Plasma and Fecal Metabolites Combined with Gut Microbiome Reveal Systemic Metabolic Shifts in 60Co Gamma-Irradiated Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animals and Experimental Design
2.3. Sample Collection, Complete Blood Count, and Pretreatment
2.4. Untargeted Metabolomics Assay of Plasma Metabolites
2.5. DNA Sequencing and Gut Microbiota Analysis
2.6. Statistical Analyses
3. Results
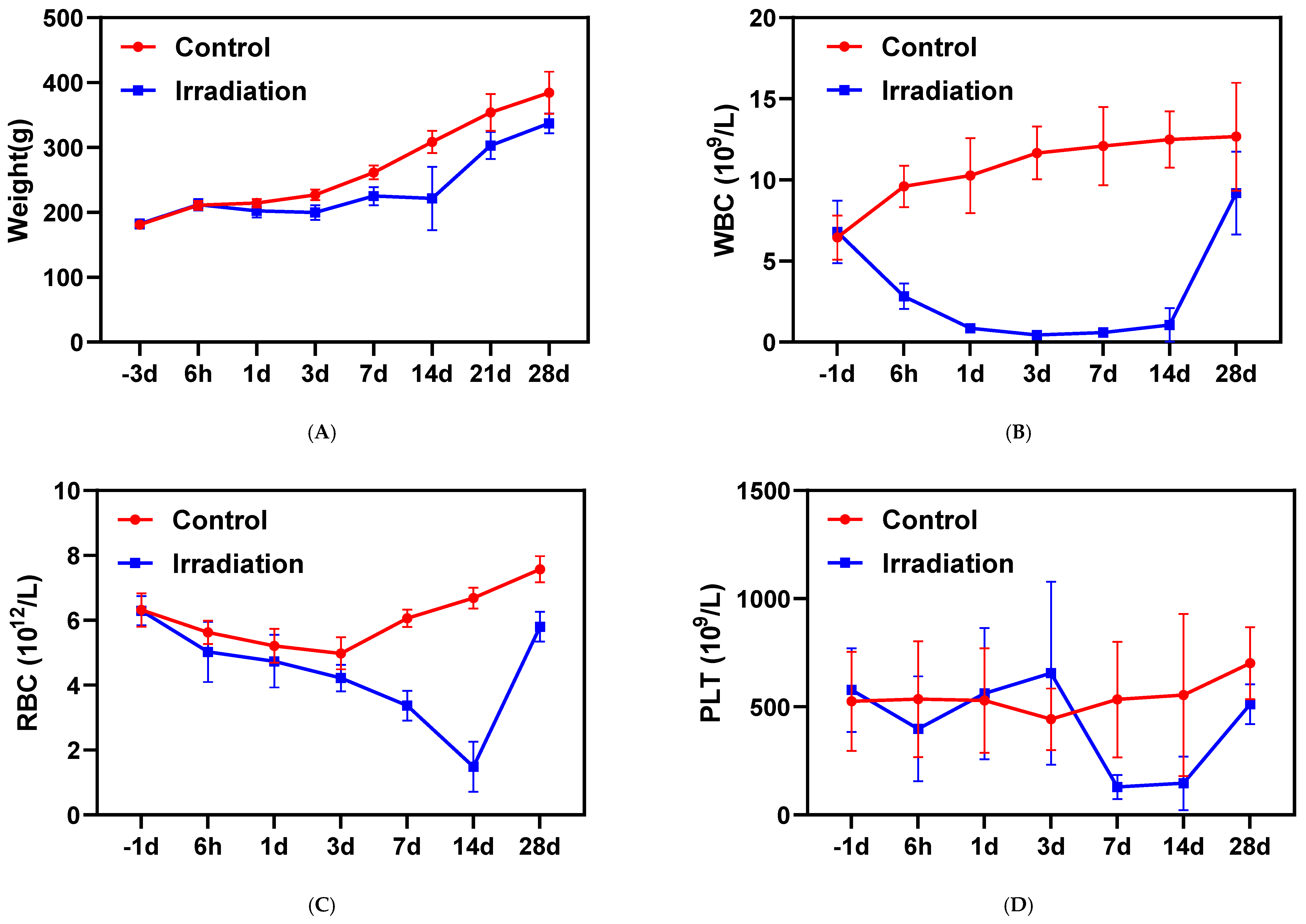
3.1. Body Weights and Complete Blood Counts
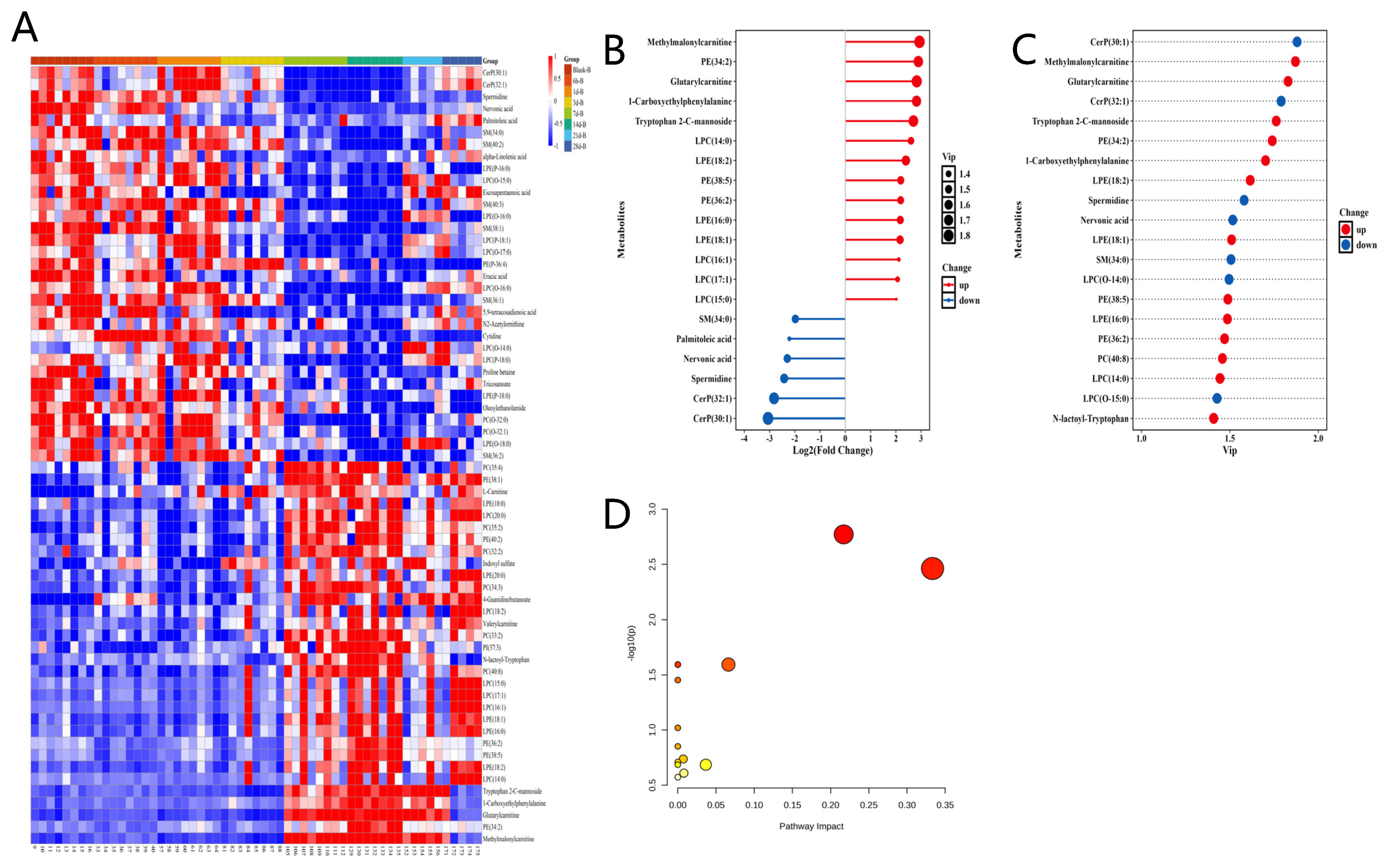
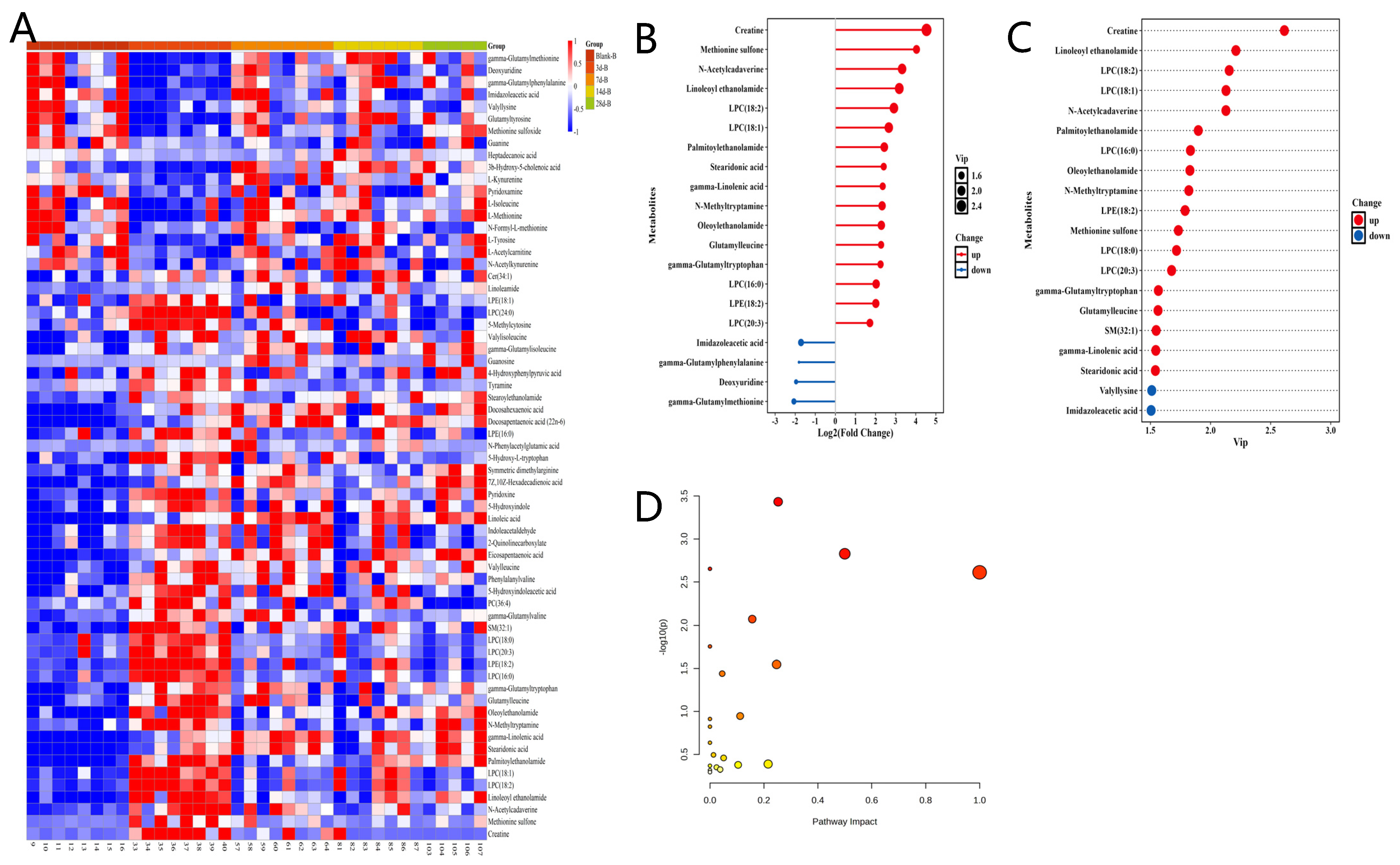
3.2. Temporal Hotspot of Radiation-Induced Metabolic Shifts
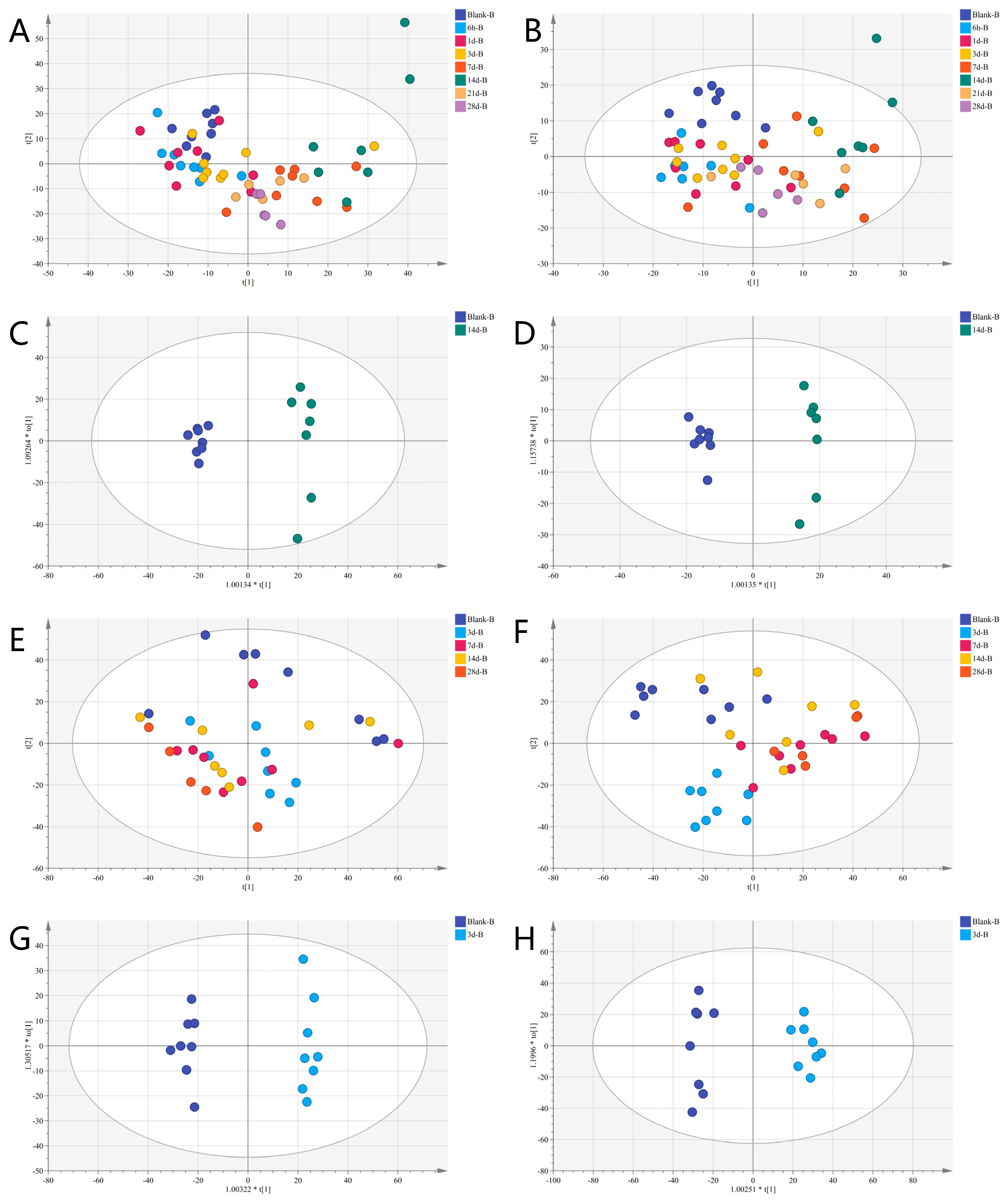
3.3. Metabolomics Analysis
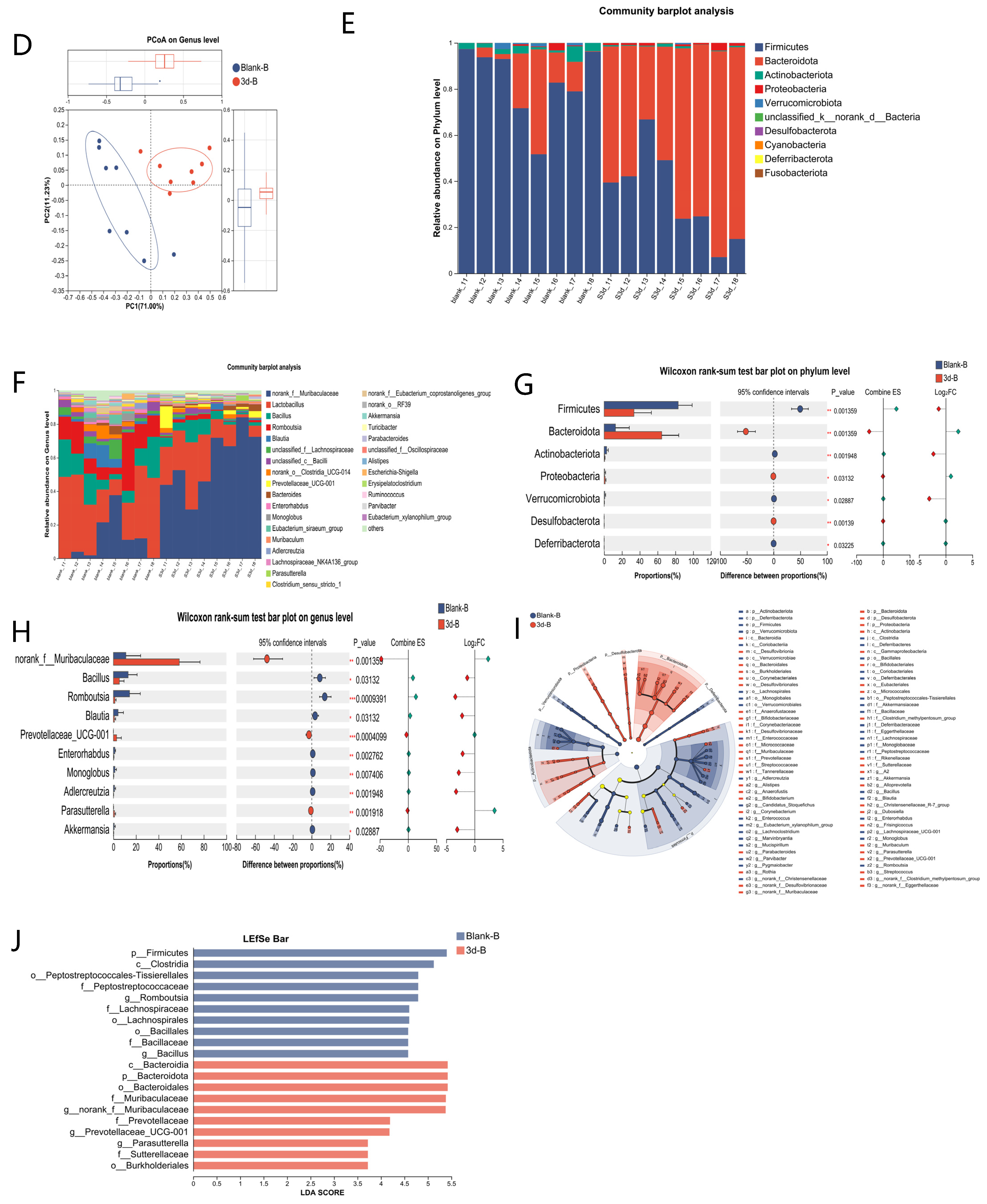
3.4. Microbiota Analysis
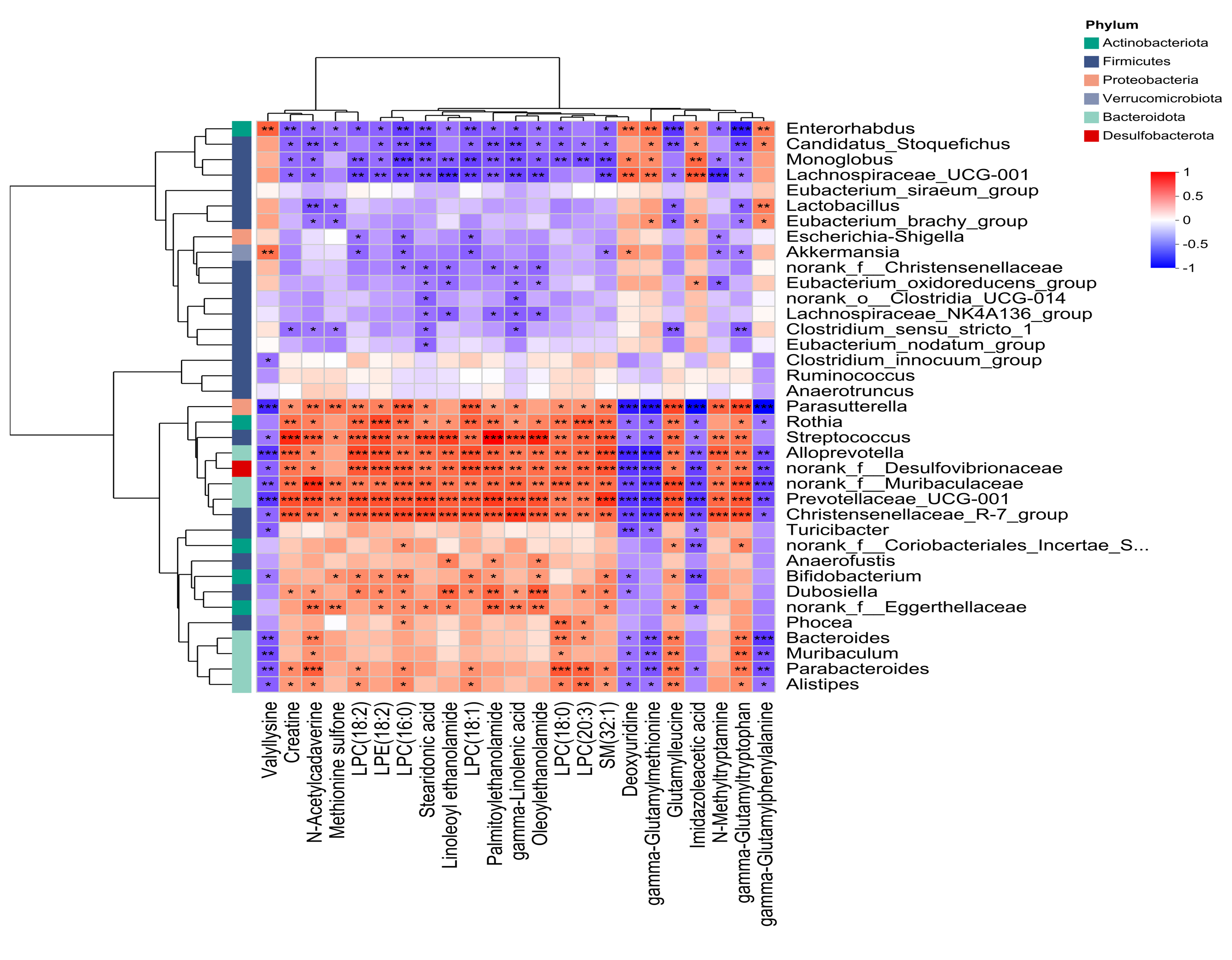
3.5. Combined Analysis of Microbiome and Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Qabandi, M.; Alshammary, J. Ionizing Radiation: Biologic Effects and Essential Cell Biology. In The Pathophysiologic Basis of Nuclear Medicine; Elgazzar, A.H., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 11–37. ISBN 978-3-030-96252-4. [Google Scholar]
- Dartnell, L.R. Ionizing Radiation and Life. Astrobiology 2011, 11, 551–582. [Google Scholar] [CrossRef] [PubMed]
- Dyk, J.V.; Battista, J.; Almond, P. A Retrospective of Cobalt-60 Radiation Therapy: “The Atom Bomb That Saves Lives”. Med. Phys. Int. J. 2020, 4, 327–350. [Google Scholar]
- Garau, M.M.I.; Calduch, A.L.; López, E.C. Radiobiology of the Acute Radiation Syndrome. Rep. Pract. Oncol. Radiother. 2011, 16, 123–130. [Google Scholar] [CrossRef]
- Plett, P.A.; Pelus, L.M.; Orschell, C.M. Establishing a Murine Model of the Hematopoietic Acute Radiation Syndrome. In Hematopoietic Stem Cells. Methods in Molecular Biology; Humana: New York, NY, USA, 2023; Volume 2567, pp. 251–262. [Google Scholar] [CrossRef]
- Leibowitz, B.J.; Wei, L.; Zhang, L.; Ping, X.; Epperly, M.; Greenberger, J.; Cheng, T.; Yu, J. Ionizing Irradiation Induces Acute Haematopoietic Syndrome and Gastrointestinal Syndrome Independently in Mice. Nat. Commun. 2014, 5, 3494. [Google Scholar] [CrossRef]
- Liu, X.; Locasale, J.W. Metabolomics: A Primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef]
- Church, D.L.; Cerutti, L.; Gürtler, A.; Griener, T.; Zelazny, A.; Emler, S. Performance and Application of 16s Rrna Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin. Microbiol. Rev. 2020, 33, 10.1128–cmr.00053. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Gill, K.; Jain, S.K.; Mehta, K.Y.; Nishita, D.; Bujold, K.; Bakke, J.; Gahagen, J.; Authier, S.; et al. Liquid Chromatography-Mass Spectrometry-Based Metabolomics of Nonhuman Primates after 4 Gy Total Body Radiation Exposure: Global Effects and Targeted Panels. J. Proteome Res. 2019, 18, 2260–2269. [Google Scholar] [CrossRef]
- Cheema, A.K.; Mehta, K.Y.; Rajagopal, M.U.; Wise, S.Y.; Fatanmi, O.O.; Singh, V.K. Metabolomic Studies of Tissue Injury in Nonhuman Primates Exposed to Gamma-Radiation. Int. J. Mol. Sci. 2019, 20, 3360. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Fornace, A.J.; Fatanmi, O.O.; Singh, V.K. A Metabolomic Serum Signature from Nonhuman Primates Treated with a Radiation Countermeasure, Gamma-Tocotrienol, and Exposed to Ionizing Radiation. Health Phys. 2018, 115, 3–11. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Pannkuk, E.L.; Diaz-Rubio, M.E.; Wang, Y.-W.; Mak, T.D.; Simbulan-Rosenthal, C.M.; Brenner, D.J.; Fornace, A.J. Implications of Genotypic Differences in the Generation of a Urinary Metabolomics Radiation Signature. Mutat. Res. Mol. Mech. Mutagen. 2016, 788, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Authier, S.; Wong, K.; Fornace, A.J., Jr. Gas Chromatography/Mass Spectrometry Metabolomics of Urine and Serum from Nonhuman Primates Exposed to Ionizing Radiation: Impacts on the Tricarboxylic Acid Cycle and Protein Metabolism. J. Proteome Res. 2017, 16, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, R.; Maan, K.; Khushu, S.; Rana, P. Urine Metabolomics Based Prediction Model Approach for Radiation Exposure. Sci. Rep. 2020, 10, 16063. [Google Scholar] [CrossRef]
- Jones, J.W.; Clifford, Z.; Li, F.; Tudor, G.L.; Farese, A.M.; Booth, C.; MacVittie, T.J.; Kane, M.A. Targeted Metabolomics Reveals Metabolomic Signatures Correlating Gastrointestinal Tissue to Plasma in a Mouse Total-Body Irradiation Model. Health Phys. 2019, 116, 473. [Google Scholar] [CrossRef]
- Wu, H.; Xu, C.; Gu, Y.; Yang, S.; Wang, Y.; Wang, C. An Improved Pseudotargeted GC-MS/MS-Based Metabolomics Method and Its Application in Radiation-Induced Hepatic Injury in a Rat Model. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020, 1152, 122250. [Google Scholar] [CrossRef]
- Xiao, X.; Hu, M.; Zhang, X.; Hu, J.Z. NMR-Based Metabolomics Analysis of Liver from C57BL/6 Mouse Exposed to Ionizing Radiation. Radiat. Res. 2017, 188, 44–55. [Google Scholar] [CrossRef]
- Goudarzi, M.; Weber, W.M.; Chung, J.; Doyle-Eisele, M.; Melo, D.R.; Mak, T.D.; Strawn, S.J.; Brenner, D.J.; Guilmette, R.; Fornace, A.J. Serum Dyslipidemia Is Induced by Internal Exposure to Strontium-90 in Mice, Lipidomic Profiling Using a Data-Independent Liquid Chromatography-Mass Spectrometry Approach. J. Proteome Res. 2015, 14, 4039–4049. [Google Scholar] [CrossRef]
- Maan, K.; Tyagi, R.; Dutta, A.; Bakhshi, R.; Rana, P. Comparative Metabolic Profiles of Total and Partial Body Radiation Exposure in Mice Using an Untargeted Metabolomics Approach. Metabolomics Off. J. Metabolomic Soc. 2020, 16, 124. [Google Scholar] [CrossRef]
- Guo, H.; Chou, W.-C.; Lai, Y.; Liang, K.; Tam, J.W.; Brickey, W.J.; Chen, L.; Montgomery, N.D.; Li, X.; Bohannon, L.M.; et al. Multi-Omics Analyses of Radiation Survivors Identify Radioprotective Microbes and Metabolites. Science 2020, 370, eaay9097. [Google Scholar] [CrossRef]
- Thandar, M.; Yang, X.; Zhu, Y.; Zhang, X.; Chen, Z.; Huang, S.; Chi, P. Dysbiosis of Gut Microbiota and Metabolites Is Associated with Radiation-Induced Colorectal Fibrosis and Is Restored by Adipose-Derived Mesenchymal Stem Cell Therapy. Life Sci. 2024, 341, 122502. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, J.; Park, S.-J. High-Throughput 16S rRNA Gene Sequencing Reveals Alterations of Mouse Intestinal Microbiota after Radiotherapy. Anaerobe 2015, 33, 1–7. [Google Scholar] [CrossRef]
- Moraitis, I.; Guiu, J.; Rubert, J. Gut Microbiota Controlling Radiation-Induced Enteritis and Intestinal Regeneration. Trends Endocrinol. Metab. 2023, 34, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Wei, K.; He, J.; Ding, N.; Hua, J.; Zhou, T.; Niu, F.; Zhou, G.; Shi, T.; et al. Review: Effect of Gut Microbiota and Its Metabolite SCFAs on Radiation-Induced Intestinal Injury. Front. Cell. Infect. Microbiol. 2021, 11, 577236. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One Fastq Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. Flash: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using Qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat. Soc 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Singh, V.K.; Newman, V.L.; Seed, T.M. Colony-Stimulating Factors for the Treatment of the Hematopoietic Component of the Acute Radiation Syndrome (h-Ars): A Review. Cytokine 2015, 71, 22–37. [Google Scholar] [CrossRef]
- Dainiak, N. Hematologic Consequences of Exposure to Ionizing Radiation. Exp. Hematol. 2002, 30, 513–528. [Google Scholar] [CrossRef]
- Azzam, E.I.; Jay-Gerin, J.-P.; Pain, D. Ionizing Radiation-Induced Metabolic Oxidative Stress and Prolonged Cell Injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Averbeck, D.; Rodriguez-Lafrasse, C. Role of Mitochondria in Radiation Responses: Epigenetic, Metabolic, and Signaling Impacts. Int. J. Mol. Sci. 2021, 22, 11047. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, A.; Nasreen, A.; Lenchyk, L.; Lysiuk, R.; Peana, M.; Shapovalova, N.; Piscopo, S.; Komisarenko, M.; Shanaida, M.; Smetanina, K.; et al. An Update on Glutathione’s Biosynthesis, Metabolism, Functions, and Medicinal Purposes. Curr. Med. Chem. 2024, 31, 4579–4601. [Google Scholar] [CrossRef]
- Xi, C.; Liu, H.-X.; Xiang, J.-Q.; Lu, X.; Cai, T.-J.; Li, S.; Gao, L.; Tian, X.-L.; Liu, K.-H.; Tian, M.; et al. Screening of Radiation Gastrointestinal Injury Biomarkers in Rat Plasma by High-Coverage Targeted Lipidomics. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2022, 27, 448–460. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Singh, V.K.; Fornace, A.J. Lipidomic Signatures of Nonhuman Primates with Radiation-Induced Hematopoietic Syndrome. Sci. Rep. 2017, 7, 9777. [Google Scholar] [CrossRef]
- Mishra, K.P. Cell Membrane Oxidative Damage Induced by Gamma-Radiation and Apoptotic Sensitivity. J. Environ. Pathol. Toxicol. Oncol. 2004, 23, 1–13. [Google Scholar] [CrossRef]
- Mathias, S.; Peña, L.A.; Kolesnick, R.N. Signal Transduction of Stress Via Ceramide. Biochem. J. 1998, 335, 465–480. [Google Scholar] [CrossRef]
- Phung, N.V.; Rong, F.; Xia, W.Y.; Fan, Y.; Li, X.Y.; Wang, S.A.; Li, F.L. Nervonic Acid and Its Sphingolipids: Biological Functions and Potential Food Applications. Crit. Rev. Food Sci. Nutr. 2024, 64, 8766–8785. [Google Scholar] [CrossRef]
- Carrozzo, R.; Dionisi-Vici, C.; Steuerwald, U.; Lucioli, S.; Deodato, F.; Di Giandomenico, S.; Bertini, E.; Franke, B.; Kluijtmans, L.A.J.; Meschini, M.C.; et al. Sucla2 Mutations Are Associated with Mild Methylmalonic Aciduria, Leigh-Like Encephalomyopathy, Dystonia and Deafness. Brain 2007, 130, 862–874. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Wang, J.; Wu, F.; Chen, Y.; Zhang, H.; Guo, Y.; Lin, Y.; Li, L.; Yu, X.; et al. Spermidine Alleviates Cardiac Aging by Improving Mitochondrial Biogenesis and Function. Aging 2020, 12, 650–671. [Google Scholar] [CrossRef]
- Guan, B.; Li, C.; Yang, Y.; Lu, Y.; Sun, Y.; Su, L.; Shi, G.; Bai, L.; Liu, J.; Meng, A. Effect of Spermidine on Radiation-Induced Long-Term Bone Marrow Cell Injury. Int. Immunopharmacol. 2023, 114, 109557. [Google Scholar] [CrossRef] [PubMed]
- Sol, J.; Obis, È.; Mota-Martorell, N.; Pradas, I.; Galo-Licona, J.D.; Martin-Garí, M.; Fernández-Bernal, A.; Ortega-Bravo, M.; Mayneris-Perxachs, J.; Borrás, C.; et al. Plasma Acylcarnitines and Gut-Derived Aromatic Amino Acids as Sex-Specific Hub Metabolites of the Human Aging Metabolome. Aging Cell 2023, 22, e13821. [Google Scholar] [CrossRef] [PubMed]
- Wyss, M.; Kaddurah-Daouk, R. Creatine and Creatinine Metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Levine, R.L. Free Radical-Mediated Oxidation of Free Amino Acids and Amino Acid Residues in Proteins. Amino Acids 2003, 25, 207–218. [Google Scholar] [CrossRef]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Lo Verme, J.; Fu, J.; Astarita, G.; La Rana, G.; Russo, R.; Calignano, A.; Piomelli, D. The Nuclear Receptor Peroxisome Proliferator-Activated Receptor-Alpha Mediates the Anti-Inflammatory Actions of Palmitoylethanolamide. Mol. Pharmacol. 2005, 67, 15–19. [Google Scholar] [CrossRef]
- Piomelli, D. The Molecular Logic of Endocannabinoid Signalling. Nat. Rev. Neurosci. 2003, 4, 873–884. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, B.; Zhang, X.; Akbar, M.T.; Wu, T.; Zhang, Y.; Zhi, L.; Shen, Q. Exploration of the Muribaculaceae Family in the Gut Microbiota: Diversity, Metabolism, and Function. Nutrients 2024, 16, 2660. [Google Scholar] [CrossRef]
- Molino, S.; Lerma-Aguilera, A.; Jiménez-Hernández, N.; Rufián Henares, J.Á.; Francino, M.P. Evaluation of the Effects of a Short Supplementation with Tannins on the Gut Microbiota of Healthy Subjects. Front. Microbiol. 2022, 13, 848611. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus Subtilis: Their Resistance to and Killing by Radiation, Heat and Chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Gao, H.; Li, X.; Chen, X.; Hai, D.; Wei, C.; Zhang, L.; Li, P. The Functional Roles of Lactobacillus Acidophilus in Different Physiological and Pathological Processes. J. Microbiol. Biotechnol. 2022, 32, 1226–1233. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, Z.; Zhao, H.; Wang, X.; Pang, J.; Li, Q.; Yang, Y.; Ling, W. Anthocyanin Supplementation Improves Anti-Oxidative and Anti-Inflammatory Capacity in a Dose-Response Manner in Subjects with Dyslipidemia. Redox Biol. 2020, 32, 101474. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Vitale, R.M. The Endocannabinoid System and PPARs: Focus on Their Signalling Crosstalk, Action and Transcriptional Regulation. Cells 2021, 10, 586. [Google Scholar] [CrossRef]
- Albenberg, L.G.; Wu, G.D. Diet and the Intestinal Microbiome: Associations, Functions, and Implications for Health and Disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and Their Metabolism in Physiology and Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism Pathways of Arachidonic Acids: Mechanisms and Potential Therapeutic Targets. Signal Transduct. Target. Ther. 2021, 6, 448–460. [Google Scholar] [CrossRef]
- Cui, W.; Hull, L.; Zizzo, A.; Wang, L.; Lin, B.; Zhai, M.; Xiao, M. The Gut Microbiome Changes in Wild Type and Il-18 Knockout Mice after 9.0 Gy Total Body Irradiation. Anim. Microbiome 2023, 5, 42. [Google Scholar] [CrossRef]
- Nikkheslat, N.; Zajkowska, Z.; Legido-Quigley, C.; Xu, J.; Manfro, P.H.; Souza, L.; Pereira, R.; Rohrsetzer, F.; Piccin, J.; Viduani, A.; et al. Sex-Specific Alterations of the Kynurenine Pathway in Association With Risk for and Remission of Depression in Adolescence. Biol. Psychiatry 2025, in press. [Google Scholar] [CrossRef]

| Pathway Name | −log(p) | Impact |
|---|---|---|
| Glycerophospholipid metabolism | 2.7719 | 0.21708 |
| Alpha-linolenic acid metabolism | 2.4622 | 0.33333 |
| Biosynthesis of unsaturated fatty acids | 1.5938 | 0 |
| Arginine and proline metabolism | 1.5938 | 0.06628 |
| Linoleic acid metabolism | 1.4539 | 0 |
| Arginine biosynthesis | 1.0194 | 0 |
| Beta-alanine metabolism | 0.85305 | 0 |
| Glutathione metabolism | 0.73783 | 0.00719 |
| Lysine degradation | 0.71063 | 0 |
| Sphingolipid metabolism | 0.68536 | 0 |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 0.68536 | 0.03665 |
| Pyrimidine metabolism | 0.60909 | 0.0079 |
| Arachidonic acid metabolism | 0.57217 | 0 |
| Pathway Name | −log(p) | Impact |
|---|---|---|
| Tryptophan metabolism | 3.4321 | 0.25291 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 2.8294 | 0.5 |
| Biosynthesis of unsaturated fatty acids | 2.653 | 0 |
| Linoleic acid metabolism | 2.6119 | 1 |
| Vitamin B6 metabolism | 2.0729 | 0.15686 |
| Alpha-linolenic acid metabolism | 1.7543 | 0 |
| Tyrosine metabolism | 1.5468 | 0.24711 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 1.4392 | 0.04545 |
| Glycerophospholipid metabolism | 0.94507 | 0.11201 |
| Valine, leucine, and isoleucine biosynthesis | 0.9112 | 0 |
| Phenylalanine metabolism | 0.82096 | 0 |
| Histidine metabolism | 0.63666 | 0 |
| Purine metabolism | 0.49398 | 0.01316 |
| One carbon pool by folate | 0.45824 | 0.05077 |
| Sphingolipid metabolism | 0.38715 | 0.21576 |
| Cysteine and methionine metabolism | 0.37694 | 0.10446 |
| Glycine, serine, and threonine metabolism | 0.36712 | 0 |
| Arginine and proline metabolism | 0.34857 | 0.02442 |
| Pyrimidine metabolism | 0.32315 | 0.03799 |
| Valine, leucine, and isoleucine degradation | 0.31526 | 0 |
| Arachidonic acid metabolism | 0.2931 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zong, J.; Wu, H.; Hu, X.; Yao, A.; Zhu, W.; Dou, G.; Liu, S.; Zhu, X.; Gu, R.; Sun, Y.; et al. Plasma and Fecal Metabolites Combined with Gut Microbiome Reveal Systemic Metabolic Shifts in 60Co Gamma-Irradiated Rats. Metabolites 2025, 15, 363. https://doi.org/10.3390/metabo15060363
Zong J, Wu H, Hu X, Yao A, Zhu W, Dou G, Liu S, Zhu X, Gu R, Sun Y, et al. Plasma and Fecal Metabolites Combined with Gut Microbiome Reveal Systemic Metabolic Shifts in 60Co Gamma-Irradiated Rats. Metabolites. 2025; 15(6):363. https://doi.org/10.3390/metabo15060363
Chicago/Turabian StyleZong, Jie, Haiyang Wu, Xuan Hu, Ami Yao, Wenhua Zhu, Guifang Dou, Shuchen Liu, Xiaoxia Zhu, Ruolan Gu, Yunbo Sun, and et al. 2025. "Plasma and Fecal Metabolites Combined with Gut Microbiome Reveal Systemic Metabolic Shifts in 60Co Gamma-Irradiated Rats" Metabolites 15, no. 6: 363. https://doi.org/10.3390/metabo15060363
APA StyleZong, J., Wu, H., Hu, X., Yao, A., Zhu, W., Dou, G., Liu, S., Zhu, X., Gu, R., Sun, Y., Wu, Z., Wang, S., & Gan, H. (2025). Plasma and Fecal Metabolites Combined with Gut Microbiome Reveal Systemic Metabolic Shifts in 60Co Gamma-Irradiated Rats. Metabolites, 15(6), 363. https://doi.org/10.3390/metabo15060363






