Abstract
RNA sequencing (RNA-Seq)-based gene expression analysis is applicable to a wide range of biological purposes in various species. Reverse transcription quantitative PCR (RT-qPCR) is also used to assess target gene expression utilizing stably expressed reference genes as internal control under a given set of conditions. However, investigations of the reference genes for RT-qPCR normalization in the process of somatic embryogenesis (SE) initial dedifferentiation in Gossypium hirsutum are rarely reported. In this study, on the basis of our previous transcriptome data of three different induction stages during SE initial dedifferentiation process in four G. hirsutum cultivars that have different SE capability, 15 candidate genes were selected during SE initial dedifferentiation process, and their expression stability was evaluated by geNorm, NormFinder, and BestKeeper. The results indicated that the two genes of endonuclease 4 (ENDO4) and 18S ribosomal RNA (18S rRNA) showed stable expression in the four different G. hirsutum cultivars, endowing them to be appropriate reference genes during three induction stages in the four cotton cultivars. In addition, the stability and reliability of the two reference genes of ENDO4 and 18S rRNA were further verified by comparing the expressions of auxin-responsive protein 22 (AUX22) and ethylene-responsive transcription factor 17 (ERF17) between RT-qPCR results and the RNA-seq data, which showed strong positive correlation coefficient (R2 = 0.8396–0.9984), validating again the steady expression of ENDO4 and 18S rRNA as the reliable reference genes. Our results provide effective reference genes for RT-qPCR normalization during SE process in different G. hirsutum cultivars.
1. Introduction
Gene expression level analysis is crucial in many fields of biological research [1]. Currently, RNA sequencing (RNA-Seq) has become the prevalent method used to analyze the transcriptional gene expression levels in various species [2,3]. Reverse transcription quantitative PCR (RT-qPCR) is the widely applied method to quantify gene expression and to validate transcriptomic data [4,5] for its prominent advantages of high sensitivity, reproducibility, and specificity [6]. However, inaccurate quantification and low-quality RNA may lead to a decrease in RT-qPCR specificity [7]. Thus, it is necessary and crucial to screen stably expressed reference genes to normalize target gene expression level under a given set of conditions. Reliable reference genes are usually selected from the stably expressed genes that are associated with basic cellular function and cell processes, including cell structure formation, cytoskeletal protein formation, and ribosomal subunit synthesis [1,8,9].
The expression levels of some reference genes fluctuate under diverse conditions in different species. The elongation factor 1-alpha (EF1α) and beta-tubulin 3 (TUB3) would be suitable for normalizing gene expression data under salt and drought stresses, respectively, while ubiquitin carrier protein 10 (UBC10) appeared to be a better option under the coaction of the two stresses in Halostachys caspica [9]. In Caragana intermedia, hypothetical protein 2 (UNK2), protein phosphatase 2A (PP2A), and sand family protein gene (SAND) were the most suitable and stable reference genes across all tested conditions, while UNK2, SAND, and EF1α were suitable for salt-treated leaves, and UNK2 and SAND were proper for salt-treated roots, respectively. Additionally, the reference genes, tonoplast intrinsic protein 41 (TIP41) and PP2A for polyethylene glycol (PEG)- treated leaves; hypothetical protein 1 (UNK1), UNK2, and PP2A for PEG-treated roots; SAND and EF1α for cold-treated leaves; SAND, TIP41, and PP2A for heat-treated leaves, were discovered as suitable candidates in different treated cotton tissues [3]. In Gracilaria lemaneiformis subjected to various temperature stimulations, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), isoleucine-tRNA synthetase (ITS2), compensatory Response (CR), and 18S rRNA were the optimal reference genes for various treatments applied at 8°C, while eukaryotic translation initiation factor (eIF) and actin (ACT) were appropriate for treated materials at 32°C, and GAPDH, EF1α, and ACT were ideal for different temperature treatments [10]. In Hedera helix, ribosomal 40S (40S) was the suitable reference gene after abscisic acid (ABA) treatment, while under various cold stress conditions in different tissues, squalene epoxidase (SE) and beta-amyrin synthase (β-AS) were perfected as reference genes [11].
Cotton fibers are major raw materials for the textile industry, and seeds are an important source of oil. Somatic embryogenesis (SE) has been considered practically as a tool for clonal propagation of cotton and is often used for the genetic modification of cotton in concert with Agrobacterium-mediated transformation, providing the effective basis for further investigating the gene function and understanding the regulatory mechanism in cotton plants at the molecular, cellular, and tissue levels [12]. Nonetheless, only a few cotton varieties have been successfully genetically modified in vitro using SE [13]. The majorities of the cotton cultivars are difficult to regenerate via SE [14], and many factors may affect SE success, including culture conditions and tissue background. Therefore, to date, most studies have focused on analyzing the molecular mechanism and identifying the genes critical for SE [15,16]. Although some miRNAs have been reported as reference genes in different cotton tissues [17,18], no systematic analysis was investigated for reference genes applied to be as a control in SE development through RT-qPCR method.
On the basis of our previous RNA-Seq data of Gossypium hirsutum, we selected 15 candidate reference genes, of which ENDO4 and 18S rRNA genes appeared to be stably expressed in the four G. hirsutum cultivars that have different SE capability after evaluation by the software geNorm [19], NormFinder [20], and BestKeeper [21] and verification of the expression consistency between the RT-qPCR results and the RNA-Seq data. In addition, the genes of ethylene-responsive transcription factor 17 (ERF17) and auxin-responsive protein 22 (AUX22) with altered expressions were further selected to validate the expression stability of ENDO4 and 18S rRNA, showing again the highly steady expression of the two genes. All these results suggest that ENDO4 and 18S rRNA can be used as appropriate reference genes for RT-qPCR normalization during SE initial dedifferentiation in different G. hirsutum cultivars.
2. Materials and Methods
2.1. Plant Materials
Four G. hirsutum cultivars, YZ1, R15, X33, and X42, were chosen as the cotton materials. Of which, YZ1 and R15 have a relatively high SE differentiation rate and, therefore, were commonly used as the main transgenic materials [22]. Although being the major commercial cultivars in Xinjiang, China, X33 and X42 have a low rate of differentiation during SE [23].
2.2. Total RNA Extraction and cDNA Synthesis
Total RNA extracted from different cotton tissues were subjected to check the purity using a NanoPhotometer spectrophotometer (IMPLEN, Calabasas, CA, USA), with the concentration and integrity measured by a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and the Bioanalyzer 2100 system (Agilent Technologies, Carlsbad, CA, USA). The cDNA was synthesized from each RNA sample using the PurelinkTM RNA Mini Kit (Life Technologies, Carlsbad, CA, USA) and the PrimerScript RT reagent Kit (Takara, Shiga, Japan) following the manufacturer’s protocols. The detailed steps were established according to our previously described method [23].
2.3. Selection of Reference Genes and Design of Primers
We performed high throughput RNA-Seq on four cultivars of G. hirsutum at three different induction stages (0h, 3h, and 3d). The four cultivars have different SE differentiation rate. Hypocotyl of 6-d sterile seedlings was cut into 1 cm segments with successive induction at 0 h, 3 h, and 3 d on callus-induction medium (MS medium plus B5 vitamins, supplemented with 0.05 mg/L IAA, 0.05 mg/L kinetin, 0.05 mg/L 2,4-D, pH 5.8) [23]. To evaluate the gene expression stability, q-value≥0.05, FPKM≥5, and |log2FoldChange|<1 were used as the criteria for screening reference genes at all sampling points [24], and 15 reference genes were selected. Gene-specific primers were designed based on the sequences of the 15 genes using the online software NCBI/Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) according to the following parameters: PCR product size of 100–150 bp, primer melting temperature (Tm) of 57°C–60°C, and primer pairs separated by at least one intron on the corresponding genomic DNA [25]. All primer pairs were synthesized by BGI TECH (BGI TECH, Shenzhen, China), and the PCR products were verified with electrophoresis on 1% agarose gels.
2.4. RT-qPCR Analysis
RT-qPCR was executed in 96-well plates on a LightCycler® 480 Real-Time PCR System (Roche Diagnostics, Mannheim, Germany) with an SYBR Green-based PCR assay. Reactions with a total volume of 10 μL, including 1 μL of a template (first-strand cDNA), 0.4 μL each of 10 μM forward and reverse gene-specific primers, 5 μL of 2×SYBR Premix Ex Taq II (TLi RanseH Plus) (Takara, Dalian, China), and 3.2 μL of ddH2O. The RT-qPCR conditions were as follows: initial denaturation at 95°C for 30 s, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s. The RT-qPCR analysis was tested in three biological replicates. Additionally, three technical replicates were used for each RT-qPCR analysis, and the R2 and amplification efficiency (E) were counted by the standard curves with the diluted series on the basis of the diluted cDNA series (version 1.5, Roche Diagnostics, Mannheim, Germany). The PCR efficiency was detected by the equation (E = (10[−1/slope] − 1) × 100%) [19].
2.5. Data Analysis
The raw Cq (quantification cycle) values are listed in Supplementary Table S1. Three common software programs, geNorm, NormFinder, and BestKeeper, were applied to calculate the expression stability of candidate reference genes. For geNorm and NormFinder, Cq values were converted into relative quantities according to the formula: 2−ΔCt (ΔCt = the corresponding Cq value − minimum Cq) [26]. The BestKeeper calculations were directly based on raw Cq values of each gene.
2.6. Validation of Reference Genes
To test the stability and reliability of the reference genes, the results of RT-qPCR were compared with the RNA-Seq data. The expression profiles of ERF17 and AUX22 were analyzed and compared with the values of Fragments Per Kilobase of transcript sequence per millions of base pairs sequenced (FPKM) obtained from the RNA-Seq data. The RT-qPCR analysis was conducted in the three biological replicates. Data were analyzed using the Origin 8 software and R package.
3. Results
3.1. Isolation of Candidate Reference Genes in Different G. hirsutum Cultivars
According to our previous RNA-Seq-based transcriptome data of three different induction stages during SE initial dedifferentiation process in four G. hirsutum cultivars that have different SE capability, the screen rules of q-value ≥ 0.05, FPKM ≥ 5, |log2FoldChange| < 1, and relatively lower coefficient of variance (CV) of FPKM were set as the higher criteria at all sampling points to select the candidate reference genes. A total of 15 candidate reference genes were obtained, with the details of all reference genes listed in Supplementary Table S2 and the heatmap of FPKM during three induction stages in four cotton cultivars shown in Figure 1.
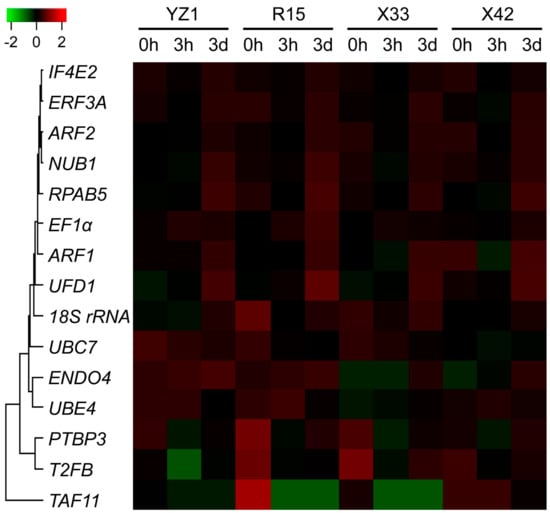
Figure 1.
FPKM-based heatmap of the 15 candidate reference genes during three induction stages in four cotton cultivars. Fifteen candidate reference genes were selected from the Gossypium hirsutum transcriptome datasets with the screening conditions of q-value ≥ 0.05, FPKM ≥ 5, and |log2FoldChange| < 1. Higher or lower FPKM of candidate reference genes were colored by red or green in four cotton cultivars, respectively. The color bar was shown at the upper apex. The visualized heatmap was generated using the R package based on the FPKM from the G. hirsutum transcriptome datasets. The sequences of the 15 candidate reference genes, including 18S rRNA, ENDO4, EF1α, ADP-ribosylation factor 1 (ARF1), ADP-ribosylation factor 2 (ARF2), eukaryotic peptide chain release factor 3A (ERF3A), eukaryotic translation initiation factor isoform 4E-2 (IF4E2), NEDD8 ultimate buster 1 (NUB1), polypyrimidine tract-binding protein homolog 3 (PTBP3), RNA polymerases I, II, and III subunit (RPAB5), transcription initiation factor (IIF), beta subunit (T2FB), TBP-associated factor 11 (TAF11), U-box domain-containing protein (UBE4), ubiquitin carrier protein 7 (UBC7), and ubiquitin fusion degradation 1 (UFD1), were obtained from NCBI GenBank nucleotide sequence database with the detailed information of the gene names and the corresponding accession numbers provided in Table 1.
3.2. Verification of Primer Specificity and PCR Amplification Efficiency
The 15 candidate reference genes were amplified by RT-qPCR using cDNA as the template to verify the specificity of the primers. One distinctive peak was observed in each melting curve, indicating that all the specific primers were appropriate for further RT-qPCR detection (Supplementary Figure S1). Gel electrophoresis results indicated that all specific primers could obtain a corresponding PCR product as designed in Table 1 successfully. The RT-qPCR amplification efficiency and R2 of the candidate reference genes were calculated based on the slopes of the standard curves in the four cotton cultivars, which showed that the RT-qPCR amplification efficiency ranged from 96.13% (TAF11) to 120.30% (UBE4), and the R2 values ranged from 0.9677 to 1.0977 (Table 1), suggesting that the primers had high specificity and amplification efficiency and were appropriate for RT-qPCR.

Table 1.
Details of the 15 candidate reference genes, primer sequences, and amplification characteristics for RT-qPCR in G. hirsutum.
3.3. Expression Profile Analysis of the Candidate Reference Genes at Different Induction Stages in Different Cotton Cultivars
RT-qPCR was used to measure the gene expression levels of the 15 candidate reference genes at three different induction stages in the four different cotton cultivars. Gene expression level was calculated based on the quantification cycle (Cq) that referred to the amplification-related fluorescence to reach a specific threshold level of test, with a smaller Cq representing a higher gene expression. Fifteen reference genes displayed a relatively wide range of mean Cq values from 17.36 to 22.61 (Figure 2, Supplementary Table S1). The most highly expressed gene was ERF3A in the X42 cultivar at the induction stage of 3 d, while END04 showed the lowest level in R15 at the induction stage of 0 h. Additionally, 18S rRNA showed the least gene expression variation, while UBE4 showed the greatest variation among the different cotton cultivars.
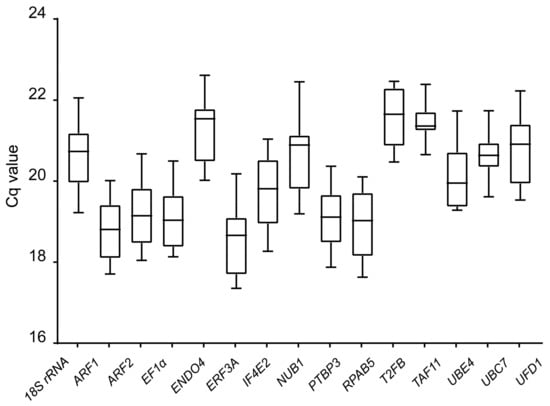
Figure 2.
Cq (quantification cycle) values of the 15 candidate reference genes across all samples. The final Cq value of each cotton sample was the mean of three biological and technical replicates. Box graph indicates the interquartile range. A line across the box represents the median. Whisker caps represent the maximum and minimum values.
3.4. Expression Stability Analysis of Reference Genes
To identify the stably expressed gene for cotton RT-qPCR normalization, the gene expression stability was assessed using three publicly available statistical tools, geNorm, NormFinder, and BestKeeper. geNorm can be used to check the most suitable number of reference genes by estimating M value that corresponds to the stability, with M = 1.5 or <1.5 indicating the standard or increased degree of stability of the candidate reference genes [19]. As shown in Table 2 and Figure 3, the results of the geNorm analysis showed that 18S rRNA, END04, and ARF1 were the most suitable reference genes in the four different cotton cultivars at three induction stages, while UBC7 and UBE4 were the least suitable reference genes. For the cotton cultivar YZ1, ARF1 had the lowest M values, and TAF11 had the highest M values under the same conditions. END04, IF4E2, and 18S rRNA in R15, END04, ARF2, and ARF1 in X33, and ARF2, T2FB, and UFD1 in X42 were detected as the more stably expressed genes, respectively. The variation V/(Vn/n+1) between gene pairs can also establish the optimal number of reference genes for normalization. Generally, Vn/n+1 is the cut-off value. If Vn/n+1 = 0.15, the optimal number of reference genes for accurate normalization should be optimal n+1; if Vn/n+1 < 0.15, the optimal number of reference genes should be n [27]. As shown in Figure 3, the V2/3 value was below 1.5; in conclusion, according to the geNorm analysis, the most stable reference genes were 18S rRNA and END04 in four cotton cultivars.

Table 2.
Expression stability of the 15 candidate reference genes calculated by geNorm (M)a.
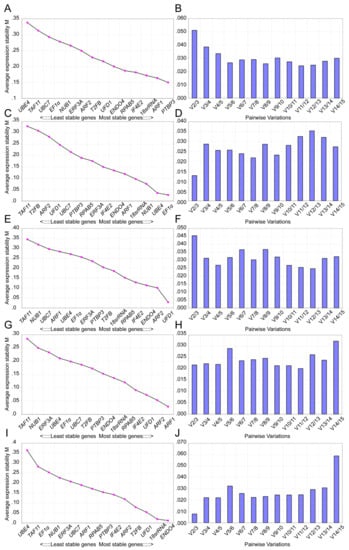
Figure 3.
geNorm analysis of the values of average expression stability (M) and pairwise variation (Vn/Vn+1) for the candidate reference genes. M and pairwise variation (Vn/Vn+1) were calculated on the basis of 2−ΔCt (ΔCt = the corresponding Cq value – minimum Cq) by geNorm. Lower M values indicate more stable gene expression. Ranking of the gene expression stability was determined in presented samples. According to the algorithm and software instructions, the cut-off V value of 0.15 was used to determine the optimal number of reference genes for RT-qPCR normalization. (A–B) Values of M and Vn/Vn+1 for the reference genes in the four cotton cultivars. (C–D) Values of M and Vn/Vn+1 for the reference genes in YZ1. (E–F) Values of M and Vn/Vn+1 for the reference genes in R15. (G–H) Values of M and Vn/Vn+1 for the reference genes in X33. (I–J) Values of M and Vn/Vn+1 for the reference genes in X42.
The NormFinder software is a Visual Basic application tool that establishes the expression stability of reference genes based on the stability value (Sv). The NormFinder analysis differs slightly from geNorm analysis, which takes into account intra- and inter-group variation when calculating the normalization factor (NF) [20]. Genes that have lower levels than an average of expression stability are more stable, and thus may be suitable as reference genes. As shown in Table 3, the results of NormFinder analysis were highly consistent with those of the geNorm analysis. The most stable genes were 18S rRNA, END04, and ARF1 in four cotton cultivars. PRAB5, ARF1, and 18S rRNA were the most stable genes in YZ1. END04, IF4E2, and 18S rRNA had high expression stabilities in R15. For the cotton cultivar X33, END04, ARF2, and 18S rRNA appeared to have the most stable expressions. ARF2, T2FB, and UFD1 were highly stably expressed in X42.

Table 3.
Expression stability of the 15 candidate reference genes calculated by NormFinder (Sv)a.
The BestKeeper software can be used to analyze the stability and expression of reference genes based on the coefficient of variance (CV), standard deviation (SD), and correlation coefficient (R) [21]. Reference genes are considered to be stable if R is high, but SD and CV are low. Additionally, if SD > 1, the gene is considered to be unacceptable [9]. As shown in Table 4, END04, 18S rRNA, and ARF1 were relatively stable expressed genes under all conditions. TAF11 was an unstable gene with a low CV± SD value and lower R value. For R15 cotton cultivar, 18S rRNA, UBC7, and ARF1 were identified as relatively suitable reference genes. END04, 18S rRNA, and PTBP3 in X33 and T2FB, ARF2, and EDD04 in X44 were the more stably expressed genes and could be considered as acceptable reference genes with high R values and low CV ± SD values. These results of comprehensive consideration of the geNorm, NormFinder, and BestKeeper analyses indicate that 18S rRNA and ENDO4 could be appropriate reference genes.

Table 4.
Expression stability of the 15 candidate reference genes calculated by BestKeeper.
3.5. Expression Stability Validation of the Reference Genes of 18S rRNA and ENDO4
Through the identity analysis between the RT-qPCR results and the RNA-Seq data, two target genes, AUX22 and ERF17, were selected to further test the stability and reliability of the validated reference genes of 18S rRNA and ENDO4. The relative expression levels of AUX22 and ERF17 were calculated based on the validated reference genes and then were compared with the relative expression levels and the FPKM values of these reference genes in the RNA-Seq data. The 18S rRNA and ENDO4 were used as reference genes for the normalization of the target gene expression in the four cotton cultivars (Figure 4). Strong positive correlation coefficients (R2 = 0.8396–0.9984) between the RT-qPCR results and the RNA-Seq data, and similar patterns between relative expression profiles and the FPKM values, were also observed (Figure 5). The results indicated that the 18S rRNA and ENDO4 were suitable as the reference genes to obtain the reliable RT-qPCR results in the four cotton cultivars.
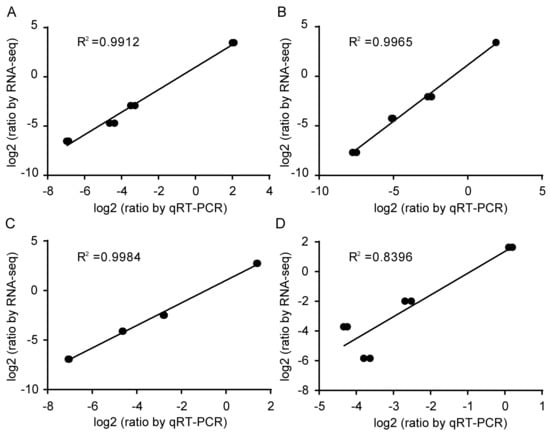
Figure 4.
Correlation analysis between RT-qPCR results and the RNA-Seq data. X-axis represented log2(ratio by RT-qPCR), and equation 2−ΔCt was applied to calculate the RT-qPCR relative expression level using selected target genes as the reference gene. Y-axis represented log2(ratio by RNA-Seq) (p-value ≤ 0.01). Correlation analysis using ENDO4 and 18S rRNA was used as the internal controls for normalization of AUX22 and ERF17 in YZ1 (A), R15 (B), X33 (C), and X42 (D).
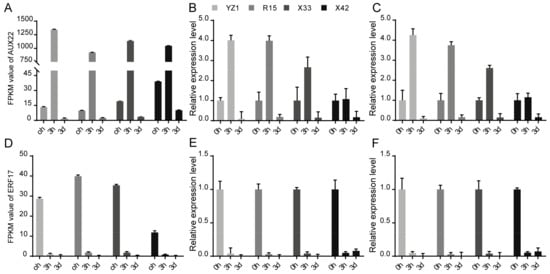
Figure 5.
Validation of the reference genes of 18S rRNA and ENDO4 as internal controls for normalization of target gene expressions of AUX22 and ERF17. The results were shown as mean fold changes in relative expression when compared to 0 h. (A) The FPKM values of AUX22 in RNA-Seq data. (B) The expression profiles of AUX22 normalized using 18S rRNA as an internal control. (C) The expression profiles of AUX22 normalized using ENDO4 as an internal control. (D) The FPKM values of ERF17 in RNA-Seq data. (E) The expression profiles of ERF17 normalized using 18S rRNA as an internal control. (F) The expression profiles of ERF17 normalized using ENDO4 as internal control.
4. Discussion
Gene expression analysis is an effective basic method to predict plant gene function [28]. RT-qPCR is also widely used for gene expression detection using proper reference gene as an internal control [6]. Whereas, the expression of the reference gene is unavoidably influenced by different tissues and treatments, thus leading to unreliable results [29]. Therefore, it is essential to select valid and reliable reference genes as internal controls for normalization to ensure the reliability and accuracy of RT-qPCR reactions, without consideration of different experimental conditions [8]. In this study, on the basis of our high throughout RNA-Seq data of four G. hirsutum cultivars at three induction stages, we analyzed 15 candidates as reference genes according to the conditions of |log2FoldChange|<1 and a small coefficient of variation (CV) of FPKM values (Figure 1), which are widely considered parameters for choosing candidate reference genes [30].
Three public statistical algorithms, geNorm, NormFinder, and BestKeeper, can be used to determine the stability of reference gene expression [19,20,21]. Some studies of the two programs of geNorm and NormFinder were mostly used to identify candidate reference genes, showing the results that differed slightly in Oxytropis ochrocephala [30]. Several reports also showed that the NormFinder results were consistent with geNorm results in a variety of organisms, including the Sapium sebiferum [31], Chrysanthemum morifolium and Chrysanthemum lavandulifolium [32], Oryza sativa [33], and Rhododendron molle [34]. In this study, the ranking of NormFinder was relatively consistent with the results of geNorm analysis (Table 2 and Table 3). However, the results differed from those obtained using BestKeeper, indicating that UBC7 was the most reliable reference gene in R15 analyzed by BestKeeper, but was the least reliable one analyzed by geNorm and NormFinder (Table 4). The divergent ranking may be caused due to the different algorithms [35]. By BestKeeper analysis, most studies utilized CV and SD of the Cq values, as well as the R values, to evaluate the stability and expression of reference genes [2,34,35]. TAF11 showed the lowest CV± SD values across all cultivars with the lowest R values in YZ1 and X42 (Table 4) and was identified as the least reliable gene by geNorm and NormFinder (Figure 3 and Table 3). Through integrated analysis of the three programs, better accuracy for each reference gene can be obtained, and our results showed that 18S rRNA and ENDO4 were appropriate and reliable candidates as reference genes for RT-qPCR normalization in the four cotton cultivars under the condition of three different induction stages. ENDO4 encodes a putative endonuclease without enzyme activity, and 18S rRNA is part of the ribosomal RNA that constitutes the basic components of the eukaryotic cells. Ribosomal genes are often recognized as suitable reference genes [9] but are observed as the least stable expressions in peaches [36]. Generally, EF1α and UBCs were often used as reference genes in plants, such as Platycladus orientalis and Caragana intermedia [3,37], while EF1α, UBC7, and UBE4 showed unstable expressions in the different cotton cultivars (Table 2 and Table 3).
The selected reference genes based on RNA-Seq were often further verified by RT-qPCR [38]. The RT-qPCR results were compared with that of RNA-Seq data to further validate the stability of ENDO4 and 18S rRNA expression, with the appearance of ENDO4 and 18S rRNA as reference genes in the four cotton cultivars showing strong positive R2 between the RT-qPCR results and the RNA-Seq data (Figure 4). When 18S rRNA and ENDO4 were used as the reference genes in the four cotton cultivars, the expression patterns of ERF17 and AUX22 indicated similar correlation between the RT-qPCR results and the RNA-Seq data (Figure 5), suggesting that ENDO4 and 18S rRNA are reliable reference genes for studies of gene expression normalization in these four cultivars of G. hirsutum under three different induction stages.
5. Conclusion
We selected 15 candidate reference genes for RT-qPCR normalization in different cotton cultivars at three different induction stages according to the transcriptome datasets of G. hirsutum. After assessment of the 15 candidates by three statistical software geNorm, NormFinder, and BestKeeper, two genes of ENDO4 and 18S rRNA were identified as appropriate reference genes during three induction stages in the four cotton cultivars. The results of further validation of 18S rRNA and ENDO4 genes by comparing the RT-qPCR results and the RNA-Seq data showed a strong positive correlation, indicating that ENDO4 and 18S rRNA are reliable reference genes for studies of gene expression normalization. Our results identify necessary and appropriate reference genes as internal controls for RT-qPCR normalization and provide an effective basis for accurate quantification of target gene expression in different G. hirsutum cultivars.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/7/497/s1, Figure S1: Melting curves of the 15 selected reference genes, Table S1: Cq value of the 15 candidate reference genes, Table S2: The details of the 15 candidate reference genes based on RNA-Seq data.
Author Contributions
Conceptualization, H.L. and J.S.; methodology, A.C.; software, A.C.; validation, D.S., X.T., and B.C.; formal analysis, A.C.; investigation, Y.Z.; resources, A.C.; data curation, A.C. and H.L.; writing—original draft preparation, A.C.; writing—review and editing, H.L. and J.S.; visualization, A.C. and D.S.; supervision, H.L.; project administration, H.L. and J.S.; funding acquisition, H.L.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31660408, and Scientific and Technological Achievement Transformation Project of Bingtuan, grant number 2016AC017.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, C.; Liu, Q.; Zhang, Z.; Zheng, B.; Bao, M. De novo comparative transcriptome analysis provides new insights into sucrose induced somatic embryogenesis in camphor tree (Cinnamomum camphora L.). BMC Genom. 2016, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L.; Li, W.; Han, S.; Yang, W.; Qi, L. Reference Gene Selection for Quantitative Real-time PCR Normalization in Caragana intermedia under Different Abiotic Stress Conditions. PLoS ONE 2013, 8, e53196. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and Validation of Appropriate Reference Genes for Quantitative Real-Time PCR Analysis of Gene Expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Hong, S.Y.; Seo, P.J.; Yang, M.S.; Xiang, F.; Park, C.M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008, 8, 112. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Zhang, S.; Zeng, Y.; Yi, X.; Zhang, Y. Selection of suitable reference genes for quantitative RT-PCR normalization in the halophyte Halostachys caspica under salt and drought stress. Sci. Rep. 2016, 6, 30363. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, H.; Zhang, R.; Yang, Q.; Liu, Y.; Zang, X. Selection of reference gene from Gracilaria lemaneiformis under temperature stress. J. Appl. Phycol. 2014, 27, 1–8. [Google Scholar] [CrossRef]
- Sun, H.P.; Li, F.; Ruan, Q.M.; Zhong, X.H. Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L. Plant Physiol. Biochem. 2016, 108, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi-Darvari, F.; Noor, N.M.; Ismanizan, I. Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell Tissue Organ Cult. 2015, 120, 407–422. [Google Scholar] [CrossRef]
- Kumria, R.; Sunnichan, V.G.; Das, D.K.; Gupta, S.K.; Reddy, V.S.; Bhatnagar, R.K.; Leelavathi, S. High-frequency somatic embryo production and maturation into normal plants in cotton (Gossypium hirsutum) through metabolic stress. Plant Cell Rep. 2003, 21, 635–639. [Google Scholar] [PubMed]
- Yang, X.; Zhang, X.; Yuan, D.; Jin, F.; Zhang, Y.; Jiao, X. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012, 12, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Zhu, H.G.; Tian, W.G.; Zhu, S.H.; Xiong, X.P.; Sun, Y.Q.; Zhu, Q.H.; Sun, J. De novo transcriptome analysis reveals insights into dynamic homeostasis regulation of somatic embryogenesis in upland cotton (G. hirsutum L.). Plant Mol. Biol. 2016, 92, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Zhang, X.L. Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 2010, 29, 36–57. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Grossi-de-Sa, M.F.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Fausto, A.K.S.; Silva, T.D.F.; Romanel, E.; Vaslin, M.F.S. microRNAs as reference genes for quantitative PCR in cotton. PLoS ONE 2017, 12, e0174722. [Google Scholar] [CrossRef] [PubMed]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, X.Y.; Guo, X.; Liang, S.; Zhu, H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plant. 2006, 50, 519–524. [Google Scholar] [CrossRef]
- Cao, A.; Zheng, Y.; Yu, Y.; Wang, X.; Shao, D.; Sun, J.; Cui, B. Comparative Transcriptome Analysis of SE initial dedifferentiation in cotton of different SE capability. Sci. Rep. 2017, 7, 8583. [Google Scholar] [CrossRef] [PubMed]
- Jonge, H.; Fehrmann, R.; Bont, E.; Hofstra, R.; Gerbens, F.; Kamps, W.; Vries, E.; Zee, A.; Meerman, G.; Elst, A. Evidence Based Selection of Housekeeping Genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, B.; Tan, Z.; Liu, J.; Yang, Z.; Li, Z.; Huang, B. Selection of reference genes for quantitative real-time PCR normalization in creeping bentgrass involved in four abiotic stresses. Plant Cell Rep. 2015, 34, 1825–1834. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yan, W.; Huang, X.; Jiang, L.; Wang, H.; Li, J. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 2015, 15, 439. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–153. [Google Scholar] [CrossRef]
- Zhuang, H.; Fu, Y.; He, W.; Wang, L.; Wei, Y. Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef]
- Xue, C.; Yingji, M.; Shengwei, H.; Jun, N.; Weili, L.; Jinyan, H. Selection of suitable reference genes for quantitative real-time PCR in Sapium sebiferum. Front. Plant Sci. 2017, 8, 637. [Google Scholar]
- Qi, S.; Yang, L.; Wen, X.; Hong, Y.; Song, X.; Zhang, M.; Dai, S. Reference Gene Selection for RT-qPCR Analysis of Flower Development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 2016, 11, 287. [Google Scholar] [CrossRef] [PubMed]
- Jain, M. Genome-wide identification of novel internal control genes for normalization of gene expression during various stages of development in rice. Plant Sci. 2009, 176, 702–706. [Google Scholar] [CrossRef]
- Xiao, Z.; Sun, X.; Liu, X.; Li, C.; He, L.; Chen, S.; Su, J. Selection of Reliable Reference Genes for Gene Expression Studies on Rhododendron molle G. Don. Front. Plant Sci. 2016, 18, 1547. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Qi, J.; Zhang, G.; Xu, J.; Tao, A.; Fang, P.; Su, J. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 2015, 14, 848. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 20, 71. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.; Shi, S.; Liu, J.; Cheng, T.; Xue, L.; Yang, X.; Yang, W.; Lan, Q.; Jiang, Z. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using real-time PCR. PLoS ONE 2012, 7, e33278. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.M.; Sá, P.H.; Castro, T.L.; Carvalho, R.D.; Pinto, A.; Gil, D.J.P.; Silva, A. Reference genes for RT-qPCR studies in Corynebacterium pseudotuberculosisi dentified through analysis of RNA-seq data. Antonie Leeuwenhoek 2014, 106, 605–614. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).