Simple Summary
A large population of bacteria, protozoa, fungi, and algae colonizes an animal’s body surface. The complex community of microbes that occupies a specific area of the organism and develops symbiotic relationships with the host is referred to as the microbiota. In this context, the gut microbiota plays an important physiological role as it influences the digestion and absorption of nutrients, the development and maturation of the immune system, and thus the growth, resistance to disease, and welfare of the animal. For these reasons, it is important to know the role of the microbiota in these specific functions, as well as which factors can impact the composition of bacterial populations, because changes in the microbiota can result in both beneficial and detrimental effects (dysbiosis) on the host. This study, in addition to providing information on the composition of the microbiota in the jejunum and cecum of rabbits, also evaluates the effect of breed and different growth rates using a modern methodological approach. Since the effect of these factors on the modulation of the gut microbiota has been little studied, this research could be the starting point for new experimental investigations aimed at enhancing rabbit productivity and welfare.
Abstract
This study aimed to evaluate the productive performance and microbiota variation in the jejunum and cecum of two rabbit breeds with different growth rates. This study was carried out on Native Middle-Egypt Breed (NMER) and Giant Flanders (GF) rabbits from 5 weeks to 12 weeks of age. Twenty NMER (NM) and GF male rabbits were slaughtered, and the jejunum and cecum tracts were collected to assay gut microbiota composition via 16S ribosomal RNA (rRNA) gene sequencing and histology examination. At 12 weeks of age, daily weight gain, villus height in the jejunum, total protein, and albumin were higher in GF rabbits than in NMER rabbits. Also, the jejunal villi of GF were well arranged in their dense borders. The microbiota between the jejunum and cecum was significantly different in terms of Beta-diversity. A significant correlation between Enterococcus (jejunum NM samples) and Lactobacillus (cecum GF samples) with body weight and weight gain was found (p < 0.05). Moreover, Escherichia-Shigella in the cecum of NM was significantly correlated with weight gain (p < 0.05). The most abundant genera identified in the jejunal and cecal contents of GF were generally beneficial microbiota. They may also play a role in reducing the pathogenic effects of Escherichia coli in these rabbits.
Keywords:
rabbits; microbiota; 16S rRNA gene sequencing; jejunum; cecum; breeds; growth; NMER breed; Giant Flander breed 1. Introduction
The microbial population that inhabits specific niches in the body, developing symbiotic relationships with the host, is called the microbiota. This complex ecosystem of microbial communities is involved in important physiological processes in the host, such as nutrient digestion and absorption [1], immune system development [2], intestinal permeability [3], and both direct and indirect protection against the development of dangerous or pathogenic bacteria [4]. The gut microbiota plays an important role in healthy animals but can also be a cause of pathology [5]. In recent decades, the development of high-throughput sequencing technologies, such as 16S ribosomal RNA (rRNA) amplicon sequencing, has reduced execution costs and facilitated knowledge of the microbiota composition of the different organs of the gastrointestinal tract (stomach, small, and large intestine) in various animal species such as pigs [6], poultry [7], and horses [8]. Also, 16S rRNA sequencing can be used to study the changes induced by managerial, environmental, and individual factors in microbial populations [9,10,11]. Recently, this technology has also been used in domestic rabbits (Oryctolagus cuniculus) that are monogastric, hindgut-fermenting herbivores and rely on cecotrophy. The rabbit is a very interesting species because it is considered, at the same time, a farm animal, a pet, and also a laboratory animal [12]. The composition of the microbiota of different intestinal tracts has been evaluated in livestock [13], laboratory [14], pet, and shelter rabbits [15], finding a different microbiota composition in the diverse gastrointestinal tracts. In particular, the large intestine shows the highest richness and diversity in bacterial species, while the small gut has the highest variability in the gastrointestinal tract of rabbits [13]. Several studies have been performed to evaluate the effects of the quality and levels of the diet [16,17], dietary supplementation with nutraceuticals [18,19,20], the temperature of the drinking water [21], age [22], and weaning period [23]. The fecal microbiota and its functional capacity associated with weaning weight in meat rabbits [24], hygiene of the environment [25], season [15], and drug treatments [26,27] have been investigated. It was established that these factors influencing the microbiota composition can also affect the quality of the carcass and meat of the rabbit [28,29,30]. Actually, only a few studies evaluated the effect of the breeds and the growth rate on the microbiota composition [31]. Significant differences in the gut microbiota observed in two rabbit breeds and some families, Ruminococcus and Lachnospiraceae, could be considered biomarkers for improving the health and production performance of meat rabbits [31]. The present study aimed at comparing, using a 16S rRNA-based analysis, the microbial populations in the jejunum and cecum of two breeds of rabbits: the Native Middle Egypt Rabbit—NMER (NM) as a local breed with a light body weight and the Giant Flander (GF) as an exotic breed with a heavy body weight. In addition, histology, scanning electron microscope (SEM) examinations, and biochemical parameters were used to compare NM with GF rabbits.
2. Materials and Methods
2.1. Experimental Design and Sampling
The experimental protocols were approved by the Animal Production Research Institute (APRI)’s animal care and use committee (ethical approval number: 2021920153429). The current research was conducted by APRI, the Agricultural Research Center (ARC), at the Rabbitry Research Farm in Sakha, Kafr el-Sheikh Governorate, which is located in the north of Egypt, during the period from June to September 2020.
A total of 200 Native Middle-Egypt Rabbit-NMER (NM) and Giant Flanders (GF) breeds, NM (n = 107) and GF (n = 93), rabbits in growing stages (5–12 weeks) were used in the present study. At 5 weeks of age, the young rabbits were weaned and housed in individual cages with 35 × 35 × 25 cm dimensions and equipped with feeding hoppers made of galvanized steel and nipples for automatic drinking. The animals were bred under the same environmental conditions and were watered and fed ad libitum from a commercial pelleted diet without probiotics or antibiotics. The ingredients and chemical composition of the diet used in the experiment are presented in Table 1.

Table 1.
Ingredients and chemical composition of the diet.
At 12 weeks, 20 male rabbits of the NM and GF breeds with similar weight at weaning (from 490.5 g to 542.5 g, average = 516.5 g) were selected for the study to avoid the effect of sex on studied traits. The rabbits were euthanized at 12 weeks. At euthanasia, the gastrointestinal tracts were removed shortly after death, and the contents of the jejunum and cecum were collected and immediately stored at −80 °C until genomic DNA extraction.
2.2. Blood Biochemical Parameters
Before euthanasia, blood samples were collected from all the rabbits (n = 10 for each breed) and centrifuged at 1500× g for 20 min; the serum samples were then kept at −80 °C for further biochemical parameter analyses: glucose, total protein, albumin, globulin, triglycerides (TG), and urea were analyzed according to the manufacturing instructions of the Biodiagnostic company kits (Dokki, Giza, Egypt; www.bio-diagnostic.com, accessed on 22 March 2021). The total protein and albumin differences in the collected samples were used to determine the globulin levels. The biochemical parameters were measured using a UV-VIS spectrometer (model T60UV, PG Instruments Limited, Lutterworth, UK).
2.3. Histological Characteristics
Immediately after the excision, the samples from the jejunum and cecum (n = 10 for each breed, NM and GF) were preserved in neutral buffered formalin. Then, the tissues were dehydrated in an ascending grade of ethanol and embedded in paraffin wax. Serial sections were cut at 5 μm with a microtome (Galileo SEMI, Diapath, Martinengo, Italy). The cross-sections of both the jejunum and cecum—three sections for each organ per rabbit—were stained with Mayer’s hematoxylin and eosin (H&E). Histological morphometric characteristics in the jejunum and cecum included villus height (VH), villus width (VW), and tunica muscularis (TM), according to [32]. Ten individual villus and ten loci in the tunica muscularis were measured for each rabbit. Histological characteristics were studied using a Leica light microscope and imaging software from Leica Microsystems (Application Suite 3.1.0 software, Leica, Wetzlar, Germany).
2.4. Electron Microscopic Examination
The collected jejunum and cecum samples (n = 10 for each breed, NM and GF, and one sample for each organ) were fixed in 3% glutaraldehyde and 0.1 M sodium cacodylate buffer (pH 7.0) for 2 h at room temperature, then rinsed in the same buffer, and finally post-fixed in 1% osmium tetroxide for another 2 h at room temperature. The scanning electron microscope (SEM) samples were dehydrated in an ethanol series ranging from 10% to 90% for 15 min in each alcohol dilution, followed by 30 min in absolute ethanol. The SEM samples were critical-point dried by using liquid carbon dioxide. Specimens were mounted on aluminum stubs with silver paint and coated with gold/palladium in a SPI-Module Sputter Coater device (SPI Supplies, West Chester, PA, USA). The SEM was performed in the electron microscope JEM-2100 (JEOL, Ltd., Tokyo, Japan) at a 20 kV accelerating voltage for studying the villi of the cecum and jejunum.
2.5. DNA Extraction, Library Generation, and Sequencing
Total bacterial genomic DNA was isolated from the content of the jejunum and cecum (n = 10 for each breed, NM and GF) by using the Easy Pure Stool genomic kit protocol (TransGen Biotech, Haidian District, Beijing, China) following the manufacturer’s instructions. The 16S ribosomal RNA (rRNA) gene was amplified using primers targeting the V3-V4 hypervariable regions according to the 16S Metagenomic Sequencing Library Preparation (https://support.illumina.com/documents/documentation/chemistrydocumentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf, accessed on 3 July 2023, Illumina, San Diego, CA, USA). All of the PCR amplifications were performed in 25 µL. A total of 12.5 µL of KAPA HIFI Master Mix 2× (Kapa Biosystems, Inc., Wilmington, MA, USA) and 0.2 µL of each primer (100 µM) were added to 2 µL of genomic DNA (5 ng/µL). Blank controls (no DNA template added to the reaction) were also performed. The PCR conditions were as follows: 94 °C for 5 min, 98 °C for 30 s, 56 °C for 60 s, and 72 °C for 60 s for a total of 25 cycles, with a final extension step at 72 °C for 10 min.
Amplicons were cleaned with Agencourt AMPure XP (Beckman, Coulter Brea, CA, USA), and libraries were prepared following the 16S Metagenomic Sequencing Library Preparation Protocol (Illumina, San Diego, CA, USA). The libraries obtained were quantified by real-time PCR with KAPA Library Quantification Kits (Kapa Biosystems, Inc., Wilmington, MA, USA), pooled in equimolar proportion, and sequenced in one MiSeq (Illumina) run with 2 × 250-base paired-end reads.
2.5.1. Bioinformatics—Sequence Processing
Raw paired reads from each sample were merged into one single sequence per fragment by PandaSeq [33]; then, low-quality bases (Phred quality score < 3) were trimmed from the 3′-end and fragments having a length < 75% of the initial fragment length were discarded. Filtered reads were clustered into zero-radius operational taxonomic units (zOTUs) by USEARCH (v. 11.0.667, Edgar), retaining only those supported by 5 or more reads. All downstream analyses were performed in the QIIME 1.9.0 suite [34]. Taxonomic assignment of zOTUs was performed by the RDP classifier [35] against the SILVA 132 database [36], using 0.5 as a confidence threshold.
The dataset comprised a total of 40 samples, derived from 2 organs (jejunum and cecum) and 2 rabbit breeds (NM and GF). Each combination had 10 independent replicates. The comparison among the experimental categories comprised three levels of analysis: (a) Breed: comparison between microbiota from GF and NM rabbits (as a whole, considering both organs); (b) Organ: comparison between microbiota from caecum and jejunum (as a whole, considering both breeds); and (c) stratified analyses comparing organs within the same breed and breeds in samples from the same organ.
2.5.2. Statistical Analysis Diversity
In order to have a comparable representation of the bacterial communities, read counts per sample were normalized to the least-sequenced one, at 18,561 reads per sample. The analysis of the sample biodiversity (i.e., alpha-diversity) was based on different metrics (i.e., Shannon’s diversity, chao1 diversity index, observed species, and Faith’s phylogenetic diversity index (PD whole tree)). A non-parametric permutation-based t-test (equivalent to the Mann–Whitney U-test) with 999 random permutations was made to assess whether the samples belonging to one experimental class were statistically different from those of a different class.
Beta-diversity analysis was performed according to the unweighted and weighted UniFrac distances among samples and represented via a Principal Coordinate Analysis (PCoA). A statistical test (i.e., the “Adonis” test, Permutational Multivariate Analysis of Variance Using Distance Matrices, using pseudo-F ratios) was used in order to define whether there was a significant difference among the experimental groups using 999 random permutations. Statistical analyses and graphs were performed in Matlab (v. 2008a, Natick, MA, USA).
2.5.3. Co-Abundance Analysis
This analysis was aimed at identifying groups of bacterial genera whose abundance was correlated with each other. Spearman’s rank correlation was calculated for all the bacterial genera having abundance > 0.5% in at least 50% of the samples in each experimental category: cecum NM, cecum GF, jejunum NM, and jejunum GF. On the basis of the correlations, bacterial genera were clustered in co-abundance groups (CAGs), following the procedure originally developed by Claesson and co-workers [37] and using Euclidean distance and average linkage; Cytoscape v. 3.0 [38] was used to graphically represent CAGs, as well as the relative abundance of bacterial genera and strength of correlation.
2.5.4. Microbiota-Body Weight Correlations
Spearman’s rank correlation was used to analyze the correlation between bacterial genera, the body weight, and the weight gain of the animals measured at 12 weeks of age.
2.5.5. Microbial Function Prediction
Metabolic functional capacities of gut bacteria were predicted from 16S rRNA data using the Tax4Fun R package [39] and the Kyoto Encyclopedia of Genes and Genomes (KEGG). Tax4Fun transformed the SILVA-classified zOTUs into prokaryotic KEGG organisms and normalized them according to the 16S rRNA copy number. The output tables provided KEGG orthology (KO) numbers for gene annotations and Enzyme Commission (EC) numbers.
2.6. Statistical Analysis
The individual animal was considered the experimental unit, and data on body weight, weight gain, and blood biochemical and histological parameters were represented as means with standard deviations (SEM) by using SAS 2002 software’s GLM technique (SAS, Cary, NC, USA). A one-way ANOVA was used to compare the means, and the differences were considered significant at p < 0.05.
3. Results
3.1. Body Weight and Weight Gain
The mean body weight at 5 and 12 weeks of age, as well as the daily weight gain, are presented in Table 2. The body weight at 12 weeks of age and daily weight gain were greater in GF than those in the NM breed. The daily weight gain was significantly (p < 0.05) higher in GF rabbits than in NM rabbits (p = 0.048).

Table 2.
Body weight and weight gain in GF and NM rabbits.
3.2. Blood Biochemical Parameters
The analysis of blood biochemical parameters in GF and NM rabbits is presented in Figure 1. Total Protein (g/dL) and albumin (g/dL) were significantly higher in GF than in NM rabbits (p = 0.02 and 0.034, respectively).
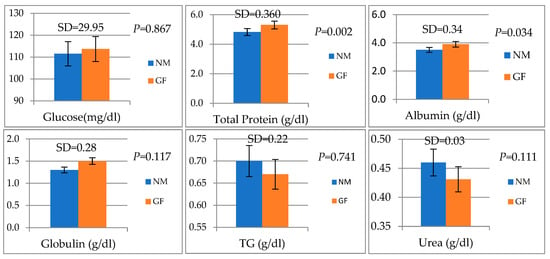
Figure 1.
Blood biochemical parameters in GF and NM rabbits. Means and standard deviations (SD), and the differences were considered significant at p < 0.05.
The comparison among the histological morphometric parameters between NM and GF rabbits in the jejunum and cecum is reported in Figure 2. In cecum, the villus width was higher in the NM group compared to the GF group (p = 0.001), while in the jejunum, the GF rabbits had a higher villus height with respect to the NM group (p ≤ 0.001).
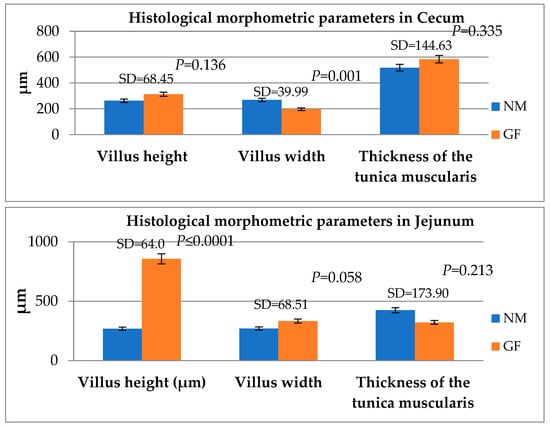
Figure 2.
Comparing the histological morphometric parameters between GF and NM rabbits in the cecum and jejunum. Means and standard deviations (SD), and the differences were considered significant at p < 0.05.
3.3. Electron Microscopic Examination
The photographs of the scanning electron microscope (SEM) showed that villi on the epithelial cecum in the rabbit had a semi-zigzag pattern (Figure 3, photographs A–D). The villi on the epithelial cecum of NM showed a lower density (Photograph A) than those in GF (Photograph B). The villi tips were round and smoothed in the GF breed (Photograph D), while they were marked by irregular edges in the NM breed (Photograph C).
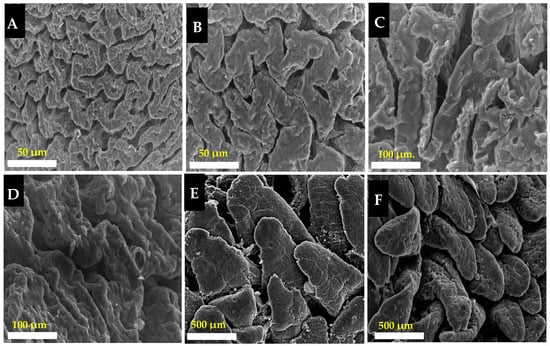
Figure 3.
SEM photographs of villi of the cecum and jejunum in NM and GF breeds. (A,C) photographs showed the villi in the cecum of NM. (B,D) photographs showed the villi on the cecum of the GF breed. (E,F) photographs of the villi on the jejunum of the NM and GF breeds, respectively. Scale bars: (A,B) 50 µm; (C,D) 100 µm; (E,F) 500 µm.
SEM examination of the villi on the jejunal epithelium in the rabbit revealed a tongue-like shape (Photographs E and F). The jejunal villi of NM and GF were well arranged in their dense borders, but without neat borders in NM (Photograph E), whereas in GF there were clear and neat borders (Photograph F).
3.4. Microbial Profile
The overall composition of the microbiota belonged to kingdom bacteria for 99.97%, with a residual of 0.03% Archaea. The main bacterial phyla in all experimental conditions included Firmicutes (average relative abundance: 58.04%), Proteobacteria (13.88%), Patescibacteria (6.54%), Bacteroidetes (4.64%), and Verrucomicrobia (3.56%), while the unclassified bacteria were 1.47% on average. With regard to bacterial families, the most abundant were: Ruminococcaceae (23.76%), Enterobacteriaceae (10.71%), Eubacteriaceae (8.62%), Saccharimonadaceae (6.54%), Lactobacillaceae (6.35%), Enterococcaceae (6.09%), Lachnospiraceae (4.26%), and Akkermansiaceae (3.56%). Finally, the main bacterial genera found in the samples were: Escherichia-Shigella (9.26%), uncultured Eubacteriaceae (8.60%), Ruminococcaceae NK4A214 group (6.66%), Candidatus Saccharimonas (6.54%), Lactobacillus (6.35%), Enterococcus (6.08%), Ruminococcaceae UCG-014 (5.66%), Akkermansia (3.56%), Christensenellaceae R-7 group (3.10%), and Subdoligranulum (3.08%). About 37% of the average relative abundance was due to genera whose rel. ab. was <3% on average.
Alpha-diversity estimations for cecum and jejunum samples in both rabbit breeds (NM and GF) showed that the diversity across the different experimental conditions was comparable for all the metrics analyzed (with the only exception of a significant difference between jejunum and cecum samples of the GF breed, p = 0.011), as shown in Figure 4. For beta-diversity analysis, a significant difference was found for the intestine tract experimental variable, with cecum and jejunum samples being different (regardless of the rabbit breed) for both the unweighted and the weighted UniFrac distances (p = 0.022 and p = 0.001, respectively, data not shown). Stratifying by organ and breed, a significant difference was found between cecum and jejunum samples of the GF breed. No difference was found for the NM breed when comparing the two breeds for jejunum and cecum samples separately (Figure 5), this statement is right
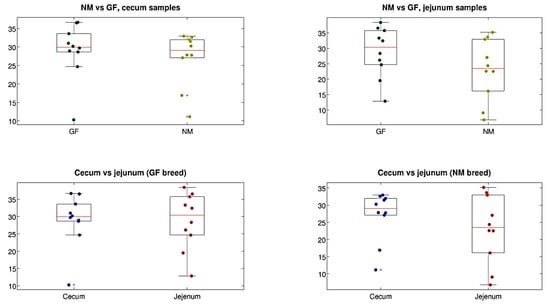
Figure 4.
Boxplots of alpha-diversity estimations (PD whole tree metric) for rabbit breeds and organs separated. Each point represents a sample, whereas the median value is in the red line.
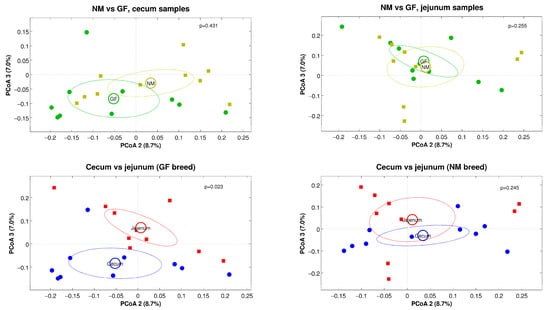
Figure 5.
Principal Coordinate Analysis (PCoA) plots derived from unweighted UniFrac distances comparing the two breeds and organs separately. In the plots, each point represents a sample, colored according to the experimental group. Ellipses represent the 95% SEM-based confidence, and the p-values reported are those deriving from the “adonis” test on the distance matrices. The second and third principal coordinates are plotted.
3.5. Comparison between Breeds in Cecum Samples
For microbiota composition, the most abundant phyla were Firmicutes, with an average relative abundance of 54.0% and 65.4% in NM and GF breeds, respectively, followed by Patescibacteria (7.3% and 10.3%) and Bacteroidetes (7.2% and 3.5%). The average relative abundances for genera having an average abundance > 1% in the cecum of NM and GF breeds are reported in Table 3. At the genus level, the p-value indicated no significant difference (p < 0.05) for any genera between GF and NM breeds.

Table 3.
Average of the relative abundance and standard deviation (between parentheses) at genus level for microbiota composition in the cecum of NM and GF breeds. The average abundance over the replicates (n = 10) of the same condition, as well as (between parenthesis) the standard deviation, are the results of the Mann–Whitney U-test. Only genera with an average abundance > 1% were shown. Average abundances at higher levels are calculated by summing up the genera belonging to each order and phylum.
3.6. Comparison between Breeds in Jejunum Samples
In jejunum samples, a high relative abundance at phylum level was found for Firmicutes (44.0% and 40.2% in NM and GF breeds, respectively), followed by Proteobacteria (19.6% and 24.2%). The average relative abundances for main genera (average abundance > 1%) in the jejunum of NM and GF breeds are shown in Table 4. At genus level, no different significances (p < 0.05) between the GF and NM breeds were found. However, GF showed a tendency towards an increase in Bacteroides and towards a depletion of unclassified members of the Eubacteriaceae family and the Christensenellaceae R-7 group.

Table 4.
Average of the relative abundance and standard deviation (between parentheses) at genus level for microbiota in the jejunum for NM and GF breeds. The average abundance over the replicates (n = 10) of the same condition, as well as (between parenthesis) the standard deviation, are shown. The p-value refers to that of a two-tailed Mann–Whitney U-test. Only genera with an average abundance > 1% were shown. Average abundances at higher levels are calculated by summing up the genera belonging to each order and phylum.
3.7. Comparisons between Organs in NM Breed
The phylum-level composition of the microbiota in cecum and jejunum samples from the NM breed was dominated by Firmicutes (average rel ab.: 52.9% and 45.8%, respectively), followed by Proteobacteria (0.1% vs. 18.9%), and Bacteroidetes (7.2% vs. 2.4%). Seven out of the 17 identified genera in the Firmicutes phylum were significantly (p < 0.05) different between the examined organs. Among them, bacteria from the Lactobacillales order were more abundant in jejunum samples than in cecum (avg. rel. ab.: 23.0% vs. 9.9%) and, in particular, the Enterococcus genus. On the other hand, uncultured Clostridiales vadinBB60 group, Ruminococcaceae NK4A214 group, Ruminiclostridium 5, and members of the family Ruminococcaceae (all belonging to the Firmicutes phylum) were significantly (p < 0.05) higher in the cecum than the jejunum (Table 5). The Escherichia-Shigella genus (phylum Proteobacteria) was also significantly higher in jejunum samples (Table 5).

Table 5.
Average relative abundances and the standard deviation (between parentheses) at genus level for microbiota in the cecum and jejunum in the NM breed. The average abundance over the replicates (n = 10) of the same condition, as well as (between parenthesis) the standard deviation, are shown. The p-value refers to that of a two-tailed Mann–Whitney U-test. “*” indicated statistical significance (p < 0.05). Only genera with an average abundance > 1% were shown. Average abundances at higher levels are calculated by summing up the genera belonging to each order and phylum.
3.8. Comparisons between Samples in GF Breed
In the GF rabbit breed, at the phylum level, the average relative abundance of Firmicutes was higher in the cecum than the jejunum (61.8% vs. 41.3%, respectively), while Proteobacteria were present in the jejunum (average rel. ab.: 24.2%) and nearly absent in cecum (average rel. ab.: 0.8%), as shown in Table 6. At the genus level, 12 out of 16 genera identified in the Firmicutes phylum were significantly (p < 0.05) different between organ samples. In particular, nine genera (i.e., Bacillus, Lactobacillus, Christensenellaceae R-7 group, Lachnospiraceae (other), Ruminococcaceae NK4A214 group, Ruminococcaceae UCG-013, Ruminococcaceae UCG-014, Ruminococcus 2, and Subdoligranulum) were significantly higher in the cecum than the jejunum of the GF breed (Table 6). On the other hand, Enterococcus, Sarcina, and Dubosiella genus were higher in the jejunum than the cecum (Table 6).

Table 6.
Average relative abundances and the standard deviation (between parentheses) at genus level for microbiota in the cecum and jejunum in the GF breed. The average abundance over the replicates (n = 10) of the same condition, as well as (between parenthesis) the standard deviation, are shown. The p-value refers to that of a two-tailed Mann–Whitney U-test. “*” indicated statistical significance (p < 0.05). Only genera with an average abundance > 1% were shown. Average abundances at higher levels are calculated by summing up the genera belonging to each order and phylum.
Regarding bacteria belonging to other phyla, the genus Candidatus Saccharimonas (phylum: Patescibacteria) was significantly higher in the cecum, whereas the Escherichia-Shigella genus (phylum: Proteobacteria) was significantly higher in the jejunum (Table 6).
3.9. Correlation and Co-Abundance Analysis
Co-abundance analysis aimed to infer interactions between bacteria genera in the gut microbiota of NM and GF breeds and organ sampling places in the cecum and jejunum. Clustering the matrix of the pairwise Spearman’s correlations between the main bacterial genera in the experiment highlighted four main co-abundant groups (CAGs), i.e., genera whose abundance was concordantly increased/decreased throughout all the 40 samples. CAG1 included genera from the families of Ruminococcaceae (Ruminococcus 5, Ruminococcus 1, other Ruminococcus, Ruminococcaceae UCG-014, Ruminococcaceae UCG-013, and Ruminococcaceae NK4A214), and Lachnospiraceae (unclassified Lachnospiraceae, Lachnospiraceae NK4A136), plus other genera such as Akkemansia, Subdoligranulum, Christenellaceae R-7 group, uncultured Gastroaerophilales and Candidatus Saccharimonas, all highly correlated one to each other; CAG2 comprised the uncultured members of the Eubacteriaceae family and some unclassified Atopobiaceae; CAG3 was composed by bacteria unclassified at lower phylogenetic levels; the fourth (CAG4) was made only by Escherichia-Shigella genus, which was correlated negatively with CAGs1-3 (Supplementary, Figure S1).
The samples from NM breed cecum were characterized by the involvement of Atopobiaceae and uncultured members of Eubacteriaceae in the correlations with the main cluster of genera and by relatively few connections between Lachnospiraceae NK4A136, Ruminococcus 1, and Lachnospiraceae (Figure 6). In cecum samples from GF breeds, the analyzed bacterial groups had few correlations and seemed to be more independent of each other (Figure 6). In cecum samples, regardless of the rabbit breed, Escherichia-Shigella (even at low abundance) was inversely correlated to the other genera.
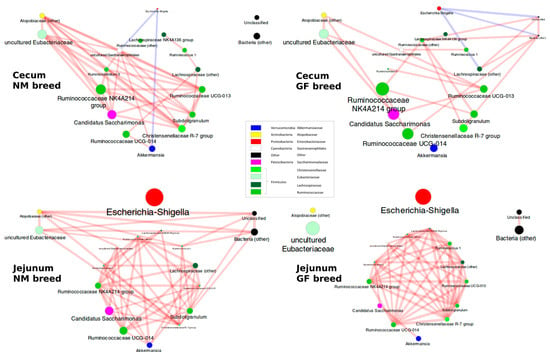
Figure 6.
The Co-Occurrence patterns among the bacterial genera in the cecum and jejunum in NM and GF breeds, separately. Patterns here depict the significant (p < 0.05) correlations among the bacterial genera for the four experimental classes. Node size is proportional to the average abundance of the genera in the experimental condition, and the fill color follows the taxonomic classification of the genera. The edges size is proportional to the strength of correlation, whereas the edges color is blue for negative correlations and red for positive correlations.
In jejunum samples, the network of correlations among the genera of the main cluster involves many or quite all the bacteria. Even though Escherichia-Shigella was relatively abundant, it was uncorrelated with the other bacteria. In the jejunum samples from the NM breed, a correlation was evident also between the members of the Atopobiaceae and Eubacteriaceae families and other unclassified bacteria (Figure 6). In the jejunum of GF breed samples, these further correlations were not significant, and the network involved only the main cluster of bacteria (Figure 6).
3.10. Microbiota-Body Weight Correlations
Figure 7 reports the correlations estimated between the twenty most abundant genera in the gut microbiota, the body weight at 12 weeks, and the weight gain, separated for the two organs and the two breeds. In cecum samples from the NM breed, Escherichia-Shigella (relative abundance: 0.8%) was significantly (p < 0.05) correlated with the weight gain.
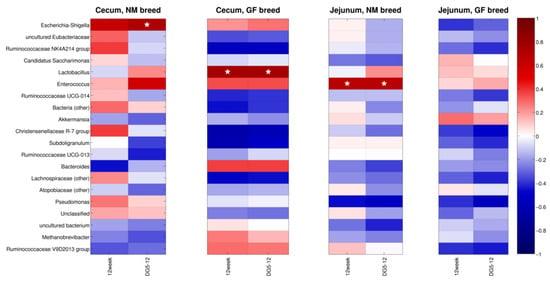
Figure 7.
Heatmap of the Spearman’s correlation between the top twenty most abundant genera, the body weight, and the weight gain in the experimental classes. Blue is for negative correlations, red for positive correlations, and “★” indicates significant (p < 0.05) correlations.
In GF cecum samples, the abundances of the Lactobacillus genus (average 8.3%) were significantly (p < 0.05) correlated with both the weight at 12 weeks and the weight gain. On the other hand, in the jejunum of NM samples, a significant correlation was found between the Enterococcus abundances (average: 13.7%) and both the weight at 12 weeks and the weight gain.
3.11. Microbial Function Prediction
To explore the effects of host genetics on the potential functional capacities of the gut microbiome, functional profiles of cecum and jejunum bacterial communities in NM and GF breeds were predicted based on 16S rRNA sequencing data. Predictions of the functional capacities of gut bacteria were based on the Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways, identifying a total of 6419 KEGG Orthologies (KOs) in all samples (Supplementary Table S1). Functional predictions in the gut microbiome belonged to signaling and cellular processes (secretion system and transporters), genetic information processing (chaperones and folding catalysts, transcription, transfer RNA biogenesis, and translation), cellular processes (growth factor and mobility), and metabolism (amino acids, carbohydrates, lipids, cofactors, and vitamins) categories, as presented in Figure 8A–D, respectively. The functional predictions were not significantly different between NM and GF breeds in both jejunum and cecum except for the growth factor and mobility pathway (cellular processes) of the jejunum samples (Figure 8C), which resulted in higher levels in the GF breed than the NM breed. On the other hand, significant differences were reported for the jejunum vs. cecum comparison: both NM and GF breeds showed differences in transporters, transcription, carbohydrate metabolism, and lipid metabolism; the GF breed was also characterized by a differential abundance in the pathways of the secretion system, chaperones and folding catalysts, translation, and metabolism of cofactors and vitamins; and finally, NM breed samples showed a significant difference in the pathway of amino acid metabolism. In all comparisons, the functional potential of cecum samples was higher than that of jejunum samples.
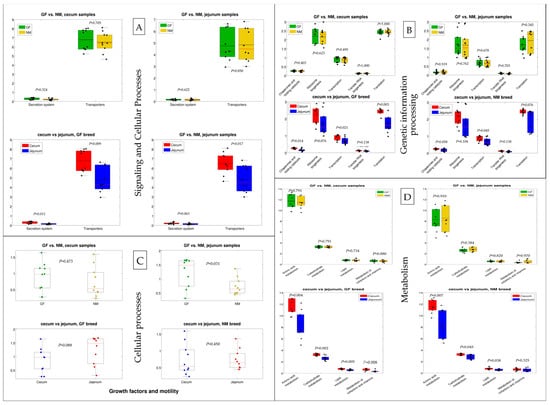
Figure 8.
The functional prediction of gut microbiome in rabbit breeds NM and GF, separated by 0.008 organs (cecum and jejunum). (A) Signaling and cellular processes; (B) Genetic information processing; (C) Cellular processes; (D) Metabolism.
4. Discussion
The body weight and weight values in NM and GF rabbits were similar to the findings (NM, 1169.4 vs. GF, 1327.7) of our previous work [40]. Moreover, we already reported that GF rabbits were significantly heavier and had a higher relative growth rate as compared to NM rabbits. NM is a rabbit breed established in three governorates in Middle Egypt by the Animal Production Research Institute (APRI) [41] and, according to the general classification used in the European Rabbit Breed Standards Book [42], belongs to the small-sized breed, while GF belongs to the medium-sized breed.
Total proteins and globulins were significantly affected by the breed, as documented in previous studies in four breeds [43] and three breeds [44] under Egyptian environmental conditions. High globulin concentrations in GF rabbits could be attributed to increasing the gamma globulin fraction, which indicates immunity status [45]. Previous studies have indicated that an altered serum component profile could reflect differences in the gut microbiome of rabbits [31], which is consistent with our findings as shown in Figure 4. This study suggested that host rabbit breeds can shape the gut microbiome and serum metabolome, including the interactions among the host-gut microbiome and serum metabolome that are important. Thus, total proteins and globulins may be indicators of the health status and production traits of meat rabbits.
The villi on the jejunum and cecum were higher in GF than NM rabbits in addition, the villi of the cecum in MN were less dense and their tips were marked by irregular edges, as shown in the SEM examination. These observations may be due to some kind of toxins produced in the intestine and cecum [46] by bacteria that were in close contact only with epithelial cells that had lost their brush border [47]. In addition, the fermentation operated by bacteria could produce hydrogen ions that can induce damage in the cecum and jejunum mucosa [46].
The microbiota composition of the samples was not significantly different when comparing the two rabbit breeds (NM and GF) within the same intestinal tract. In fact, among the major constituents of the microbiota (relative abundances > 1%), none of them were found to be significantly diverse. On the other hand, differences were more evident when comparing the jejunum vs. cecum microbiota, in particular in the GF breed, where both alpha- and beta-diversity estimations were found to be significantly different. This was also reflected when considering the functional predictions deriving from the microbial profiles, with jejunum and cecal samples appearing very different from one another. The predominant phylum in microbiota samples for both intestinal tracts (jejunum and caecum) was Firmicutes, followed by Patescibacteria and Bacteroidetes (in cecum) and Proteobacteria and Actinobacteria (in jejunum), in accordance with the fact that Firmicutes was found to be the most dominant phylum in the rabbit microbiota, regardless of source, age, and season [14]. Our results are also in harmony with the findings of Fu et al. [48], who reported that Firmicutes was the most dominant phylum in the foregut and hindgut of rabbits, while the second most dominant one was Proteobacteria (in the foregut) or Bacteroidetes (in the hindgut). Another study reported that Firmicutes were the most abundant phylum in all of the sections of the gastro-intestinal tract examined (45.9%), such as the stomach, duodenum, jejunum, ileum, cecum, and colon, followed by Bacteroidetes in the large intestine (38.9%), Euryarchaeota (29.6%), and Patescibacteria (13.8%) in the foregut, especially in jejunum [13]. The abundance of members of the Firmicutes phylum was higher in GF than NM rabbits, both in the cecum (73.5% GF vs. 63.9% NM) and jejunum (44.0% GF vs. 40.2% NM) samples. Previous studies have shown that a high abundance of Firmicutes, as shown in GF, can enhance intestinal mucosa and reduce oxidative stress in the intestinal tract in piglets [49,50]. This concept was further supported by histological examinations and scanning electron microscopy (SEM) in the cecum and jejunum villi of GF rabbits.
Within the Firmicutes phylum, the most abundant order was Clostridiales and, among genera, those from the Ruminococcaceae and the Lachnospiraceae families. As highlighted in the co-abundance groups’ analysis, the abundance of the members of these two families was highly correlated, establishing a sort of “core” microbiota. A high abundance of Clostridiales, uncultured Clostridiales vadinBB60 group, and Ruminiclostridium 5, such as that highlighted in the jejunum of GF, may reduce the effect of Escherichia coli as a pathogenic agent through remodeling the signaling pathway [51]. The high abundance of Bacteroides and Ruminococcus in the cecum of GF breed rabbits could be related to a healthy gut, in accordance with the findings of previous studies on rabbits [31].
In cecum samples, members of the Eubacteriaceae family occupied a central position in the interaction network, with positive correlations observed between Lachnospiraceae, Ruminiclostridium, and Eubacteriaceae, possibly due to their functional potential in the specific metabolic pathways. Eubacteriaceae and Lachnospiraceae bacteria exhibited metabolic specificities for pyruvate and carbohydrate degradation [24]. Members of Ruminococcaceae seemed to be highly specialized in pyruvate-to-lactate fermentation [52,53], as happens in the degradation of plant material such as pectin and cellulose in the colonic fermentation of dietary fibers in mammals [54,55]. These results could be confirmed by Ye et al. [31], who reported that families Ruminococcus and Lachnospiraceae could be considered biomarkers for improving the health and production performance of meat rabbits. Moreover, other low-abundance microorganisms, such as the Akkermansia genus (Verrucomicrobiales order), could play a key role in the hydrolysis of diverse ingested polysaccharides and contribute to a more complete digestion of dietary cellulose [51,56]. The significant decrease in the proportion of Verrucomicrobia in the jejunum of the NM breed as compared to the cecum suggests less optimal jejunum health as well as a more pro-inflammatory state, as already reported in mice [57]. The positive correlations among Ruminiclostridium, Lachnospiraceae, and Eubacteriaceae that were observed in the cecum with increasing Verrucomicrobia in the jejunum of the GF could explain the significant difference in the microbiota composition between organs in the GF breed.
The abundance of the Escherichia-Shigella genus in the cecum of NM breed rabbits was positively correlated to body weight gain, as were Enterococcus and both body weight at 12 weeks and body weight gain in the jejunum of the same animals. Escherichia-Shigella should be considered a pathogen in hosts [9]; meanwhile, Enterococcus could be considered a beneficial bacteria that could produce bacteriocins active against bacteria such as Listeria and indigenous clostridia in the gut of rabbits [58]. On the other hand, in the cecum of the GF rabbits, there were significant correlations between Lactobacillus and both the body weight at 12 weeks and the body weight gain. Lactobacillus can promote the fermentation of carbohydrates into lactic acid and intestinal health [51], increase the concentration of short-chain fatty acids (SCFAs) in the intestines of mice, promote the growth of intestinal epithelial cells [59], and have a biological antagonistic effect on pathogenic bacteria such as E. coli. These results confirmed that the cecum is the main organ harboring the microbial fermentation processes in the gut of the rabbit [16], and hosting Lactobacillus, as occurred in the GF breed, could cause increased growth.
The functional potential of cecum samples was higher than that of jejunum samples, in particular for the secretion system, transporters, amino acids and carbohydrate metabolism, ribosome biogenesis, and translation. This could be attributed to the fact that cecum is the richest and most diverse microbial community in the rabbit gut [14,60]. The significantly different carbohydrates and lipid metabolic activity between the cecum and jejunum we observed could be due to the difference in microbial communities and their capacity to ferment to obtain metabolic energy [9].
The limited number of replicates per condition (n = 10) and the high variability observed in the microbial profiles of the samples could have negatively influenced a better characterization of the differences in the microbiota composition inherent to the two breeds. On the other hand, we were able to highlight the different bacterial communities inhabiting the jejunum and cecum tracts of both rabbit breeds and gain insight into the specific functions they preside over.
5. Conclusions
This study characterized the composition of the rabbit jejunal and cecal microbiota as well as their potential influence on rabbit growth. Our study provides that Patescibacetria and Bacteroides were major constituents of the microbiota in the rabbit hindgut, as well as Proteobacteria (mostly made up by members of the Escherichia-Shigella genus) in the foregut. Firmicutes was the most abundant phylum in both intestinal tracts. Within the Firmicutes phylum, the most abundant genera were all members of the Ruminococcaceae and Lachnospiraceae families, which are generally beneficial key members of the gut ecosystem, have multiple interactions with the other members of the gut microbiota, and may also reduce the pathogenetic effect of Escherichia coli. The high abundance of Bacteroides and Ruminococcus in the cecum of GF-bred rabbits could indicate a more healthy gut, as shown by the rounding and smoothing of the villi tips and the high density in the epithelium, which reflected in a high growth rate. A better understanding of the relationship between gut microbiota and the factors influencing its composition could improve the management and health of the rabbit.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani13142294/s1 Table S1: The KEGG Orthology (KO) numbers for gene annotations and Enzyme Commission (EC) numbers in all samples of gut bacteria were predicted from 16S rRNA sequence data using the Tax4Fun R package; Figure S1: Co-abundant groups (CAGs) for interactions between bacteria genera in the gut microbiota in NM and GF breeds and organ sampling places in the cecum and jejunum.
Author Contributions
Conceptualization, E.-S.M.A.-K. and H.M.A.S.; methodology, K.I.K.; software, S.S.G.; validation, S.H.A.M., N.I.A. and S.S.G.; formal analysis, M.S. and E.-S.M.A.-K.; investigation, K.I.K. and G.B.; resources, P.C.; data curation, S.H.A.M., N.I.A., Y.Z.A.-G. and W.A.H.A.; writing—original draft preparation, E.-S.M.A.-K.; writing—review and editing, W.A.H.A., P.C. and G.B.; visualization, M.S. and Y.Z.A.-G.; project administration K.I.K.; funding acquisition, H.M.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science, Technology, and Innovation Funding Authority (STDF), Egypt, grant number 38182.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved the Animal Production Research Institute (APRI)’s animal care and use committee (ethical approval number: 2021920153429).
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw data from microbial genome sequencing have been uploaded to the National Center for Biotechnology Information (NCBI) and can be found under BioProject ID: PRJNA992887 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA992887, accessed on 3 July 2023).
Acknowledgments
We would like to express our thanks to the staffs in the Rabbitry Research Farm in Sakha, Kafr el-Sheikh Governorate, Egypt.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 2018, 23, 458–469.e455. [Google Scholar] [CrossRef] [PubMed]
- Gaboriau-Routhiau, V.; Cerf-Bensussan, N. Gut microbiota and development of the immune system. Med. Sci. M/S 2016, 32, 961–967. [Google Scholar]
- Allam-Ndoul, B.; Castonguay-Paradis, S.; Veilleux, A. Gut microbiota and intestinal trans-epithelial permeability. Int. J. Mol. Sci. 2020, 21, 6402. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J. Microbiota in health and diseases. Signal Transduct. Target Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Piazuelo, D.; Estellé, J.; Revilla, M.; Criado-Mesas, L.; Ramayo-Caldas, Y.; Óvilo, C.; Fernández, A.I.; Ballester, M.; Folch, J.M. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci. Rep. 2018, 8, 12727. [Google Scholar] [CrossRef]
- Zhou, Q.; Lan, F.; Li, X.; Yan, W.; Sun, C.; Li, J.; Yang, N.; Wen, C. The spatial and temporal characterization of gut microbiota in broilers. Front. Vet. Sci. 2021, 8, 712226. [Google Scholar] [CrossRef]
- Stewart, H.L.; Pitta, D.; Indugu, N.; Vecchiarelli, B.; Engiles, J.B.; Southwood, L.L. Characterization of the fecal microbiota of healthy horses. Am. J. Vet. Res. 2018, 79, 811–819. [Google Scholar] [CrossRef]
- Wang, J.; Xia, S.; Fan, H.; Shao, J.; Tang, T.; Yang, L.; Sun, W.; Jia, X.; Chen, S.; Lai, S. Microbiomics revealed the disturbance of intestinal balance in rabbits with diarrhea caused by stopping the use of an antibiotic diet. Microorganisms 2022, 10, 841. [Google Scholar] [CrossRef]
- Richards, P.; Fothergill, J.; Bernardeau, M.; Wigley, P. Development of the caecal microbiota in three broiler breeds. Front. Vet. Sci. 2019, 6, 201. [Google Scholar] [CrossRef]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci. Rep. 2018, 8, 8510. [Google Scholar] [CrossRef]
- AVMA. Avma Pet Ownership and Demographics Sourcebook: 2017–2018 Edition; AVMA: Schaumburg, IL, USA, 2018. [Google Scholar]
- Cotozzolo, E.; Cremonesi, P.; Curone, G.; Menchetti, L.; Riva, F.; Biscarini, F.; Marongiu, M.L.; Castrica, M.; Castiglioni, B.; Miraglia, D. Characterization of bacterial microbiota composition along the gastrointestinal tract in rabbits. Animals 2020, 11, 31. [Google Scholar] [CrossRef]
- Hu, X.; Wang, F.; Yang, S.; Yuan, X.; Yang, T.; Zhou, Y.; Li, Y. Rabbit microbiota across the whole body revealed by 16s rrna gene amplicon sequencing. BMC Microbiol. 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Kylie, J.; Weese, J.S.; Turner, P.V. Comparison of the fecal microbiota of domestic commercial meat, laboratory, companion, and shelter rabbits (oryctolagus cuniculi). BMC Vet. Res. 2018, 14, 1–15. [Google Scholar] [CrossRef]
- Velasco-Galilea, M.; Piles, M.; Ramayo-Caldas, Y.; Sánchez, J.P. The value of gut microbiota to predict feed efficiency and growth of rabbits under different feeding regimes. Sci. Rep. 2021, 11, 19495. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Deng, F.; Jia, X.; Liu, H.; Zhang, G.-W.; Lai, S.-J. Gut microbiota profiling with differential tolerance against the reduced dietary fibre level in rabbit. Sci. Rep. 2019, 9, 288. [Google Scholar] [CrossRef]
- Agradi, S.; Cremonesi, P.; Menchetti, L.; Balzaretti, C.; Severgnini, M.; Riva, F.; Castiglioni, B.; Draghi, S.; Di Giancamillo, A.; Castrica, M. Bovine colostrum supplementation modulates the intestinal microbial community in rabbits. Animals 2023, 13, 976. [Google Scholar] [CrossRef]
- Curone, G.; Biscarini, F.; Cotozzolo, E.; Menchetti, L.; Dal Bosco, A.; Riva, F.; Cremonesi, P.; Agradi, S.; Mattioli, S.; Castiglioni, B. Could dietary supplementation with different sources of n-3 polyunsaturated fatty acids modify the rabbit gut microbiota? Antibiotics 2022, 11, 227. [Google Scholar] [CrossRef]
- Cremonesi, P.; Curone, G.; Biscarini, F.; Cotozzolo, E.; Menchetti, L.; Riva, F.; Marongiu, M.L.; Castiglioni, B.; Barbato, O.; Munga, A. Dietary supplementation with goji berries (lycium barbarum) modulates the microbiota of digestive tract and caecal metabolites in rabbits. Animals 2022, 12, 121. [Google Scholar] [CrossRef]
- Wang, Q.; Fu, W.; Guo, Y.; Tang, Y.; Du, H.; Wang, M.; Liu, Z.; Li, Q.; An, L.; Tian, J. Drinking warm water improves growth performance and optimizes the gut microbiota in early postweaning rabbits during winter. Animals 2019, 9, 346. [Google Scholar] [CrossRef]
- Combes, S.; Michelland, R.J.; Monteils, V.; Cauquil, L.; Soulié, V.; Tran, N.U.; Gidenne, T.; Fortun-Lamothe, L. Postnatal development of the rabbit caecal microbiota composition and activity. FEMS Microbiol. Ecol. 2011, 77, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Chen, X.; Zhou, L.; Wang, C.; Chen, Q.; Lin, R.; Xiao, T.; Gan, Q. Faecal microbiota and functional capacity associated with weaning weight in meat rabbits. Microb. Biotechnol. 2019, 12, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Read, T.; Fortun-Lamothe, L.; Pascal, G.; Le Boulch, M.; Cauquil, L.; Gabinaud, B.; Bannelier, C.; Balmisse, E.; Destombes, N.; Bouchez, O. Diversity and co-occurrence pattern analysis of cecal microbiota establishment at the onset of solid feeding in young rabbits. Front. Microbiol. 2019, 10, 973. [Google Scholar] [CrossRef] [PubMed]
- Bennegadi, N.; Gidenne, T.; Licois, D. Impact of fibre deficiency and sanitary status on non-specific enteropathy of the growing rabbit. Anim. Res. 2001, 50, 401–413. [Google Scholar] [CrossRef]
- Abecia, L.; Fondevila, M.; Balcells, J.; Lobley, G.; McEwan, N. The effect of medicated diets and level of feeding on caecal microbiota of lactating rabbit does. J. Appl. Microbiol. 2007, 103, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Zou, F.; Zeng, D.; Wen, B.; Sun, H.; Zhou, Y.; Yang, M.; Peng, Z.; Xu, S.; Wang, H.; Fu, X. Illumina miseq platform analysis caecum bacterial communities of rex rabbits fed with different antibiotics. AMB Express 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Liu, B.; Cui, Y.; Ali, Q.; Zhu, X.; Li, D.; Ma, S.; Wang, Z.; Wang, C.; Shi, Y. Gut microbiota modulate rabbit meat quality in response to dietary fiber. Front Nutr. 2022, 9, 849429. [Google Scholar] [CrossRef]
- Castrica, M.; Menchetti, L.; Agradi, S.; Curone, G.; Vigo, D.; Pastorelli, G.; Di Giancamillo, A.; Modina, S.C.; Riva, F.; Serra, V. Effect of bovine colostrum dietary supplementation on rabbit meat quality. Foods 2022, 11, 3433. [Google Scholar] [CrossRef]
- Menchetti, L.; Brecchia, G.; Branciari, R.; Barbato, O.; Fioretti, B.; Codini, M.; Bellezza, E.; Trabalza-Marinucci, M.; Miraglia, D. The effect of goji berries (lycium barbarum) dietary supplementation on rabbit meat quality. Meat Sci. 2020, 161, 108018. [Google Scholar] [CrossRef]
- Ye, X.; Zhou, L.; Zhang, Y.; Xue, S.; Gan, Q.F.; Fang, S. Effect of host breeds on gut microbiome and serum metabolome in meat rabbits. BMC Vet. Res. 2021, 17, 1–13. [Google Scholar] [CrossRef]
- Alshamy, Z.; Richardson, K.C.; Hunigen, H.; Hafez, H.M.; Plendl, J.; Al Masri, S. Comparison of the gastrointestinal tract of a dual-purpose to a broiler chicken line: A qualitative and quantitative macroscopic and microscopic study. PLoS ONE 2018, 13, e0204921. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. Pandaseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 1–7. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’connor, E.M.; Cusack, S.; Harris, H.; Coakley, M.; Lakshminarayanan, B.; O’sullivan, O. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4fun: Predicting functional profiles from metagenomic 16s rrna data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]
- Abdel-Kafy, E.-S.; El-Deighadi, A.S.; Shabaan, H.M.; Ali, W.H.; Sabra, Z.E.-A.A.; Farid, A. Genetic evaluation for growth traits in new synthetic rabbit line in egypt. Open J. Agric. Res. 2021, 1, 62–73. [Google Scholar] [CrossRef]
- Abdel-Kafy, E.M.G.I.S.; Benlarbi, M.; Ahmed, S.S.; Badawi, Y.K.; Hassan, N.S. Genetic diversity and phenotype characterization of native rabbitin middle-egypt. J. New Sci. 2016, 16, 1312–1320. [Google Scholar]
- Zigo, F.; Pyskatý, O.; Ondrašovičová, S.; Zigová, M.; Šimek, V.; Supuka, P. Comparison of exterior traits in selected giant and medium rabbit breeds. World Rabbit Sci. 2020, 28, 251–266. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.; Abdel-Azim, A.; Darwish, A.; Omar, E. Haematological and biochemical observations in four pure breeds of rabbits and their crosses under egyptian environmental conditions. World Rabbit Sci. 2010, 18, 103–110. [Google Scholar] [CrossRef]
- Abdel-Hamid, T.M.; Dawod, A. Breed effects on growth performance, blood parameters and the levels of metabolic hormones in rabbits under heat stress in egypt. Zagazig Vet. J. 2020, 48, 284–295. [Google Scholar] [CrossRef]
- Ramakrishnan, S. Textbook of Medical Biochemistry; Orient Blackswan: Hyderabad, India, 2004. [Google Scholar]
- Abdel-Khalek, A.E.; Kalaba, Z.M.; El-Gogary, M.R. Functional, anatomical and histological development of caecum in rabbits. Curr. Res. Poult. Sci. 2011, 1, 54–65. [Google Scholar] [CrossRef]
- Takeuchi, A.I.L.R.; O’Hanley, P.D.; Cantey, J.R.; Lushbaugh, W.B. Scanning and transmission electron microscopic study of escherichia coli 015 (rdec-1) enteric infection in rabbit. Infect. Immun. 1978, 19, 686–694. [Google Scholar]
- Fu, X.; Zeng, B.; Wang, P.; Wang, L.; Wen, B.; Li, Y.; Liu, H.; Bai, S.; Jia, G. Microbiome of total versus live bacteria in the gut of rex rabbits. Front. Microbiol. 2018, 9, 733. [Google Scholar] [CrossRef]
- Xiang, X.-D.; Deng, Z.-C.; Wang, Y.-W.; Sun, H.; Wang, L.; Han, Y.-M.; Wu, Y.-Y.; Liu, J.-G.; Sun, L.-H. Organic acids improve growth performance with potential regulation of redox homeostasis, immunity, and microflora in intestines of weaned piglets. Antioxidants 2021, 10, 1665. [Google Scholar] [CrossRef]
- Xu, C.; Yang, S.; Zhu, L.; Cai, X.; Sheng, Y.; Zhu, S.; Xu, J. Regulation of n-acetyl cysteine on gut redox status and major microbiota in weaned piglets. J. Anim. Sci. 2014, 92, 1504–1511. [Google Scholar] [CrossRef]
- Li, H.; Shang, Z.; Liu, X.; Qiao, Y.; Wang, K.; Qiao, J. Clostridium butyricum alleviates enterotoxigenic escherichia coli k88-induced oxidative damage through regulating the p62-keap1-nrf2 signaling pathway and remodeling the cecal microbial community. Front. Immunol. 2021, 12, 771826. [Google Scholar] [CrossRef]
- Adewole, D.; Akinyemi, F. Gut microbiota dynamics, growth performance, and gut morphology in broiler chickens fed diets varying in energy density with or without bacitracin methylene disalicylate (bmd). Microorganisms 2021, 9, 787. [Google Scholar] [CrossRef]
- Hollister, E.B.; Gao, C.; Versalovic, J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014, 146, 1449–1458. [Google Scholar] [CrossRef]
- Venegas, D.P.; De La Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (scfas)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 1486. [Google Scholar] [CrossRef]
- Gosalbes, M.J.; Vázquez-Castellanos, J.F.; Angebault, C.; Woerther, P.-L.; Ruppé, E.; Ferrús, M.L.; Latorre, A.; Andremont, A.; Moya, A. Carriage of enterobacteria producing extended-spectrum β-lactamases and composition of the gut microbiota in an amerindian community. Antimicrob. Agents Chemother. 2016, 60, 507–514. [Google Scholar] [CrossRef]
- Wertz, J.T.; Kim, E.; Breznak, J.A.; Schmidt, T.M.; Rodrigues, J.L. Genomic and physiological characterization of the verrucomicrobia isolate diplosphaera colitermitum gen. Nov., sp. Nov., reveals microaerophily and nitrogen fixation genes. Appl. Environ. Microbiol. 2012, 78, 1544–1555. [Google Scholar] [CrossRef]
- Schneeberger, M.; Everard, A.; Gómez-Valadés, A.G.; Matamoros, S.; Ramírez, S.; Delzenne, N.M.; Gomis, R.; Claret, M.; Cani, P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643. [Google Scholar] [CrossRef]
- Linaje, R.; Coloma, M.; Pérez-Martínez, G.; Zúñiga, M. Characterization of faecal enterococci from rabbits for the selection of probiotic strains. J. Appl. Microbiol. 2004, 96, 761–771. [Google Scholar] [CrossRef]
- Andrejčáková, Z.; Sopková, D.; Vlčková, R.; Hertelyová, Z.; Gancarčíková, S.; Nemcová, R. The application of lactobacillus reuteri ccm 8617 and flaxseed positively improved the health of mice challenged with enterotoxigenic E. coli o149: F4. Probiotics Antimicrob. Proteins 2020, 12, 937–951. [Google Scholar] [CrossRef]
- Velasco-Galilea, M.; Piles, M.; Viñas, M.; Rafel, O.; González-Rodríguez, O.; Guivernau, M.; Sánchez, J.P. Rabbit microbiota changes throughout the intestinal tract. Front. Microbiol. 2018, 9, 2144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).