- Review
Transplacental Antibody Transfer: Mechanisms, Pregnancy-Related Disruptions, and Emerging Experimental Models
- Qiqi Li,
- Zhengyuan Huang and
- Nishel M. Shah
- + 3 authors
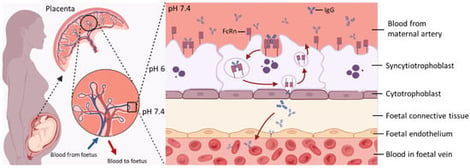
The transplacental transfer of maternal immunoglobulin G from the mother to the foetus is central for providing immunity in early life, resulting in full-term newborns having IgG repertoires and levels similar to those of their mothers. The neonatal Fc receptor is recognised as the primary transporter of IgGs across the placental epithelium. Understanding the mechanisms of transplacental antibody transfer and factors that affect them is essential in optimising maternal vaccination strategies, ultimately protecting infants from various environmental pathogens. This review first outlines the biological mechanisms governing transplacental IgG transfer, followed by a discussion of how this process may be disrupted by physiological and pathological conditions during pregnancy, including preterm birth, hypergammaglobulinemia, maternal pathogenic IgG, maternal infections, hyperglycaemia, and exposure to biological therapies. We also summarise currently available models used to study transplacental IgG transfer, highlighting existing knowledge gaps and future directions for research in this field.
6 February 2026