808. [Google Scholar] [CrossRef]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The Traditional Medicine and Modern Medicine from Natural Products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef]
- Mahomoodally, M.F. Evidence Based Complementary and Alternative Medicine Traditional Medicines in Africa: An Appraisal of Ten Potent African Medicinal Plants. Evid.-Based Complement. Altern. Med. 2013, 2013, 617459. [Google Scholar] [CrossRef] [PubMed]
- Mutombo, P.N.; Kasilo, O.M.J.; James, P.B.; Wardle, J.; Kunle, O.; Katerere, D.; Wambebe, C.; Matsabisa, M.G.; Rahmatullah, M.; Nikiema, J.B.; et al. Experiences and Challenges of African Traditional Medicine: Lessons from COVID-19 Pandemic. BMJ Glob. Health 2023, 8, e010813. [Google Scholar] [CrossRef] [PubMed]
- Meghana, N.; Sunil, A.; Patil, S. Review on ’Swastya’-Principles of Charaka Samhita as Explained in Dinacharya and Rutucharya. J. Ayurveda Integr. Med. Sci. 2022, 7, 122–126. [Google Scholar]
- Yang, D.; Yang, X. Historical Background of Ben Cao Gang Mu (Compendium of Materia Medica). Chin. Med. Cult. 2019, 2, 32–35. [Google Scholar] [CrossRef]
- Bhavana, K. Shreevathsa Medical Geography in Charaka Samhita. AYU (Int. Q. J. Res. Ayurveda) 2014, 35, 371. [Google Scholar] [CrossRef]
- Leung, A.Y. Traditional Toxicity Documentation of Chinese Materia Medica—An Overview. Toxicol. Pathol. 2006, 34, 319–326. [Google Scholar] [CrossRef]
- Afolayan, A.J.; Aladejana, A.E.; Bradley, G. In Vitro Evaluation of the Anti-Diabetic Potential of Helichrysum Petiolare Hilliard & B.L. Burtt Using HepG2 (C3A) and L6 Cell Lines. F1000Research 2021, 9, 1240. [Google Scholar] [CrossRef]
- van Wyk, B.E.; Albrecht, C. A Review of the Taxonomy, Ethnobotany, Chemistry and Pharmacology of Sutherlandia Frutescens (Fabaceae). J. Ethnopharmacol. 2008, 119, 620–629. [Google Scholar] [CrossRef]
- Liu, N.Q.; Van der Kooy, F.; Verpoorte, R. Artemisia Afra: A Potential Flagship for African Medicinal Plants? S. Afr. J. Bot. 2009, 75, 185–195. [Google Scholar] [CrossRef]
- Zulak, K.G.; Weljie, A.M.; Vogel, H.J.; Facchini, P.J. Quantitative 1H NMR Metabolomics Reveals Extensive Metabolic Reprogramming of Primary and Secondary Metabolism in Elicitor-Treated Opium Poppy Cell Cultures. BMC Plant Biol. 2008, 8, 1–19. [Google Scholar] [CrossRef]
- Ramasamy, J.; Kandasamy, R.; Palanisamy, S.; Nadesan, S. Optimization of Ultrasonic-Assisted Extraction of Flavonoids and Anti-oxidant Capacity from the Whole Plant of Andrographis echioides (L.) Nees by Response Surface Methodology and Chemical Composition Analysis. Pharmacogn. Mag. 2019, 15, 547–556. [Google Scholar] [CrossRef]
- Magnini, R.D.; Hilou, A.; Millogo-Koné, H.; Pagès, J.M.; Davin-Regli, A. Acacia Senegal Extract Rejuvenates the Activity of Phenicols on Selected Enterobacteriaceae Multi Drug Resistant Strains. Antibiotics 2020, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Gherbon, A.; Frandes, M.; Timar, R.; Nicula, M. Beneficial Effects of Aloe Ferox on Lipid Profile, Blood Pressure, and Glycemic Control in Obese Persons A CONSORT-Clinical Study. Medicine 2021, 100, E28336. [Google Scholar] [CrossRef] [PubMed]
- Nigam, M.; Atanassova, M.; Mishra, A.P.; Pezzani, R.; Devkota, H.P.; Plygun, S.; Salehi, B.; Setzer, W.N.; Sharifi-Rad, J. Bioactive Compounds and Health Benefits of Artemisia Species. Nat. Prod. Commun. 2019, 14. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, B.; Singh, R. Catharanthus roseus (L.) G. Don: A Review of Its Ethnobotany, Phytochemistry, Ethnopharmacology and Toxicities. J. Ethnopharmacol. 2022, 284, 114647. [Google Scholar] [CrossRef]
- Povšnar, M.; Koželj, G.; Kreft, S.; Lumpert, M. Rare Tradition of the Folk Medicinal Use of Aconitum spp. Is Kept Alive in Solčavsko, Slovenia. J. Ethnobiol. Ethnomed. 2017, 13, 45. [Google Scholar] [CrossRef]
- Park, S.M.; Jeon, S.K.; Kim, O.H.; Ahn, J.Y.; Kim, C.H.; Park, S.D.; Lee, J.H. Anti-Tumor Effects of the Ethanolic Extract of Trichosanthes Kirilowii Seeds in Colorectal Cancer. Chin. Med. 2019, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Škoro, N.; Živković, S.; Jevremović, S.; Puač, N. Treatment of Chrysanthemum Synthetic Seeds by Air SDBD Plasma. Plants 2022, 11, 907. [Google Scholar] [CrossRef]
- Wang, H.; Peng, D.; Xie, J. Ginseng Leaf-Stem: Bioactive Constituents and Pharmacological Functions. Chin. Med. 2009, 4, 20. [Google Scholar] [CrossRef]
- Cheong, D.H.J.; Tan, D.W.S.; Wong, F.W.S.; Tran, T. Anti-Malarial Drug, Artemisinin and Its Derivatives for the Treatment of Respiratory Diseases. Pharmacol. Res. 2020, 158, 104901. [Google Scholar] [CrossRef]
- Mikaili, P.; Maadirad, S.; Moloudizargari, M.; Aghajanshakeri, S. Ijbms-16-1031- Garlic and Shallot.Pdf. Iran. J. Basic. Med. Sci. 2013, 16, 1031–1048. [Google Scholar] [PubMed]
- Oliveira, A.I.; Pinho, C.; Sarmento, B.; Dias, A.C.P. Neuroprotective Activity of Hypericum Perforatum and Its Major Components. Front. Plant Sci. 2016, 7, 1004. [Google Scholar] [CrossRef]
- Tabeshpour, J.; Imenshahidi, M.; Hosseinzadeh, H. A Review of the Effects of Berberis Vulgaris and Its Major Component, Berberine, in Metabolic Syndrome. Iran. J. Basic. Med. Sci. 2017, 20, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Manayi, A.; Vazirian, M.; Saeidnia, S. Echinacea Purpurea: Pharmacology, Phytochemistry and Analysis Methods. Pharmacogn. Rev. 2015, 9, 63–72. [Google Scholar] [CrossRef]
- Ketchuma, R.E.B.; Rithner, C.D.; Qiu, D.; Kim, Y.S.; Williams, R.M.; Croteaua, R.B. Taxus Metabolomics: Methyl Jasmonate Preferentially Induces Production of Taxoids Oxygenated at C-13 in Taxus x Media Cell Cultures. Phytochemistry 2003, 62, 901–909. [Google Scholar] [CrossRef]
- Ravi, B.G.; Guardian, M.G.E.; Dickman, R.; Wang, Z.Q. High-Resolution Tandem Mass Spectrometry Dataset Reveals Fragmentation Patterns of Cardiac Glycosides in Leaves of the Foxglove Plants. Data Brief. 2020, 30, 105464. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.; Chopra, D. Revisiting Terminalia Arjuna-an Ancient Cardiovascular Drug. J. Tradit. Complement. Med. 2014, 4, 224–231. [Google Scholar] [CrossRef]
- Hossain, M.S.; Urbi, Z.; Sule, A.; Rahman, K.M.H. Andrographis Paniculata (Burm. f.) Wall. Ex Nees: A Review of Ethnobotany, Phytochemistry, and Pharmacology. Sci. World J. 2014, 2014, 274905. [Google Scholar] [CrossRef] [PubMed]
- Pathania, R.; Chawla, P.; Khan, H.; Kaushik, R.; Khan, M.A. An Assessment of Potential Nutritive and Medicinal Properties of Mucuna Pruriens: A Natural Food Legume. 3 Biotech. 2020, 10, 261. [Google Scholar] [CrossRef]
- Sunil, M.A.; Sunitha, V.S.; Radhakrishnan, E.K.; Jyothis, M. Immunomodulatory Activities of Acacia Catechu, a Traditional Thirst Quencher of South India. J. Ayurveda Integr. Med. 2019, 10, 185–191. [Google Scholar] [CrossRef]
- Pham, H.N.T.; Van Vuong, Q.; Bowyer, M.C.; Scarlett, C.J. Phytochemicals Derived from Catharanthus Roseus and Their Health Benefits. Technology 2020, 8, 80. [Google Scholar] [CrossRef]
- Lo, H.-Y.; Li, C.-C.; Huang, H.-C.; Lin, L.-J.; Hsiang, C.-Y.; Ho, T.-Y. Application of Transcriptomics in Chinese Herbal Medicine Studies. J. Tradit. Complement. Med. 2012, 2, 105–114. [Google Scholar] [CrossRef]
- Li, S.; Fan, T.P.; Jia, W.; Lu, A.; Zhang, W. Network Pharmacology in Traditional Chinese Medicine. Evid.-Based Complement. Altern. Med. 2014, 2014, 138460. [Google Scholar] [CrossRef]
- Narayana, D.B.A.; Durg, S. Ayurveda: (W)Here Is the Evidence. J. Ayurveda Integr. Med. 2021, 12, 408–411. [Google Scholar] [CrossRef]
- Chauhan, A.; Semwal, D.; Mishra, S.; Semwal, R. Ayurvedic Research and Methodology: Present Status and Future Strategies. AYU (Int. Q. J. Res. Ayurveda) 2015, 36, 364. [Google Scholar] [CrossRef]
- Gouws, C.; Smit, T.; Willers, C.; Svitina, H.; Calitz, C.; Wrzesinski, K. Anticancer Potential of Sutherlandia Frutescens and Xysmalobium Undulatum in Ls180 Colorectal Cancer Mini-Tumors. Molecules 2021, 26, 605. [Google Scholar] [CrossRef]
- Agbor, A.M.; Naidoo, S. A Review of the Role of African Traditional Medicine in the Management of Oral Diseases. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 133–142. [Google Scholar] [CrossRef]
- Ghorbani, A.; Esmaeilizadeh, M. Pharmacological Properties of Salvia Officinalis and Its Components. J. Tradit. Complement. Med. 2017, 7, 433–440. [Google Scholar] [CrossRef]
- Kmail, A. Mitigating Digestive Disorders: Action Mechanisms of Mediterranean Herbal Active Compounds. Open Life Sci. 2024, 19, 20220857. [Google Scholar] [CrossRef]
- Nieto, G. A Review on Applications and Uses of Thymus in the Food Industry. Plants 2020, 9, 961. [Google Scholar] [CrossRef]
- Ekor, M. The Growing Use of Herbal Medicines: Issues Relating to Adverse Reactions and Challenges in Monitoring Safety. Front. Neurol. 2014, 4, 177. [Google Scholar] [CrossRef]
- Su, L.; Pan, Y. Ethical Considerations in the Use of Endangered Species for Traditional Chinese Medicine Practices: A Case Study of the Pangolin. J. Res. Soc. Humanit. 2024, 3, 24–30. [Google Scholar] [CrossRef]
- Dresselhaus, T. Biotic and Abiotic Stress Responses in Crop Plants. Agronomy 2018, 8, 267. [Google Scholar] [CrossRef]
- Pimm, S.L.; Joppa, L.N. How Many Plant Species Are There, Where Are They, and at What Rate Are They Going Extinct? Ann. Mo. Bot. Gard. 2015, 100, 170–176. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The Biomass Distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Reher, R.; Kim, H.W.; Zhang, C.; Mao, H.H.; Wang, M.; Nothias, L.F.; Caraballo-Rodriguez, A.M.; Glukhov, E.; Teke, B.; Leao, T.; et al. A Convolutional Neural Network-Based Approach for the Rapid Annotation of Molecularly Diverse Natural Products. J. Am. Chem. Soc. 2020, 142, 4114–4120. [Google Scholar] [CrossRef]
- Krishnamurti, C.; Rao, S.S.C.C. The Isolation of Morphine by Serturner. Indian J. Anaesth. 2016, 60, 861–862. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Kendall, M.J.; Wade, O.L. William Withering and Digitalis, 1785 to 1985. Br. Med. J. (Clin. Res. Ed.) 1985, 290, 7–8. [Google Scholar] [CrossRef]
- Krikler, D.M. The Foxglove, “the Old Woman from Shropshire” and William Withering. J. Am. Coll. Cardiol. 1985, 5, 3A–9A. [Google Scholar] [CrossRef]
- Littler, W.A. William Withering digitalis and the Pulse. Int. J. Med. 2019, 112, 565–566. [Google Scholar] [CrossRef]
- Su, X.Z.; Miller, L.H. The discovery of artemisinin and the Nobel Prize in Physiology or Medicine. Sci. China Life Sci. 2015, 58, 1175–1179. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Huang, E.; Li, B.; Zhang, S.; Wang, W.; Guo, Z.; Wu, K.; Zhang, Y.; Zhao, B.; et al. Metabolomics Analysis of Three Artemisia Species in the Tibet Autonomous Region of China. BMC Plant Biol. 2022, 22, 208. [Google Scholar] [CrossRef]
- Brendler, T.; Abdel-Tawab, M. Buchu (Agathosma betulina and A. crenulata): Rightfully Forgotten or Underutilized? Front. Pharmacol. 2022, 13, 813142. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Savka, M.A.; Hudson, A.O. The Synthesis and Role of β-Alanine in Plants. Front. Plant Sci. 2019, 10, 921. [Google Scholar] [CrossRef]
- Lone, S.H.; Bhat, K.A.; Khuroo, M.A. Arglabin: From Isolation to Antitumor Evaluation. Chem. Biol. Interact. 2015, 240, 180–198. [Google Scholar] [CrossRef]
- Huestis, M.A.; Solimini, R.; Pichini, S.; Pacifici, R.; Carlier, J.; Busardò, F.P. Cannabidiol Adverse Effects and Toxicity. Curr. Neuropharmacol. 2019, 17, 974–989. [Google Scholar] [CrossRef]
- Sharma, S.K.; Vij, A.S.; Sharma, M. Mechanisms and Clinical Uses of Capsaicin. Eur. J. Pharmacol. 2013, 720, 55–62. [Google Scholar] [CrossRef]
- Dalbeth, N.; Lauterio, T.J.; Wolfe, H.R. Mechanism of Action of Colchicine in the Treatment of Gout. Clin. Ther. 2014, 36, 1465–1479. [Google Scholar] [CrossRef]
- Tuli, H.S.; Tuorkey, M.J.; Thakral, F.; Sak, K.; Kumar, M.; Sharma, A.K.; Sharma, U.; Jain, A.; Aggarwal, V.; Bishayee, A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front. Pharmacol. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Keshmiri-Neghab, H.; Goliaei, B. Therapeutic Potential of Gossypol: An Overview. Pharm. Biol. 2014, 52, 124–128. [Google Scholar] [CrossRef]
- Ferraz da Costa, D.C.; Rangel, L.P.; Duarte da Cunha Martins-Dinis, M.M.; da Silva Ferretti, G.D.; Ferreira, V.F.; Silva, J.L. Anticancer Potential of Resveratrol, β-Lapachone and Their Analogues. Molecules 2020, 25, 893. [Google Scholar] [CrossRef]
- Ardalani, H.; Avan, A.; Ghayour-Mobarhan, M. Podophyllotoxin: A Novel Potential Natural Anticancer Agent. Avicenna J. Phytomed. 2017, 7, 285–294. [Google Scholar]
- Stoll, D.R.; Carr, P.W. Two-Dimensional Liquid Chromatography: A State of the Art Tutorial. Anal. Chem. 2017, 89, 519–531. [Google Scholar] [CrossRef]
- Boyce, M.C.; Lawler, N.G.; Tu, Y.; Reinke, S.N.; Lawler, N.G.; Tu, Y.; Reinke, S.N. Introducing Undergraduate Students to Metabolomics Using Liquid Chromatography–High Resolution Mass Spectrometry Analysis of Horse Blood. J. Chem. Educ. 2019, 96, 745–750. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Lin, L.G.; Ye, W.C. Techniques for Extraction and Isolation of Natural Products: A Comprehensive Review. Chin. Med. 2018, 13, 20. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Litaudon, M.; Touboul, D.; Queiroz, E.F. Innovative Omics-Based Approaches for Prioritisation and Targeted Isolation of Natural Products-New Strategies for Drug Discovery. Nat. Prod. Rep. 2019, 36, 855–868. [Google Scholar] [CrossRef]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef]
- van der Hooft, J.J.J.; Mihaleva, V.; De Vos, R.C.H.; Bino, R.J.; Vervoort, J. A Strategy for Fast Structural Elucidation of Metabolites in Small Volume Plant Extracts Using Automated MS-Guided LC-MS-SPE-NMR. Magn. Reson. Chem. 2011, 49, S55–S60. [Google Scholar] [CrossRef]
- Guenard, D.; Guiritte-Voegelein, F.; Potier, P. Taxol and Taxotere: Discovery, chemistry, and structure-activity relationships. Acc. Chem. Res. 1993, 26, 160–167. [Google Scholar] [CrossRef]
- Choudhari, A.S.; Mandave, P.C.; Deshpande, M.; Ranjekar, P.; Prakash, O. Phytochemicals in Cancer Treatment: From Preclinical Studies to Clinical Practice. Front. Pharmacol. 2020, 10, 1614. [Google Scholar] [CrossRef]
- Dias, D.A.; Urban, S.; Roessner, U. A Historical Overview of Natural Products in Drug Discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef]
- Roopashree, K.M.; Naik, D. Advanced Method of Secondary Metabolite Extraction and Quality Analysis. J. Pharmacogn. Phytochem. 2019, 8, 1829–1842. [Google Scholar]
- Hegazi, N.M.; Radwan, R.A.; Bakry, S.M.; Saad, H.H. Molecular Networking Aided Metabolomic Profiling of Beet Leaves Using Three Extraction Solvents and in Relation to Its Anti-Obesity Effects. J. Adv. Res. 2020, 24, 545–555. [Google Scholar] [CrossRef]
- Panzella, L.; Moccia, F.; Nasti, R.; Marzorati, S.; Verotta, L.; Napolitano, A. Bioactive Phenolic Compounds From Agri-Food Wastes: An Update on Green and Sustainable Extraction Methodologies. Front. Nutr. 2020, 7, 60. [Google Scholar] [CrossRef]
- Kurita, K.L.; Glassey, E.; Linington, R.G. Integration of High-Content Screening and Untargeted Metabolomics for Comprehensive Functional Annotation of Natural Product Libraries. Proc. Natl. Acad. Sci. USA 2015, 112, 11999–12004. [Google Scholar] [CrossRef]
- Jang, M.H.; Piao, X.L.; Kim, J.M.; Kwon, S.W.; Park, J.H. Inhibition of Cholinesterase and Amyloid-&bgr; Aggregation by Resveratrol Oligomers from Vitis Amurensis. Phytother. Res. 2008, 22, 544–549. [Google Scholar]
- Barnes, S.; Benton, H.P.; Casazza, K.; Cooper, S.J.; Cui, X.; Du, X.; Engler, J.; Kabarowski, J.H.; Li, S.; Pathmasiri, W.; et al. Special Feature: Tutorial Training in Metabolomics Research. I. Designing the Experiment, Collecting and Extracting Samples and Generating Metabolomics Data. J. Mass Spectrom. 2016, 51, 461–475. [Google Scholar] [CrossRef]
- Choudhury, S.; Sharma, P.; Moulick, D.; Mazumder, M.K. Unrevealing Metabolomics for Abiotic Stress Adaptation and Tolerance in Plants. J. Crop Sci. Biotechnol. 2021, 24, 479–493. [Google Scholar] [CrossRef]
- Weller, M.G. A Unifying Review of Bioassay-Guided Fractionation, Effect-Directed Analysis and Related Techniques. Sensors 2012, 12, 9181–9209. [Google Scholar] [CrossRef]
- Louwen, J.J.R.; Medema, M.H.; Van Der Hooft, J.J.J. Enhanced Correlation-Based Linking of Biosynthetic Gene Clusters to Their Metabolic Products through Chemical Class Matching. Res. Sq. 2022, 11, 13. [Google Scholar] [CrossRef]
- Tinte, M.M.; Chele, K.H.; van der Hooft, J.J.J.; Tugizimana, F. Metabolomics-Guided Elucidation of Plant Abiotic Stress Responses in the 4IR Era: An Overview. Metabolites 2021, 11, 445. [Google Scholar] [CrossRef]
- Tinte, M.M.; Masike, K.; Steenkamp, P.A.; Huyser, J.; van der Hooft, J.J.J.; Tugizimana, F. Computational Metabolomics Tools Reveal Metabolic Reconfigurations Underlying the Effects of Biostimulant Seaweed Extracts on Maize Plants under Drought Stress Conditions. Metabolites 2022, 12, 487. [Google Scholar] [CrossRef]
- Ebbels, T.M.D.; van der Hooft, J.J.J.; Chatelaine, H.; Broeckling, C.; Zamboni, N.; Hassoun, S.; Mathé, E.A. Recent Advances in Mass Spectrometry-Based Computational Metabolomics. Curr. Opin. Chem. Biol. 2023, 74, 102288. [Google Scholar] [CrossRef]
- Gaquerel, E.; Kuhl, C.; Neumann, S. Computational Annotation of Plant Metabolomics Profiles via a Novel Network-Assisted Approach. Metabolomics 2013, 9, 904–918. [Google Scholar] [CrossRef]
- Wainwright, C.L.; Teixeira, M.M.; Adelson, D.L.; Buenz, E.J.; David, B.; Glaser, K.B.; Harata-Lee, Y.; Howes, M.J.R.; Izzo, A.A.; Maffia, P.; et al. Future Directions for the Discovery of Natural Product-Derived Immunomodulating Drugs: An IUPHAR Positional Review. Pharmacol. Res. 2022, 177, 106076. [Google Scholar] [CrossRef]
- Volkova, S.; Matos, M.R.A.; Mattanovich, M.; de Mas, I.M. Metabolic Modelling as a Framework for Metabolomics Data Integration and Analysis. Metabolites 2020, 10, 303. [Google Scholar] [CrossRef]
- Alexandrov, T. Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annu. Rev. Biomed. Data Sci. 2020, 3, 61–87. [Google Scholar] [CrossRef]
- Rubtsov, D.V.; Jenkins, H.; Ludwig, C.; Easton, J.; Viant, M.R.; Günther, U.; Griffin, J.L.; Hardy, N. Proposed Reporting Requirements for the Description of NMR-Based Metabolomics Experiments. Metabolomics 2007, 3, 223–229. [Google Scholar] [CrossRef]
- Perez de Souza, L.; Alseekh, S.; Naake, T.; Fernie, A. Mass Spectrometry-Based Untargeted Plant Metabolomics. Curr. Protoc. Plant Biol. 2019, 4, e20100. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances and Future Approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. ScienceDirect Integrated Metabolomics for Abiotic Stress Responses in Plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef]
- Kell, D.B. Metabolomics, Machine Learning and Modelling: Towards an Understanding of the Language of Cells. Biochem. Soc. Trans. 2005, 33, 520–524. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [CrossRef]
- Fürtauer, L.; Pschenitschnigg, A.; Scharkosi, H.; Weckwerth, W.; Nägele, T. Combined Multivariate Analysis and Machine Learning Reveals a Predictive Module of Metabolic Stress Response in Arabidopsis Thaliana. Mol. Omics 2018, 14, 437–449. [Google Scholar] [CrossRef]
- Da Silva, R.R.; Dorrestein, P.C.; Quinn, R.A. Illuminating the Dark Matter in Metabolomics. Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Seven Golden Rules for Heuristic Filtering of Molecular Formulas Obtained by Accurate Mass Spectrometry. BMC Bioinform. 2007, 8, 105. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Pfeuffer, J.; Bielow, C.; Wein, S.; Jeong, K.; Netz, E.; Walter, A.; Alka, O.; Nilse, L.; Colaianni, P.D.; McCloskey, D.; et al. OpenMS 3 Enables Reproducible Analysis of Large-Scale Mass Spectrometry Data. Nat. Methods 2024, 21, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Mildau, K.; van der Hooft, J.J.J.; Flasch, M.; Warth, B.; Abiead, Y.E.; Koellensperger, G.; Zanghellini, J.; Büschl, C. Homologue Series Detection and Management in LC-MS Data with HomologueDiscoverer. bioRxiv 2022, 30, 2022.07.20.500749. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Carlin, D.E.; Demchak, B.; Pratt, D.; Sage, E.; Ideker, T. Network Propagation in the Cytoscape Cyberinfrastructure. PLoS Comput. Biol. 2017, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models. Genome Res. 1971, 13, 426. [Google Scholar] [CrossRef]
- van Der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic Modeling for Untargeted Substructure Exploration in Metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef]
- Van Der Hooft, J.J.J.; Wandy, J.; Young, F.; Padmanabhan, S.; Gerasimidis, K.; Burgess, K.E.V.; Barrett, M.P.; Rogers, S. Unsupervised Discovery and Comparison of Structural Families Across Multiple Samples in Untargeted Metabolomics. Anal. Chem. 2017, 89, 7569–7577. [Google Scholar] [CrossRef]
- Wandy, J.; Zhu, Y.; Van Der Hooft, J.J.J.; Daly, R.; Barrett, M.P.; Rogers, S. Ms2lda.Org: Web-Based Topic Modelling for Substructure Discovery in Mass Spectrometry. Bioinformatics 2018, 34, 317–318. [Google Scholar] [CrossRef]
- Nephali, L.; Steenkamp, P.; Burgess, K.; Huyser, J.; Brand, M.; van der Hooft, J.J.J.; Tugizimana, F. Mass Spectral Molecular Networking to Profile the Metabolome of Biostimulant Bacillus Strains. Front. Plant Sci. 2022, 13, 920963. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.F.; Wandy, J.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; van der Hooft, J.J.J. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Huber, F.; Ridder, L.; Verhoeven, S.; Spaaks, J.H.; Diblen, F.; Rogers, S.; Van Der Hooft, J.J.J. Spec2Vec: Improved Mass Spectral Similarity Scoring through Learning of Structural Relationships. PLoS Comput. Biol. 2021, 17, 1008724. [Google Scholar] [CrossRef] [PubMed]
- Huber, F.; van der Burg, S.; van der Hooft, J.J.J.; Ridder, L. MS2DeepScore: A Novel Deep Learning Similarity Measure to Compare Tandem Mass Spectra. J. Cheminform. 2021, 13, 84. [Google Scholar] [CrossRef]
- da Silva, R.R.; Wang, M.; Nothias, L.F.; van der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating Annotations of Molecular Networks Using in Silico Fragmentation. PLoS Comput. Biol. 2018, 14, 1006089. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of Microbial Metabolites through Database Search of Mass Spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef]
- Cao, L.; Guler, M.; Tagirdzhanov, A.; Lee, Y.Y.; Gurevich, A.; Mohimani, H. MolDiscovery: Learning Mass Spectrometry Fragmentation of Small Molecules. Nat. Commun. 2021, 12, 3718. [Google Scholar] [CrossRef] [PubMed]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform. 2016, 8, 1–20. [Google Scholar] [CrossRef]
- Nothias, L.F.; Nothias-Esposito, M.; Da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef]
- Mildau, K.; Büschl, C.; Zanghellini, J.; Van Der Hooft, J.J. Combined LC-MS/MS Feature Grouping, Statistical Prioritization, and Interactive Networking in MsFeaST. Bioinformatics 2024, 40, btae584. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Maldonado, L.M.B.; Augustijn, H.E.; Soldatou, S.; de Jonge, N.; Jaspars, M.; van Wezel, G.P.; Medema, M.H.; Hooft, J.J.J. van der FERMO: A Dashboard for Streamlined Rationalized Prioritization of Molecular Features from Mass Spectrometry Data. bioRxiv 2022. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid Identification, Annotation and Analysis of Secondary Metabolite Biosynthesis Gene Clusters in Bacterial and Fungal Genome Sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Suarez Duran, H.G.; Blin, K.; Osbourn, A.; Medema, M.H. PlantiSMASH: Automated Identification, Annotation and Expression Analysis of Plant Biosynthetic Gene Clusters. Nucleic Acids Res. 2017, 45, W55–W63. [Google Scholar] [CrossRef] [PubMed]
- Eldjarn, G.H.; Ramsay, A.; Van Der Hooft, J.J.J.; Duncan, K.R.; Soldatou, S.; Rousu, J.; Daly, R.; Wandy, J.; Rogers, S. Ranking Microbial Metabolomic and Genomic Links in the NPLinker Framework Using Complementary Scoring Functions. PLoS Comput. Biol. 2021, 17, 1008920. [Google Scholar] [CrossRef]
- Schorn, M.A.; Verhoeven, S.; Ridder, L.; Huber, F.; Acharya, D.D.; Aksenov, A.A.; Aleti, G.; Moghaddam, J.A.; Aron, A.T.; Aziz, S.; et al. A Community Resource for Paired Genomic and Metabolomic Data Mining. Nat. Chem. Biol. 2021, 17, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T.; et al. RIKEN Tandem Mass Spectral Database (ReSpect) for Phytochemicals: A Plant-Specific MS/MS-Based Data Resource and Database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef]
- Wang, K.; Deng, J.; Damaris, R.N.; Yang, M.; Xu, L.; Yang, P. LOTUS-DB: An Integrative and Interactive Database for Nelumbo Nucifera Study. Database 2015, 2015, bav023. [Google Scholar] [CrossRef]
- Duan, F.L.; Duan, C.B.; Xu, H.L.; Zhao, X.Y.; Sukhbaatar, O.; Gao, J.; Zhang, M.Z.; Zhang, W.H.; Gu, Y.C. AI-Driven Drug Discovery from Natural Products. Adv. Agrochem. 2024, 3, 185–187. [Google Scholar] [CrossRef]
- Bayona, L.M.; de Voogd, N.J.; Choi, Y.H. Metabolomics on the Study of Marine Organisms. Metabolomics 2022, 18, 17. [Google Scholar] [CrossRef]
- Ishii, C.; Nakanishi, Y.; Murakami, S.; Nozu, R.; Ueno, M.; Hioki, K.; Aw, W.; Hirayama, A.; Soga, T.; Ito, M.; et al. A Metabologenomic Approach Reveals Changes in the Intestinal Environment of Mice Fed on American Diet. Int. J. Mol. Sci. 2018, 19, 4079. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A Public Repository for Sharing Mass Spectral Data for Life Sciences. J. Mass. Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Elapavalore, A.; Kondic’, T.; Singh, R.R.; Shoemaker, A.B.; Thiessen, A.P.; Zhang, J.; Bolton, E.E.; Schymanski, L.E. Adding Open Spectral Data to MassBank and PubChem Using Open Source Tools to Support Nontargeted Exposomics of Mixtures. Environ. Sci. Process. Impacts 2023, 25, 1788–1801. [Google Scholar] [CrossRef]
- de Jonge, N.F.; Louwen, J.J.R.; Chekmeneva, E.; Camuzeaux, S.; Vermeir, F.J.; Jansen, R.S.; Huber, F.; van der Hooft, J.J.J. MS2Query: Reliable and Scalable MS2 Mass Spectra-Based Analogue Search. Nat. Commun. 2023, 14, 1752. [Google Scholar] [CrossRef]
- Russo, F.F.; Nowatzky, Y.; Jaeger, C.; Parr, K.M.; Benner, P.; Muth, T.; Lisec, J. Machine Learning Methods for Compound Annotation in Non-targeted Mass Spectrometry—A Brief Overview of Fingerprinting, in silico Fragmentation and de novo Methods. Rapid Commun. Mass Spectrom. 2024, 38, E9876. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Prihoda, D.; Palicka, A.; Soukup, J.; Klempir, O.; Rampula, L.; Durcak, J.; Wurst, M.; Kotowski, J.; Chang, D.; et al. A Deep Learning Genome-Mining Strategy for Biosynthetic Gene Cluster Prediction. Nucleic Acids Res. 2019, 47, E110. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-Omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 7–9. [Google Scholar] [CrossRef]
- Sirangelo, T.M. Multi-Omics Approaches in the Study of Plants. Int. J. Adv. Res. Bot. 2019, 5, 1–7. [Google Scholar] [CrossRef]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins dos Santos, V.A.P.; Saccenti, E. From Correlation to Causation: Analysis of Metabolomics Data Using Systems Biology Approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef]
- Filippou, P.; Zarza, X.; Antoniou, C.; Obata, T.; Villarroel, C.A.; Ganopoulos, I.; Harokopos, V.; Gohari, G.; Aidinis, V.; Madesis, P.; et al. Systems Biology Reveals Key Tissue-Specific Metabolic and Transcriptional Signatures Involved in the Response of Medicago Truncatula Plant Genotypes to Salt Stress. Comput. Struct. Biotechnol. J. 2021, 19, 2133–2147. [Google Scholar] [CrossRef]
- Majumder, E.L.W.; Billings, E.M.; Benton, H.P.; Martin, R.L.; Palermo, A.; Guijas, C.; Rinschen, M.M.; Domingo-Almenara, X.; Montenegro-Burke, J.R.; Tagtow, B.A.; et al. Cognitive Analysis of Metabolomics Data for Systems Biology. Nat. Protoc. 2021, 16, 1376–1418. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Kumar, N.; Mukhtar, M.S. Systems Biology and Machine Learning in Plant–Pathogen Interactions. Mol. Plant-Microbe Interact. 2019, 32, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Wolters, F.C.; Del Pup, E.; Singh, K.S.; Bouwmeester, K.; Schranz, M.E.; van der Hooft, J.J.J.; Medema, M.H. Pairing Omics to Decode the Diversity of Plant Specialized Metabolism. Curr. Opin. Plant. Biol. 2024, 82, 102657. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Yamazaki, M.; Saito, K. A New Era in Plant Functional Genomics. Curr. Opin. Syst. Biol. 2019, 15, 58–67. [Google Scholar] [CrossRef]
- Balyan, C.S.; Mutum, D.R.; Kansal, S.; Kumar, S.; Mathur, S.; Raghuvanshi, S. Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Pandey, G.K., Ed.; Springer: New York, NY, USA, 2015; Volume 2, Chapter 1; pp. 45–91. [Google Scholar] [CrossRef]
- Abdin, M.Z.; Khan, M.A.; Ali, A.; Alam, P.; Ahmad, A.; Sarwat, M. Stress Signaling in Plants: Genomics and Proteomics Perspective; Sarwat, M., Ahmad, A., Abdin, M.Z., Eds.; Springer: New York, NY, USA, 2013; Volume 1, Chapter 4; pp. 69–90. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Bayram, E.; Lukić Bilela, L.; Cueto, M.; Díaz-Marrero, A.R.; Haznedaroglu, B.Z.; Jimenez, C.; Mandalakis, M.; Pereira, F.; Reyes, F.; et al. Advanced Methods for Natural Products Discovery: Bioactivity Screening, Dereplication, Metabolomics Profiling, Genomic Sequencing, Databases and Informatic Tools, and Structure Elucidation. Mar. Drugs 2023, 21, 308. [Google Scholar] [CrossRef]
- Singh, K.S.; van der Hooft, J.J.J.; van Wees, S.C.M.; Medema, M.H. Integrative Omics Approaches for Biosynthetic Pathway Discovery in Plants. Nat. Prod. Rep. 2022, 39, 1876–1896. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Benton, H.P.; Siuzdak, G. Bioinformatics: The next Frontier of Metabolomics. Anal. Chem. 2015, 87, 147–156. [Google Scholar] [CrossRef]
- Johnston, C.W.; Skinnider, M.A.; Wyatt, M.A.; Li, X.; Ranieri, M.R.M.; Yang, L.; Zechel, D.L.; Ma, B.; Magarvey, N.A. An Automated Genomes-to-Natural Products Platform (GNP) for the Discovery of Modular Natural Products. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Cao, J.; Li, C.; Cui, Z.; Deng, S.; Lei, T.; Liu, W.; Yang, H.; Chen, P. Spatial Transcriptomics: A Powerful Tool in Disease Understanding and Drug Discovery. Theranostics 2024, 14, 2946–2968. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Li, Z.; Dai, S.; Ding, H.; Wang, Q.; Li, X.; Ding, G.; Wang, P.; Guan, Y.; Liu, W. Integrative Analyses of Transcriptomics and Metabolomics upon Seed Germination of Foxtail Millet in Response to Salinity. Sci. Rep. 2020, 10, 13660. [Google Scholar] [CrossRef]
- Singh, K.S.; Suarez Duran, H.; Del Pup, E.; Zafra-Delgado, O.; Van Wees, S.C.M.; van der Hooft, J.J.J.; Medema, M.H. MEANtools: Multi-Omics Integration towards Metabolite Anticipation and Biosynthetic Pathway Prediction. Biorxiv 2024. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant Abiotic Stress Proteomics: The Major Factors Determining Alterations in Cellular Proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef]
- Lin, B.B.; Scientific, T.C. Proteomics Potential and Its Contribution Toward Sustainable Agriculture. Agroecology, Ecosystems, and Sustainability; Benkeblia, N., Ed.; CRC Press: Boca Raton, FL, USA, 2014; Volume 1, Chapter 8; p. 170. [Google Scholar] [CrossRef]
- Pedde, R.D.; Li, H.; Borchers, C.H.; Akbari, M. Microfluidic-Mass Spectrometry Interfaces for Translational Proteomics. Trends Biotechnol. 2017, 35, 954–970. [Google Scholar] [CrossRef] [PubMed]
- Bumpus, S.B.; Evans, B.S.; Thomas, P.M.; Ntai, I.; Kelleher, N.L. A Proteomics Approach to Discovering Natural Products and Their Biosynthetic Pathways. Nat. Biotechnol. 2009, 27, 951–956. [Google Scholar] [CrossRef]
- Evans, B.S.; Ntai, I.; Chen, Y.; Robinson, S.J.; Kelleher, N.L. Proteomics-Based Discovery of Koranimine, a Cyclic Imine Natural Product. J. Am. Chem. Soc. 2011, 133, 7316–7319. [Google Scholar] [CrossRef]
- Winter, J.M.; Behnken, S.; Hertweck, C. Genomics-Inspired Discovery of Natural Products. Curr. Opin. Chem. Biol. 2011, 15, 22–31. [Google Scholar] [CrossRef]
- Chawla, K.; Barah, P.; Kuiper, M.; Bones, A.M. Systems Biology: A Promising Tool to Study Abiotic Stress Responses. Omics Plant Abiotic Stress Toler. 2011, 1, 163–172. [Google Scholar] [CrossRef]
- Sun, G.; Yang, Y.; Xie, F.; Wen, J.F.; Wu, J.; Wilson, I.W.; Tang, Q.; Liu, H.; Qiu, D. Deep Sequencing Reveals Transcriptome Re-Programming of Taxus × Media Cells to the Elicitation with Methyl Jasmonate. PLoS ONE 2013, 8, e62865. [Google Scholar] [CrossRef]
- Van Der Hooft, J.J.J.; Mohimani, H.; Bauermeister, A.; Dorrestein, P.C.; Duncan, K.R.; Medema, M.H. Linking Genomics and Metabolomics to Chart Specialized Metabolic Diversity. Chem. Soc. Rev. 2020, 49, 3297–3314. [Google Scholar] [CrossRef] [PubMed]
- Leão, T.; Wang, M.; Moss, N.; da Silva, R.; Sanders, J.; Nurk, S.; Gurevich, A.; Humphrey, G.; Reher, R.; Zhu, Q.; et al. A Multi-Omics Characterization of the Natural Product Potential of Tropical Filamentous Marine Cyanobacteria. Mar. Drugs 2021, 19, 20. [Google Scholar] [CrossRef]
- Qiao, X.; Houghton, A.; Reed, J.; Steuernagel, B.; Zhang, J.; Owen, C.; Leveau, A.; Orme, A.; Louveau, T.; Melton, R.; et al. Comprehensive mutant chemotyping reveals embedding of a lineage-specific biosynthetic gene cluster in wider plant metabolism. Proc. Natl. Acad. Sci. USA 2025, 122, e2417588122. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Choi, B.S.; Lee, S.C.; Kim, N.H.; Park, J.Y.; Jang, W.; Lakshmanan, M.; Mohan, S.V.G.; Lee, D.Y.; Yang, T.J. Ginseng Genome Database: An Open-Access Platform for Genomics of Panax Ginseng. BMC Plant Biol. 2018, 18, 62. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; He, S.; Gao, Y.; Ma, X.; Gao, Y.; Zhang, G.; Kui, L.; Wang, W.; Wang, Y.; et al. HMOD: An Omics Database for Herbal Medicine Plants. Mol. Plant 2018, 11, 757–759. [Google Scholar] [CrossRef]
- He, S.; Yang, L.; Ye, S.; Lin, Y.; Li, X.; Wang, Y.; Chen, G.; Liu, G.; Zhao, M.; Zhao, X.; et al. MPOD: Applications of Integrated Multi-Omics Database for Medicinal Plants. Plant Biotechnol. J. 2022, 20, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.; Hu, H.; Xiao, S.; Zhou, G.; Sun, W.; Chu, Y.; Meng, X.; Wei, J.; Zhang, H.; Xu, J.; et al. Global Pharmacopoeia Genome Database Is an Integrated and Mineable Genomic Database for Traditional Medicines Derived from Eight International Pharmacopoeias. Sci. China Life Sci. 2022, 65, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.R.; Liao, B.S.; Li, Q.S.; Xu, J.; Chen, S.L. Establishing a Genomic Database for the Medicinal Plants in the Brazilian Pharmacopoeia. Chin. Med. 2021, 16, 71. [Google Scholar] [CrossRef]
- Akiyama, M. Multi-Omics Study for Interpretation of Genome-Wide Association Study. J. Hum. Genet. 2021, 66, 3–10. [Google Scholar] [CrossRef]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-Omics Data Integration Considerations and Study Design for Biological Systems and Disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef]
- Du, P.; Fan, R.; Zhang, N.; Wu, C.; Zhang, Y. Advances in Integrated Multi-Omics Analysis for Drug-Target Identification. Biomolecules 2024, 14, 692. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, Y.; Li, H. Deep Learning to Predict the Biosynthetic Gene Clusters in Bacterial Genomes. J. Mol. Biol. 2022, 434, 167597. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Scott-Boyer, M.P.; Bodein, A.; Périn, O.; Droit, A. Integration Strategies of Multi-Omics Data for Machine Learning Analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chele, K.H.; Piater, L.A.; van der Hooft, J.J.J.; Tugizimana, F. Bridging Ethnobotanical Knowledge and Multi-Omics Approaches for Plant-Derived Natural Product Discovery. Metabolites 2025, 15, 362. https://doi.org/10.3390/metabo15060362
Chele KH, Piater LA, van der Hooft JJJ, Tugizimana F. Bridging Ethnobotanical Knowledge and Multi-Omics Approaches for Plant-Derived Natural Product Discovery. Metabolites. 2025; 15(6):362. https://doi.org/10.3390/metabo15060362
Chicago/Turabian StyleChele, Kekeletso H., Lizelle A. Piater, Justin J. J. van der Hooft, and Fidele Tugizimana. 2025. "Bridging Ethnobotanical Knowledge and Multi-Omics Approaches for Plant-Derived Natural Product Discovery" Metabolites 15, no. 6: 362. https://doi.org/10.3390/metabo15060362
APA StyleChele, K. H., Piater, L. A., van der Hooft, J. J. J., & Tugizimana, F. (2025). Bridging Ethnobotanical Knowledge and Multi-Omics Approaches for Plant-Derived Natural Product Discovery. Metabolites, 15(6), 362. https://doi.org/10.3390/metabo15060362







