Abstract
Common bean is one of the most important legume crops for human consumption. Its yield is adversely affected by environmental stress. Plant non-specific lipid transfer proteins (nsLTPs) are essential for plant growth, development, and resistance to abiotic stress, such as salt, drought, and alkali. However, changes in nsLTP family genes responding to drought stress are less known. The PvLTP gene family in the common bean was identified by a comprehensive genome-wide analysis. Molecular weights, theoretical isoelectric points, phylogenetic tree, conserved motifs, gene structures, gene duplications, chromosome localization, and expression profiles were analyzed by SignalP 5.0, ExPASy, ClustalX 2.1, MEGA 7.0, NCBI-CDD, MEME, Weblogo, and TBtools 1.09876, respectively. Heatmap and qRT-PCR analyses were performed to validate the expression profiles of PvLTP genes in different organs. In addition, the expression patterns of nine PvLTP genes in common beans treated with drought stress were investigated by qRT-PCR. We obtained 58 putative PvLTP genes in the common bean genome via genome-wide analyses. Based on the diversity of the eight-cysteine motif (ECM), these genes were categorized into five types (I, II, IV, V, and VIII). The signal peptides of the PvLTP precursors were predicted to be from 16 to 42 amino acid residues. PvLTPs had a predicated theoretical isoelectric point of 3.94–10.34 and a molecular weight of 7.15–12.17 kDa. The phylogenetic analysis showed that PvLTPs were closer to AtLTPs than OsLTPs. Conserved motif and gene structure analyses indicated that PvLTPs were randomly distributed on all chromosomes except chromosome 9. In addition, 23 tandem duplicates of PvLTP genes were arranged in 10 gene clusters on chromosomes 1 and 2. The heatmap and qRT-PCR showed that PvLTP expression significantly varied in different tissues. Moreover, 9 PvLTP genes were up-regulated under drought treatment. Our results reveal that PvLTPs play potentially vital roles in plants and provide a comprehensive reference for studies on PvLTP genes and a theoretical basis for further analysis of regulatory mechanisms influencing drought tolerance in the common bean.
1. Introduction
The common bean (Phaseolus vulgaris L.) is an important legume used as food because it is a major source of proteins, minerals, and vitamins [1]. Thus, it is consumed as part of the traditional diets in Europe (e.g., the Mediterranean region) and the Middle East [2]. The increase in living standards and growing demand for healthy food have necessitated increases in common bean yields. Because of its significant economic and production value, the common bean has become one of the most profitable cultivated legumes in the worldwide market over the last two decades. However, its growth and yield are restricted by biotic and abiotic stresses, such as anthracnose, fusarium wilt, drought, and salt [3]. The yield of common bean decreased the most among all legumes under drought stress [2,4,5]. According to Daryanto et al., the common bean needs to be improved for drought resistance due to its significance in world production and human nutrition [4]. A few studies identified the functions and mechanisms of drought resistance-related genes in the common bean [6,7,8]. The transcription of PvXIP1;2, which encoded a drought-related aquaporin, was strongly induced by drought stress. Moreover, the overexpression of PvXIP1;2 in Arabidopsis increased the survival rate and proline content but decreased ion leakage and the malondialdehyde content. Furthermore, compared with control roots, PvXIP1;2-overexpressing common bean hairy roots reportedly had significantly higher water contents and growth rates [6]. The expression of another aquaporin gene (PvTIP1;1) was quickly down-regulated in drought-stressed leaves but returned to normal or higher levels after rehydration, suggesting PvTIP1;1 might be associated with high water permeability. Hence, the down-regulated expression of this gene may help prevent water loss, thereby protecting plants from the detrimental effects of drought [7,8]. Therefore, there is an urgent need to screen for drought-resistance genes, which upon modification, could improve the common bean’s resistance to drought stress by adjusting its growth, metabolism, and cellular structures.
Plant non-specific lipid transfer proteins (nsLTPs) are small molecular proteins that transport hydrophobic compounds, including acyl-coenzyme A, glycolipids, and fatty acids [9,10,11,12]. The N-terminus of nsLTP contains a signal peptide sequence, which guides nsLTP proteins that are being secreted to the cytoplasm [13]. Eight-cysteine motifs (ECM, C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C) in mature nsLTP proteins form four internal disulfide bonds, producing a stable hydrophobic tertiary structure [14]. This structure can bind different hydrophobic compounds and lipid molecules in cells [15,16].
Plant nsLTP belongs to a multi-gene family and is classified into two types: LTP1 and LTP2 [9]. LTP1 has a molecular weight of about 9 kDa, 90–95 amino acid residues in mature proteins, and a signal peptide with 21–27 amino acid residues. The molecular weight of LTP2 is about 7 kDa, with approximately 70 and 27–35 amino acid residues for mature proteins and the signal peptide, respectively [17,18]. Increasing nsLTPs have been identified, thus rendering their traditional classification methods insufficient. Boutrot et al. reclassified nsLTP into nine types (I–IX) based on the ECM interval characteristics and amino acid sequence similarity [19]. The X and XI types were also added recently [20,21]. Following the development of genome sequencing technology, the nsLTP gene family has been characterized in various plants. For instance, Arabidopsis, rice, and Nicotiana tabacum have 45 members in eight types (I–VI, VIII, and IX), 49 in eight types (I–VIII), and 100 in six types (I, II, IV, V, VII, and VIII), respectively [19,22].
Studies showed that nsLTPs participated in various biochemical and physiological pathways of plant growth and development, including signal transduction, keratin synthesis, anther development, and seed maturation [23,24,25]. Additionally, nsLTPs also played regulatory roles in plant response to abiotic stresses and defense against pathogens. Some researchers attributed nsLTPs to plant pathogenesis-related proteins of the PR-14 family [26,27,28,29]. Recently, researchers have focused on plant resistance to abiotic stresses. Overexpression of AZI1, a member of the nsLTP family in Arabidopsis, resulted in a salt-tolerant phenotype. Further research showed that AZI1 combined with mitogen-activated protein kinase 3 (MPK3) forms a complex, which MPK3 positively regulates to resist salt stress [30]. Rice OsDIL1 and foxtail millet SiLTP were induced by drought, salt, and abscisic acid (ABA) [31,32]. Overexpressing SiLTP could enhance drought and high-salt stress resistance, and the RNA interference of SiLTP led to sensitive phenotypes [32]. In drought-tolerant wheat lines, the expression of the TdLTP4 gene was significantly higher than that of drought-sensitive lines. The heterologous expression of TdLTP4 in Arabidopsis led to tolerance to abiotic and biotic treatments [33]. Because of their association with stress resistance-related transcription factors, researchers believe that nsLTP can enhance stress resistance in plants.
Until now, only four nsLTP genes have been found in the common bean. Among these, the expression of PvLTP24 increased significantly in stems and leaves following drought and ABA treatments [34]. The apical cortex specifically expressed PVR3 in the root tip, which could be used as a marker gene for cortex development [35,36]. Additionally, PvLTP1a and PvLTP1b are the two major food allergens, while the functions of the other common bean nsLTP genes are still unclear [37]. We utilized bioinformatics methods to identify the common bean’s PvLTP gene family. Chemical and physical properties, as well as a phylogenetic tree, conserved motifs, gene structure, and chromosome location, were analyzed. Moreover, a qRT-PCR assay was used to analyze the expression levels of different PvLTPs in organs and drought stress. Our findings will serve as a foundation for gene function and mechanism studies of the common bean PvLTPs in response to drought stress.
2. Materials and Methods
2.1. Identification and Bioinformatic Analyses of PvLTP Family in Common Bean
The genome and proteome sequences of common bean, Oryza sativa, and Arabidopsis thaliana were downloaded from Phytozome (http://phytozome.jgi.doe.gov/, accessed on 1 March 2021), RAP (https://rapdb.dna.affrc.go.jp/, accessed on 1 March 2021), and TAIR (https://www.arabidopsis.org/, accessed on 1 March 2021) databases, respectively [38,39,40]. The NsLTP sequence ID lists of Arabidopsis and rice were obtained from Boutrot et al. [19]. Candidate nsLTP genes of the common bean were obtained using BLASTp with AtLTP and OsLTP sequences as queries to blast against the common bean protein database with an E-value of 1 × 10−5. Proteins with the HMM domain PF00234 were further retrieved using HMMER 3.3 with default parameters to avoid missing nsLTP genes. After eliminating redundant sequences, all proteins of putative nsLTP genes lacking the essential ECM domain were removed manually. Subsequently, the signal peptide cleavage sites were analyzed using Signal 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0, accessed on 5 March 2021). Proteins without N-terminal signal sequences and the deduced hybrid proline-rich proteins were discarded. Moreover, the rest of the candidate sequences were subjected to BLASTp to exclude potential α-amylase inhibitors and cereal storage proteins using RATI and 2S-albumin as queries, respectively [41,42]. The remaining predicted proteins were uploaded to the NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd, accessed on 7 March 2021) and Pfam (http://pfam.xfam.org/, accessed on 7 March 2021) websites to identify the conserved LTP domain. Finally, the mature proteins containing more than 120 amino acid residues were eliminated. The identified PvLTPs were named according to Boutrot’s method and their orders on chromosomes [19].
2.2. Sequence Alignment and Phylogenetic Analysis
The ECM domains of PvLTPs from Arabidopsis, rice, and the common bean were subjected to multiple alignments by ClustalX 2.1 software with default parameters [43]. After that, they were utilized to build a phylogenetic tree based on the Neighbor-Joining model with MEGA 7.0 software [44,45]. The phylogenetic tree was visualized with Figtree 1.4.4 (http://tree.bio.ed.ac.uk/software/Figtree/, accessed on 8 March 2021). For statistical reliability, bootstrap tests were computed with 1000 replications.
2.3. Conserved Motifs and Gene Structure Analysis
To identify conserved motifs, PvLTPs were uploaded to the MEME (https://meme-suite.org/meme/tools/meme, accessed on 10 March 2021) website with the motif width setting as 6–50 amino acid residues and the maximum motif number setting as 10 [46]. Gene structures were analyzed and visualized using TBtools 1.09876 [47]. The WebLogo tool (http://weblogo.threeplusone.com/, accessed on 10 March 2021) was performed to draw sequence logos of the conserved ECM domain [48].
2.4. Chromosomal Localization and Gene Duplication
The common bean v2.1 gff3 file was retrieved from the Phytozome website [38,49]. The relative distances and positions of PvLTPs were obtained according to the genome annotation information and drafted on the 11 chromosomes using TBtools 1.09876 [47]. The duplication events of PvLTP gene family members in the common bean were identified using TBtools 1.09876 with the default parameters.
2.5. Plant Materials
Common bean cultivars (Pinjinyun 3) were planted in the field at the Center for Agricultural Genetic Resources Research at Shanxi Agricultural University (China, Shanxi, Latitude 112°58′ E, 37°78′ N, Figure S1). The roots, stems, leaves, flowers, seeds, and pods were collected and fast-frozen in liquid nitrogen before being preserved at −80 °C. Moreover, the cultivar seeds were cultivated at 25 °C with 14 h of light and 20 °C with 10 h of darkness in a growth chamber (Figure S2). The seedlings were treated with 20% PEG6000 for 24 h at the trifoliate leaves stage. The leaves were collected after 0, 6, 12, and 24 h under drought treatment, and then samples were stored at −80 °C before RNA extraction. For each organ, three separate biological replicates and technical replicates were performed.
2.6. Quantitative Real-Time PCR Analysis
Total RNA was extracted from roots, stems, flowers, leaves, seeds, and pods using SV Total RNA Isolation System (Promega, Madison, WI, USA) as directed by the manufacturer’s instruction. Precisely 1 μg of total RNA was reverse-transcribed by PrimeScript RT Master Mix (TaKaRa, Otsu, Japan). Then, the cDNA was amplified on a Quantstudio 6 thermal cycler (Applied Biosystems, Waltham, MA, USA) using TB Green Premix Ex Taq II (TaKaRa, Otsu, Japan). The qRT-PCR amplification procedure was 95 °C for 30 s, performed with 40 cycles of 95 °C for 5 s and 60 °C for 34 s. We used the common bean Actin gene as a reference control. For each experiment, three separate technical and biological replicates were performed. The 2−ΔΔCt analysis method was used to calculate the relative transcription levels (Table S1) [50]. All primers are listed in Table S2.
3. Results
3.1. Genome-Wide Identification of Putative nsLTPs in Common Bean
To investigate all putative nsLTPs in the common bean, we performed BLASTp using AtLTP and OsLTP sequences as queries to search against the common bean proteome database. A total of 73 candidate nsLTP genes were obtained. Another ten nsLTP genes were retrieved from the HMM domain profiles of PF00234 and PF14368. After integrating the above results, three proteins lacking the essential ECM domain were manually removed. In addition, six proteins without N-terminal signal sequences were identified using Signal 5.0 and were deleted. Five hybrid proline-rich proteins were not taken into consideration. Proteins with high similarity to α-amylase inhibitors or cereal storage proteins were not found. The remaining 69 candidate protein sequences were uploaded to the Pfam and NCBI-CDD websites to identify the conserved LTP domains. All candidates possessed the LTP domains. Because all mature nsLTPs have low molecular weights, eleven of the sixty-nine candidates with more than 120 amino acid residues were discarded. As a result, 58 genes coding nsLTPs were designated as PvLTPs.
3.2. Classification and Sequence Analysis of PvLTPs
PvLTPs were classified using the sequence similarity method. The results indicated that the 58 PvLTP genes were categorized into five types, namely types I (PvLTPI.1-45), II (PvLTPII.1-5), IV (PvLTPIV.1-5), V (PvLTPV.1-2), and VIII (PvLTPVIII.1) (Table 1). No PvLTP gene was categorized into types III, VI, or VII. Obviously, the ECM domains were highly conservative in structure, except for the number of variable inter-cysteine residues (Table 2). Similar to rice and potato, the common bean genome contained the largest proportion of type I PvLTP genes (77.59%, 45 out of 58).

Table 1.
Putative PvLTPs identified in the common bean.

Table 2.
Diversity of EMC in five types of PvLTPs.
The characteristics of PvLTP genes and proteins, such as coding sequence (CDS) length, signal peptide and protein sequences, were analyzed using biological software. The length of CDS ranged from 246 (PvLTPII.2) to 465 bp (PvLTPVIII.1). All PvLTP precursors had a signal peptide varying in length from 16 (PvLTPI.8-10, PvLTPI.14) to 42 (PvLTPVIII.1) amino acid residues. Mature PvLTPs were usually small and had a low molecular weight ranging from 7.15 (PvLTPII.5) to 12.17 kDa (PvLTPVIII.1). The predicted theoretical isoelectric point varied from 3.94 (PvLTPI.45) to 10.34 (PvLTPI.45). The mean molecular weight was 9.82 kDa, and the isoelectric point was 7.52 (Table 1).
3.3. Phylogenetic Analysis of PvLTPs
To demonstrate the evolutionary relationship of nsLTP genes among the common bean, Arabidopsis, and rice, phylogenetic analysis was completed using ClustalX 2.1 and MAGA 7.0 software. As shown in Figure 1, the 152 nsLTP genes were divided into nine distinct clades, with 77, 33, 5, 14, 9, 8, 1, 3, and 2 nsLTP genes from types I–IX in each clade, respectively. Obviously, the largest portions of common bean PvLTPs and Arabidopsis AtLTPs were clustered together in clade I, whereas rice OsLTPs were mainly gathered in clade II. In addition, PvLTPs were closer to AtLTPs than OsLTPs in each clade of the phylogenetic tree. The cluster separation revealed a closer genetic lineage-specific relationship between the common bean and Arabidopsis during dicotyledon evolution.
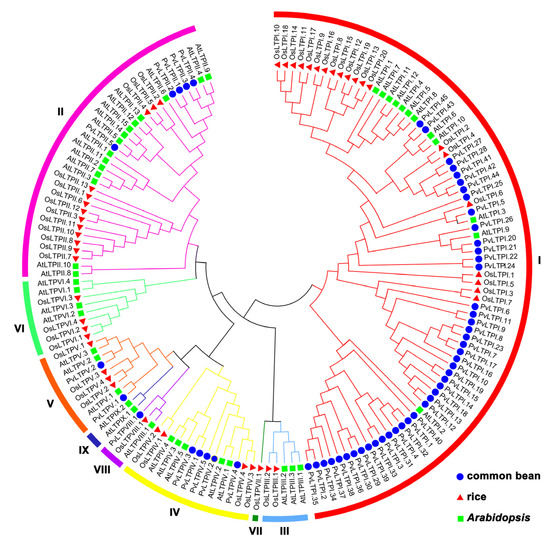
Figure 1.
Phylogenetic tree of nsLTP proteins from the common bean, rice, and Arabidopsis. Nine nsLTP types are marked using different colors. Roman numbers indicate corresponding types of nsLTP family.
3.4. Conserved Motifs and Structure of PvLTPs
CDD and Weblogo websites showed the conserved domains in the PvLTP gene family (Figure 2). PvLTP family members contained an ECM domain, which belonged to the α-amylase inhibitors and lipid transfer/seed storage proteins (AAI-LTSS) superfamily. A total of ten distinct conserved motifs (motif1–motif10) were identified using MEME tools (Figure 3) to analyze the relationship between the structural features and diversity of PvLTP members further. All PvLTP family members contained motif3 with highly conserved amino acid sequences. Moreover, motif1 and motif4 were presented in type I PvLTPs (except PvLTPI.6), while motif7 and motif10 were presented in type II PvLTPs. The results revealed that motif3 was conserved within the five PvLTP types.
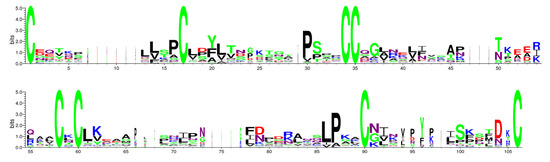
Figure 2.
Conserved domain analysis of PvLTPs using the WebLogo website. The degree of conservation is indicated by the height of each amino acid. The number on the x-axis indicates its position in the ECM. The information measured is represented in bits on the y-axis.
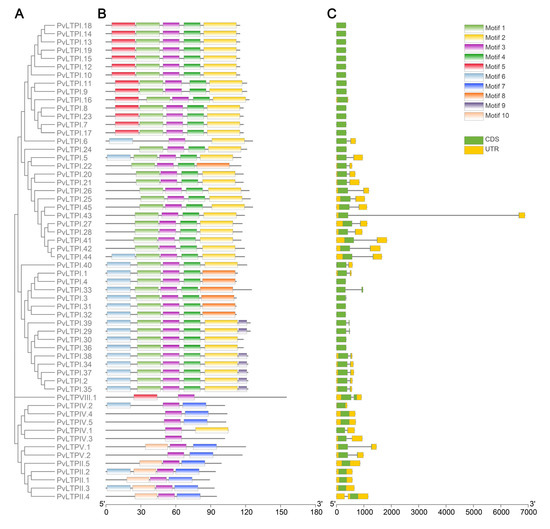
Figure 3.
Phylogenetic tree, conserved motifs and gene structures of PvLTPs. (A) A Neighbor-Joining phylogenetic tree was generated with 1000 bootstrap replicates based on ECM domains of fifty-eight PvLTP proteins. (B) The conserved motifs are shown. A total of ten distinct conserved motifs were depicted using various colored boxes. (C) The gene structures of the PvLTPs were visualized. The yellow boxes, green boxes, and black lines indicate UTRs, exons, and introns, respectively.
The amount and distribution of exons and introns in PvLTPs were examined to investigate the components of PvLTP gene structure. Among the 58 PvLTP genes, 29 contained one intron and two exons, including 25 type I, one type IV, two type V, and one type VIII PvLTPs. The other 29 had no introns. Interestingly, all type II PvLTP genes had no introns, consistent with the findings in cotton and potato. These findings indicated that the PvLTP gene family had been conserved in plant evolution.
3.5. Chromosomal Localization and Gene Duplication of PvLTPs
To better obtain the distribution and exact position of PvLTPs on chromosomes, a detailed chromosome map was constructed. The 58 PvLTP genes were unevenly dispersed on ten chromosomes (Figure 4). Among them, twenty, seventeen, five, four, four, three, one, one, and one PvLTPs were located on chromosomes 8, 6, 10, 1, 7, 4, 2, 3, and 11, respectively, while there was no PvLTP on chromosome 9.
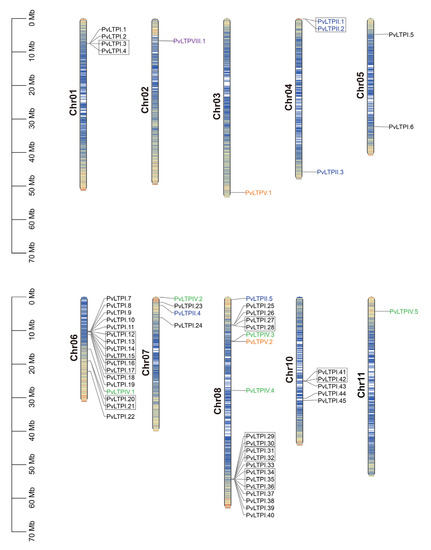
Figure 4.
Chromosomal localization and tandem-duplicate genes of PvLTP family from the common bean. PvLTP types are marked with different colors. The tandem-duplicate genes are depicted with a black rectangle. Chromosomal distances are given in megabase (Mb).
Gene duplication analysis showed that, among the 58 PvLTPs, 23 genes were arranged in ten gene tandem duplicate clusters and distributed on two chromosomes. Among the ten gene clusters, nine clusters contained 21 genes and were located on chromosome 1, and the other cluster was located on chromosome 2. The large proportion (39.66%) of tandem duplicate clusters indicated that tandem duplications might cause PvLTP gene family expansion in the common bean genome.
3.6. Expression Pattern of PvLTPs in Different Organs
The expression profile of PvLTPs in different organs was visualized in a heatmap using RNA-seq data from Phyzotome to investigate the functions of PvLTPs during common bean growth. Figure 5 showed that 19 PvLTPI genes were highly expressed in root nodules but lower in other tissues. Moreover, the rest of the PvLTP genes showed a diverse expression profile in different tissues.
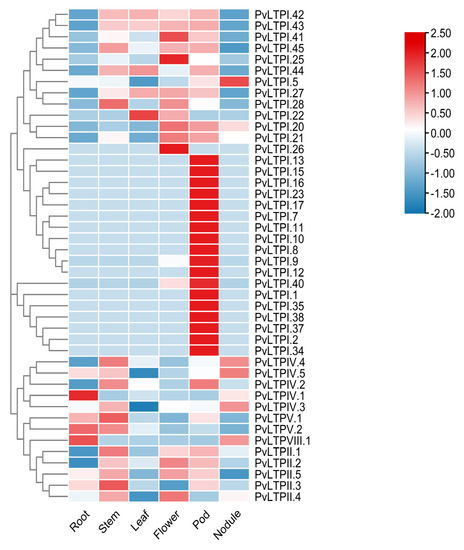
Figure 5.
Heatmap of the PvLTP genes expression profiles in different organs. Blue, light blue, colorless, light red and red are used to show the differentially expressed levels. FPKM is used as the gene expression value unit.
To further validate the above results, the expression of nine PvLTPs in roots, stems, leaves, flowers, seeds, and pods was detected using qRT-PCR (Figure 6). PvLTPI.27, PvLTPI.28, and PvLTPI.42 were highly expressed in flowers, while PvLTPI.44 and PvLTPV.1 were highly expressed in seeds, consistent with the results shown in the heatmap. In addition, PvLTPII.3 and PvLTPV.2 were predominantly expressed in stems, while PvLTPI.45 was strongly expressed in pods. These findings suggested that the differentially expressed PvLTPs might play various biological functions in the growth and development of the common bean.
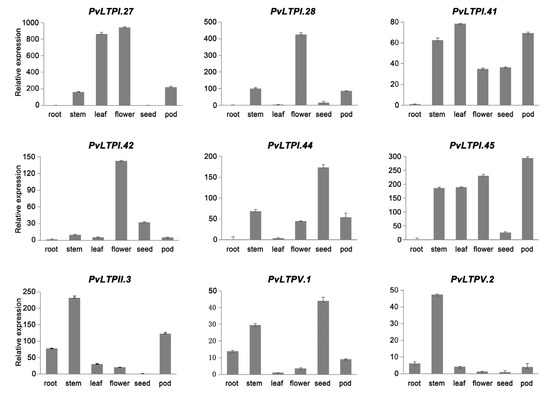
Figure 6.
Expression levels of PvLTPs in six tissues by qRT-PCR. The internal reference gene used was Actin. Error bars reflect the standard deviation..
3.7. Expression Analysis of PvLTPs under Drought Stress
Abiotic environmental conditions have direct impacts on plant growth. Therefore, it is significant to analyze how relatively resistant genes change under different stresses. The expression levels of 9 PvLTPs after drought treatment were examined using qRT-PCR to explore the functions of PvLTPs involved in drought stress. The results showed that all PvLTPs were up-regulated after drought stress (Figure 7). The relative expression levels of PvLTPI.28, PvLTPI.41, PvLTPI.42, PvLTPI.44, PvLTPI.45, and PvLTPII.3 showed a similar trend of first increasing and then decreasing, reaching the maximum at 12 h. PvLTPI.44 especially exhibited more than 115-fold up-regulation. Moreover, the relative expression levels of the other three PvLTPs, PvLTPI.27, PvLTPV.1, and PvLTPV.2 showed a trend of gradual increase and reached the highest of 37-, 78-, and 142-fold up-regulation at 24 h compared with the control, respectively. These findings displayed that PvLTPs might function as important positive regulators influencing plant tolerance towards drought stress.
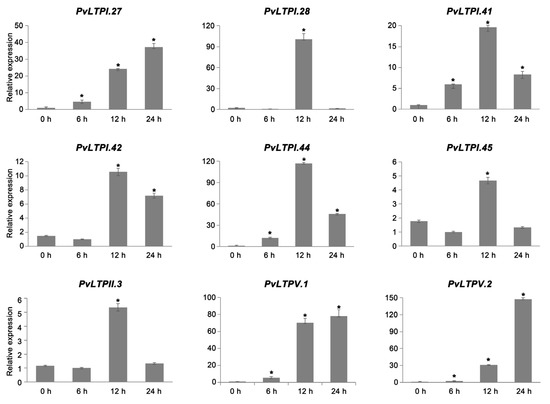
Figure 7.
Expression levels of PvLTPs under drought treatment at 0, 6, 12, and 24 h by qRT-PCR. The internal reference gene used was Actin. Error bars reflect the standard error. A significant difference is represented by an asterisk (* p < 0.05).
4. Discussion
Plants have different types and numbers of the nsLTP gene family. Brassica rapa has nine types (I–VI, VIII, IX, and XI) with 63 members, while Six Solanaceae species have five types (I, II, IV, IX, and X) with 135 members [20,21]. In this work, whole-genome identification of the common bean obtained 58 PvLTP family members unevenly distributed across ten chromosomes, except for chromosome 9. We found that these PvLTP family members could be categorized into five types (I, II, IV, V, and VIII). Nevertheless, the absence of some types indicated deletion during the evolution of the common bean. The phylogenetic tree results showed that the common bean PvLTPs were closer to Arabidopsis AtLTPs than rice OsLTPs. The relationship distance between the dicotyledons and monocotyledons in the plants’ classification was consistent with the previous studies [51], indicating that the nsLTP genes existed before the separation of monocotyledonous plants and were conserved during evolution. The internal structures of types I and II had similarities, suggesting that the same category of genes may have similar biological functions.
Studying gene expression patterns can provide a theoretical basis for gene functions. Results have shown that the nsLTP gene responded to drought and other abiotic stresses. In Arabidopsis, LTP3 was highly expressed after 6 h of drought treatment. AtLTP3 overexpression significantly enhanced tolerance to drought stress. Further studies showed that the upstream transcription factor, MYB96, regulated the expression of AtLTP3 by binding to the AtLTP3 promoter, thus enabling AtLTP3 to participate in the drought resistance signaling pathway [26]. A comparison between AtLTP3 and PvLTPI.45 revealed that their amino acid sequences are approximately 41.53% similar. Additionally, AtLTP3 was expressed at high levels in the leaves, flowers, and siliques, with peak levels detected in the siliques. The PvLTPI.45 expression pattern was similar to that of AtLTP3, with the highest expression level detected in the pods, followed by the flowers and leaves. Under drought conditions, the AtLTP3 and PvLTPI.45 expression levels increased by about 4-fold and then decreased, suggesting they may have similar functions in plant responses to drought stress. Several findings indicated that the expression levels of nsLTP genes, including OsDIL (rice), ScLTP (sugarcane), and NtLTP4 (tobacco), were up-regulated or down-regulated to varying degrees after drought, salt, and other abiotic stress treatments [31,52,53]. For example, StnsLTP1 expression in potatoes increases in response to excessive heat, salt, and drought. The overexpression of StnsLTP1 in potatoes led to enhanced tolerance to abiotic stresses because it activated antioxidative defense mechanisms and up-regulated the expression of stress-related genes [54]. These observations imply that nsLTP proteins are involved in multiple stress-induced signaling pathways. In this research, the expression levels of PvLTPs were increased to varying degrees after the drought stress treatment. For instance, PvLTPI.44 and PvLTPV.2 increased 115 and 142 times, respectively, compared with the control. Therefore, it can be speculated that PvLTPs play a vital function in drought stress response.
Gene tandem duplication during genomic DNA replication and recombination is the key driver for gene family amplifications [55,56,57]. In Arabidopsis and rice genomes, 15–20% of genes were composed of tandem repeats of gene clusters considered crucial for evolution, plant disease resistance, and abiotic stress responses [58]. Several tandem repeats of genes were found in the nsLTP gene family of angiosperms, including 47.82% (66/138) in cotton, 51.43% (36/70) in barley, and 53.01% (44/83) in potato [59,60,61]. This indicated that tandem repeats played essential roles in gene amplification during the evolution of the nsLTP gene family. In this work, chromosome mapping and gene structure revealed that the gene replication events occurred during genome amplification and evolution of the common bean. Twenty-three genes of the PvLTP gene family were categorized into 10 tandem repeats of gene clusters on chromosomes 1 and 2, accounting for 39.66% of the family members. Most of the genes exhibiting the same tandem repeats had a high similarity, suggesting that they might have functional similarities. For instance, the nucleic acid sequence similarity of PvLTPI.12/PvLTPI.13/PvLTPI.14/PvLTPI.15 was 98.62%, and the protein similarity was 96.71%. However, the nucleic acid sequence similarity of PvLTPI.41/PvLTPI.42 was only 52.18%, with a protein similarity of 58.47%. This implied that, although the PvLTP family was derived from the same ancestor, it changed when plants adapted to the external environment during evolution. Moreover, we found that PvLTPI.42 was more highly expressed in flowers than in other tissues, while the PvLTPI.41 expression was higher in the tissues except for roots. The different expression patterns suggest that the evolution of duplicate genes might lead to the emergence of functional diversity.
5. Conclusions
In summary, whole-genome identification of the common bean identified 58 members of the PvLTP gene family, which were divided into five types (I, II, IV, V, and VIII). Each member contained the conserved ECM domain. PvLTP genes were randomly distributed on ten chromosomes, except for chromosome 9, of which 23 members formed ten sets of tandemly repeating gene clusters. The organ-specific expression profiles of nine PvLTP genes differed significantly. In addition, exposure to drought stress up-regulated the expression of these genes. These findings provide important insights into the PvLTP genes in the common bean and form the theoretical basis of future research on PvLTP functions related to plant development and stress responses. Furthermore, our study will contribute to investigating the mechanisms underlying the effects of nsLTPs on plant responses to abiotic stresses.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13122394/s1, Table S1: The Ct value of qRT-PCR used in this study. Table S2: qRT-PCR primers of PvLTP genes used in this study. Figure S1: The geo-tagged photograph of the common bean cultivar in the field. Figure S2: The geo-tagged photograph of the seedlings grown in the growth chamber.
Author Contributions
Conceptualization, J.C. and X.D.; Methodology, X.D. and H.Z.; Software, X.D.; Formal analysis, X.D. and X.H.; Data curation, H.Z. and X.M.; Writing—original draft preparation, X.D. and H.Z.; Writing—review and editing, J.Z. and X.H.; Visualization, X.D., Y.W. and X.H.; Supervision, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Shanxi Province Science Foundation for Youths (201901D211561), Shanxi Academy of Agricultural Science Doctoral Research Fund (YBSJJ2003), Research Program Sponsored by National Laboratory of Minor Crops Germplasm Innovation and Molecular Breeding (in preparation) (202204010910001), China Agriculture Research System of MOF and MARA-Food Legumes (CARS-08), and the National Key Research and Development Program of China (2021YFD1600601).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gepts, P.; Aragão, F.J.L.; De Barros, E.; Blair, M.W.; Brondani, R.; Broughton, W.; Galasso, I.; Hernández, G.; Kami, J.; Lariguet, P.; et al. Genomics of Phaseolus Beans, a Major Source of Dietary Protein and Micronutrients in the Tropics. In Genomics of Tropical Crop Plants; Plant Genetics and Genomics: Crops and Models; Moore, P.H., Ming, R., Eds.; Springer: New York, NY, USA, 2018; Volume 3, pp. 113–134. [Google Scholar] [CrossRef]
- De Ron, A.M.; Kalavacharla, V.; Álvarez-García, S.; Casquero, P.A.; Carro-Huelga, G.; Gutiérrez, S.; Lorenzana, A.; Mayo-Prieto, S.; Rodríguez-González, A.; Suárez-Villanueva, V.; et al. Common Bean Genetics, Breeding, and Genomics for Adaptation to Changing to New Agri-environmental Conditions. In Genomic Designing of Climate-Smart Pulse Crops; Kole, C., Ramanna, R., Eds.; Springer Nature: Cham, Switzerland, 2019; Volume 1, pp. 1–28. [Google Scholar] [CrossRef]
- Miklas, P.N.; Singh, S.P. Common Bean. In Pulses, Sugar and Tuber Crops; Genome Mapping and Molecular Breeding in Plants; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 3, pp. 1–31. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global synthesis of drought effects on food legume production. PLoS ONE 2015, 10, e0127401. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Neelapu, N.R.R.; Wani, S.H.; Challa, S. Response of Pulses to Drought and Salinity Stress Response: A Physiological Perspective. In Pulse Improvement; Wani, S., Jain, M., Eds.; Springer: Cham, Switzerland, 2018; pp. 77–98. [Google Scholar] [CrossRef]
- Wu, L.; Chang, Y.; Wang, L.; Wang, S.; Wu, J. The aquaporin gene PvXIP1;2 conferring drought resistance identified by GWAS at seedling stage in common bean. Theor. Appl. Genet. 2022, 135, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Ariani, A.; Gepts, P. Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris L.). Mol. Genet. Genom. 2015, 290, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- Zupin, M.; Sedlar, A.; Kidrič, M.; Meglič, V. Drought-induced expression of aquaporin genes in leaves of two common bean cultivars differing in tolerance to drought stress. J. Plant Res. 2017, 130, 735–745. [Google Scholar] [CrossRef]
- Kader, J.C. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 627–654. [Google Scholar] [CrossRef]
- Carvalho, A.O.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology—A concise review. Peptides 2007, 28, 1144–1153. [Google Scholar] [CrossRef]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? JLR 2018, 59, 1374–1382. [Google Scholar] [CrossRef]
- Wong, L.H.; Gatta, A.T.; Levine, T.P. Lipid transfer proteins: The lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 2019, 20, 85–101. [Google Scholar] [CrossRef]
- Missaoui, K.; Gonzalez-Klein, Z.; Pazos-Castro, D.; Hernandez-Ramirez, G.; Garrido-Arandia, M.; Brini, F.; Diaz-Perales, A.; Tome-Amat, J. Plant non-specific lipid transfer proteins: An overview. Plant Physiol. Biochem. 2022, 15, 115–127. [Google Scholar] [CrossRef]
- José-Estanyol, M.; Gomis-Rüth, F.X.; Puigdomènech, P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 2004, 42, 355–365. [Google Scholar] [CrossRef]
- Samuel, D.; Liu, Y.J.; Cheng, C.S.; Lyu, P.C. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa). J. Biol. Chem. 2002, 277, 35267–35273. [Google Scholar] [CrossRef] [PubMed]
- Monika, M.E.; Lenita, V.; Tiina, A.S.; Johan, E. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 2011, 4, 947–964. [Google Scholar] [CrossRef]
- Castro, M.S.; Gerhardt, I.R.; Orru, S.; Pucci, P.; Bloch, C., Jr. Purification and characterization of a small (7.3 kDa) putative lipid transfer protein from maize seeds. J. Chromatogr. B 2003, 794, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.J.; Lee, C.C.; Cheng, C.S.; Lo, W.C.; Yang, W.C.; Chen, M.N.; Lyu, P.C. Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB). BMC Genom. 2012, 13, S9. [Google Scholar] [CrossRef]
- Boutrot, F.; Chantret, N.; Gautier, M.F. Genome-wide analysis of the rice and arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genom. 2008, 9, 86–105. [Google Scholar] [CrossRef]
- Liu, W.; Huang, D.; Kan, L.; Hu, S.; Yu, J.; Gang, G.; Song, S. Discovery, identification and comparative analysis of non-specific lipid transfer protein (nsLtp) family in Solanaceae. Genom. Proteom. Bioinform. 2010, 8, 229–237. [Google Scholar] [CrossRef]
- Li, J.; Gao, G.; Xu, K.; Chen, B.; Yan, G.; Li, F.; Qiao, J.; Zhang, T.; Wu, X. Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS ONE 2014, 9, e84556. [Google Scholar] [CrossRef]
- Yang, Y.; Li, P.; Liu, C.; Wang, P.; Cao, P.; Ye, X.; Li, Q. Systematic analysis of the non-specific lipid transfer protein gene family in Nicotiana tabacum reveal its potential roles in stress responses. Plant Physiol. Biochem. 2022, 172, 33–47. [Google Scholar] [CrossRef]
- Lascombe, M.B.; Bakan, B.; Buhot, N.; Marion, D.; Blein, J.P.; Larue, V.; Lamb, C.; Prangé, T. The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci. 2008, 17, 1522–1530. [Google Scholar] [CrossRef]
- Wei, K.; Zhong, X. Non-specific lipid transfer proteins in maize. BMC Plant Biol. 2014, 14, 281. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, W.; Lu, Z.; Ouyang, Y.; Yao, J. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 2015, 239, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, H.B.; Zhang, X.Y.; Yang, S.H. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, X.B.; Lu, C.M.; Zeng, X.H.; Li, Y.J.; Fu, D.H.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; De, B.C.; Cammue, B.P.; De, M.B. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gao, H.; Chu, Z.; Ji, C.; Xu, Y.; Cao, W.; Zhou, S.; Song, Y.; Liu, H.; Zhu, C. A nonspecific lipid transfer protein, StLTP10, mediates resistance to Phytophthora infestans in potato. Mol. Plant Pathol. 2021, 22, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Datta, S.; Persak, H. Salt stress in Arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 2014, 7, 722–738. [Google Scholar] [CrossRef]
- Guo, C.; Ge, X.; Ma, H. The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 2013, 82, 239–253. [Google Scholar] [CrossRef]
- Pan, Y.L.; Li, J.R.; Jiao, L.C.; Li, C.; Zhu, D.Y.; Yu, J.J. A non-specific Setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front. Plant Sci. 2016, 7, 1752. [Google Scholar] [CrossRef]
- Safi, H.; Saibi, W.; Alaoui, M.M.; Hmyene, A.; Masmoudi, K.; Hanin, M.; Brini, F. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 89, 64–75. [Google Scholar] [CrossRef]
- Colmenero-Flores, J.M.; Campos, F.; Garciarrubio, A.; Covarrubias, A.A. Characterization of Phaseolus vulgaris cDNA clones responsive to water deficit: Identification of a novel late embryogenesis abundant-like protein. Plant Mol. Biol. 1997, 35, 393–405. [Google Scholar] [CrossRef]
- Choi, D.W.; Song, J.Y.; Oh, M.H.; Lee, J.S.; Moon, J.; Suh, S.W.; Kim, S.G. Isolation of a root-specific cDNA encoding a ns-LTP-like protein from the roots of bean (Phaseolus vulgaris L.) seedlings. Plant Mol. Biol. 1996, 30, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Choi, D.W.; Lee, J.S.; Kwon, Y.M.; Kim, S.G. Cortical tissue-specific accumulation of the root-specific ns-LTP transcripts in the bean (Phaseolus vulgaris) seedlings. Plant Mol. Biol. 1998, 38, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Zoccatelli, G.; Pokoj, S.; Foetisch, K.; Bartra, J.; Valero, A.; Miguel-Moncin, M.; Vieths, S.; Scheurer, S. Identification and characterization of the major allergen of green bean (Phaseolus vulgaris) as a non-specific lipid transfer protein (Pha v 3). Mol. Immunol. 2010, 47, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Kawahara, Y.; De la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Guerche, P.; Tire, C.; De Sa, F.G.; Clercq, A.D.; Montagu, M.V.; Krebbers, E. Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 1990, 2, 469–478. [Google Scholar] [CrossRef][Green Version]
- Strobl, S.; Maskos, K.; Wiegand, G.; Huber, R.; Gomis-Rüth, F.X.; Glockshuber, R. A novel strategy for inhibition of α-amylases: Yellow meal worm α-amylase in complex with the Ragi bifunctional inhibitor at 2.5 Å resolution. Structure 1998, 6, 911. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2, 2–3. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Mei, C.; Liu, Y.; Dong, X.; Song, Q.; Wang, H.; Shi, H.; Feng, R. Genome-wide identification and characterization of the potato IQD family during development and dtress. Front. Genet. 2021, 12, 693936. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, J.; Zhang, X.; Yang, Y.; Zhou, D.; Yu, Q.; Que, Y.; Xu, L.; Guo, J. A novel non-specific lipid transfer protein gene from sugarcane (NsLTPs), obviously responded to abiotic stresses and signaling molecules of SA and MeJA. Sugar Tech 2017, 19, 17–25. [Google Scholar] [CrossRef]
- Xu, Y.; Zheng, X.; Song, Y.; Zhu, L.; Yu, Z.; Gan, L.; Zhou, S.; Liu, H.; Wen, F.; Zhu, C. NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 2018, 8, 8873. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Sajeesh, K.; Venkatesh, J.; Baskar, V.; Abhinandan, K.; Yu, J.W.; Prasad, R.; Mishra, R.K. Enhanced tolerance of transgenic potato plants over-expressing non-specific lipid transfer protein-1 (StnsLTP1) against multiple abiotic stresses. Front. Plant Sci. 2016, 7, 1228. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.S.; Yim, W.C.; Moon, J.C.; Hung, J.H.; Lee, T.G.; Lim, S.D.; Cho, S.H.; Lee, K.K.; Kim, W.; Seo, Y.W.; et al. Evolution of non-specific lipid transfer protein (nsLTP) genes in the Poaceae family: Their duplication and diversity. Mol. Genet. Genom. 2008, 279, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Yan, Y.; Liu, Z.; Chen, L.; Zhang, Y.; Li, X.; Wu, L.; Zhang, G.; Wang, X.; Ma, Z. Systematic analysisof cotton non-specific lipid transfer protein family revealed a special group that is involved in fiber elongation. Front. Plant Sci. 2018, 9, 1285. [Google Scholar] [CrossRef]
- Zhang, M.; Kim, Y.; Zong, J.; Lin, H.; Dievart, A.; Li, H.; Zhang, D.; Liang, W. Genome-wide analysis of the barley non-specific lipid transfer protein gene family. Crop J. 2019, 7, 65–76. [Google Scholar] [CrossRef]
- Li, G.; Hou, M.; Liu, Y.; Pei, Y.; Ye, M.; Zhou, Y.; Huang, C.; Zhao, Y.; Ma, H. Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genom. 2019, 20, 375. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).