Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the MYB-bHLH-WD40 Complex Gene Family Members
2.2. Phylogenetic Trees Analysis
2.3. Plant Materials and Anthocyanin Analysis
2.4. Differentially Expressed Genes Analysis
2.5. Subcellular Localization
2.6. Protein–Protein Interactions (PPIs) Analysis
2.7. Bimolecular Fluorescence Complementation (BiFC) Analysis
2.8. Quantitative Real-time Reverse Transcription PCR (RT-qPCR)
3. Results and Discussion
3.1. Identification of MYB-bHLH-WD40 Complex Gene Members in Blueberry Fruits
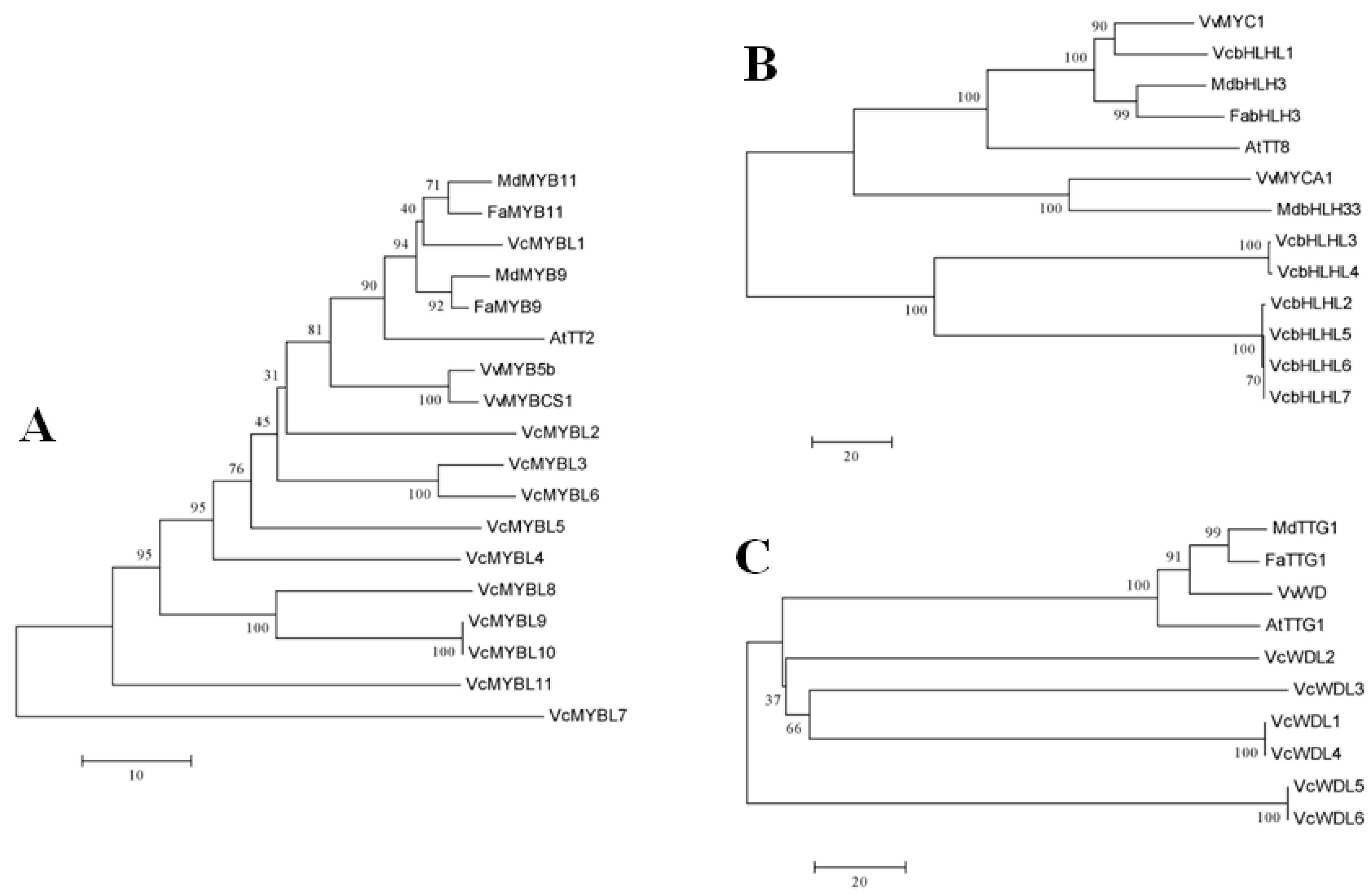
3.2. Phylogenetic Analyses of the Blueberry MYB-bHLH-WD40 Regulatory Complex Members
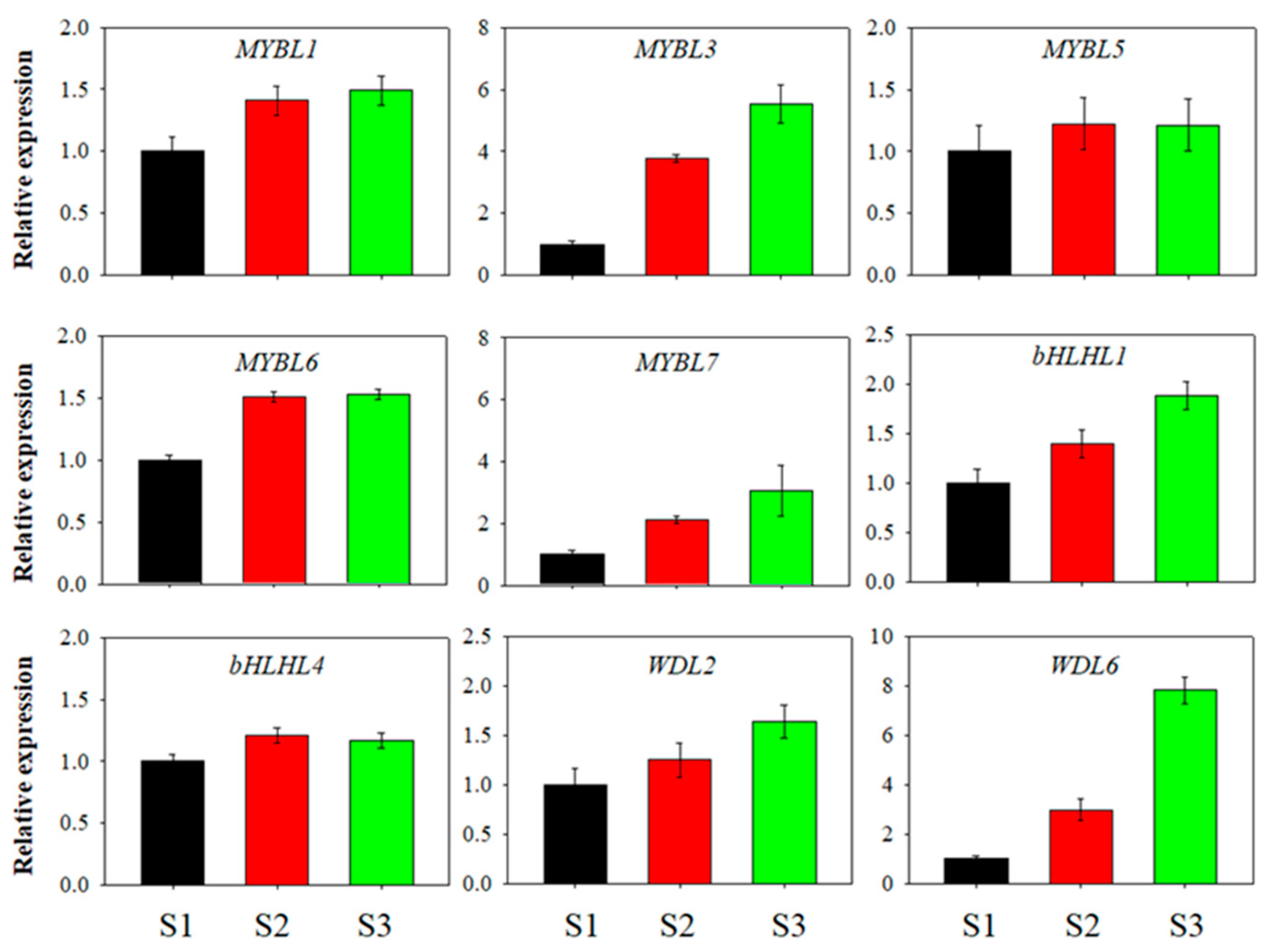
3.3. Expression Patterns of the Blueberry MYB-bHLH-WD40 Genes
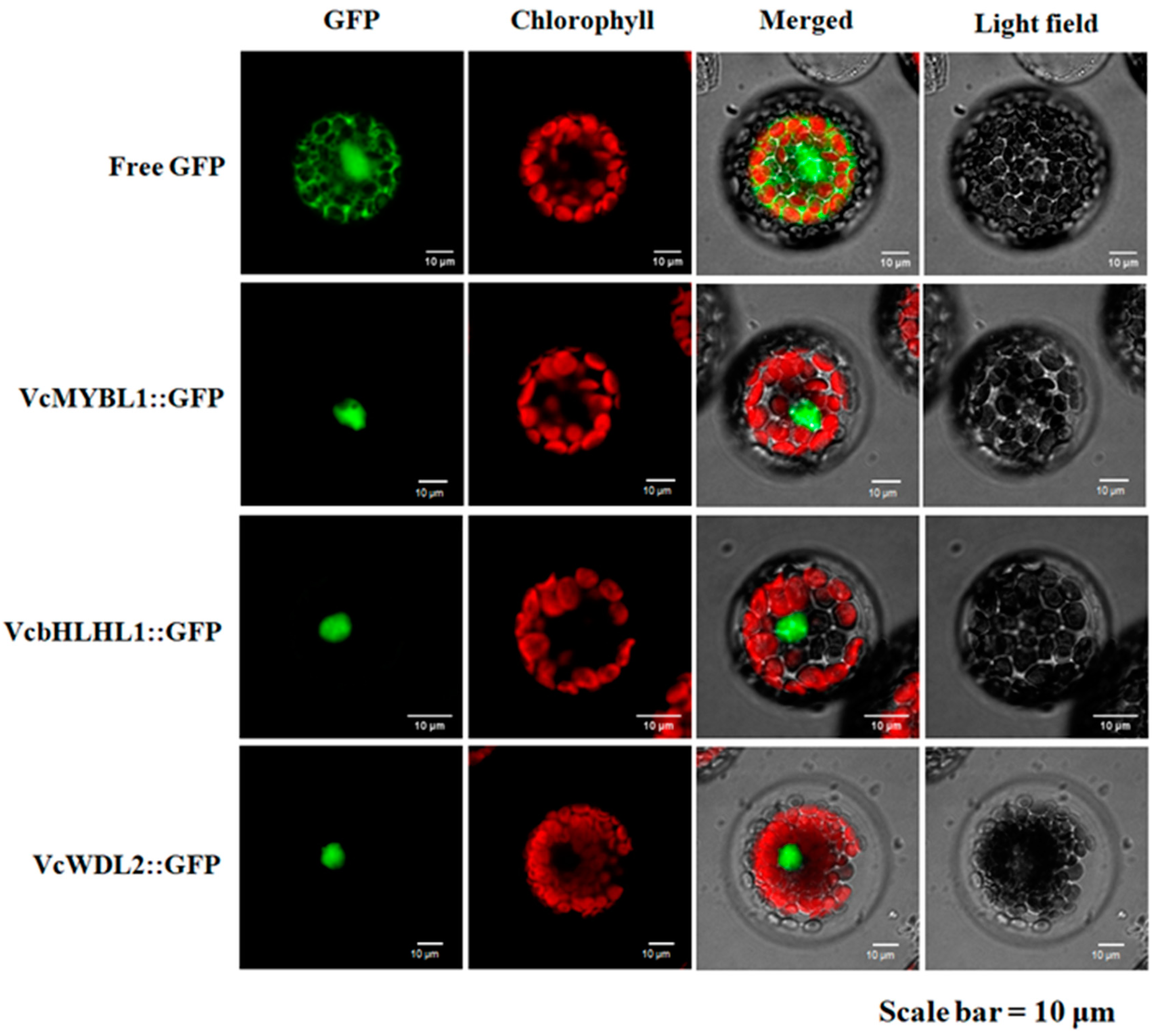
3.4. Subcellular Localization of the VcMYBL1, VcbHLHL1, and VcWD40L2 Genes
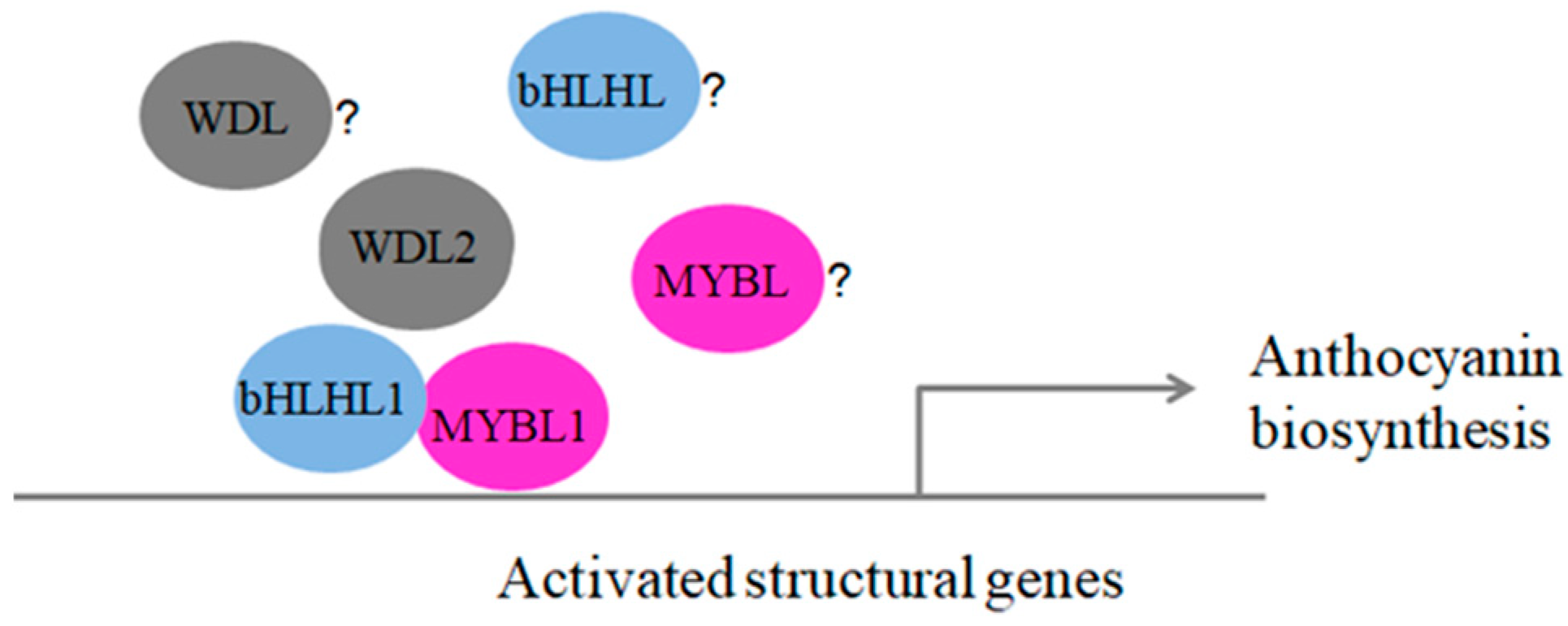
3.5. How Expression of MYB-bHLH-WD40 Complexes Can Promote Anthocyanin Biosynthesis in Blueberry Fruits
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Galletta, G.; Ballington, B. Blueberries, cranberries and lingonberries. Fruit Breeding 1996, 2, 1–107. [Google Scholar]
- Rowland, L.J.; Alkharouf, N.; Darwish, O.; Ogden, E.L.; Polashock, J.J.; Bassil, N.V.; Dorrie, M. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Takashi, A.; Ayako, I.; Tomoyuki, T.; Shozo, K.; Akihiko, S.; Atsushi, K.; Keizo, Y. Dkmyb4 is a myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009, 151, 2028–2045. [Google Scholar]
- Akagi, T.; Tsujimoto, T.; Ikegami, A.; Yonemori, K. Effects of seasonal temperature changes on dkmyb4 expression involved in proanthocyanidin regulation in two genotypes of persimmon (diospyros kaki thunb.) fruit. Planta 2011, 233, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Nijveldt, R.J.; Van Nood, E.; Van Hoorn, D.E.; Boelens, P.G.; Van Norren, K.; Van Leeuwen, P.A. Flavonoids: A review of probable mechanisms of action and potential applications. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Battino, M.; Beekwilder, J.; Denoyesrothan, B.; Laimer, M.; Mcdougall, G.J.; Mezzetti, B.; Serramajem, L.; Ngo, J.; Aranceta, J.; Solomons, N.W. Bioactive compounds in berries relevant to human health. Nutr. Rev. 2010, 67, S145–S150. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Xie, D.Y.; Sharma, S.B. Proanthocyanidins–a final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Seddon, J.M.; Rosner, B.; Willett, W.C.; Hankinson, S.E. Prospective study of intake of fruits, vegetables, vitamins, and carotenoids and risk of age-related maculopathy. Arch. Ophthalmol. 2004, 122, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Kalt, W.; Joseph, J.A.; Shukitt-Hale, B. Blueberries and human health: A review of current reseach. J. Am. Pomol. Soc. 2007, 61, 151–160. [Google Scholar]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. Plant metabolic diversity: A regulatory perspective. Trends Plant Sci. 2005, 10, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2013, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Sainz, M.B.; Tagliani, L.; Hernandez, J.M.; Bowen, B.; Chandler, V.L. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 2000, 97, 13579–13584. [Google Scholar] [CrossRef] [PubMed]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’Akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- Yongzhen, P.; Wenger, J.P.; Katie, S.; Peel, G.J.; Jiangqi, W.; David, H.; Allen, S.N.; Yuhong, T.; Xiaofei, C.; Million, T. A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 2009, 151, 1114–1129. [Google Scholar]
- Pattanaik, S.; Kong, Q.; Zaitlin, D.; Werkman, J.R.; Xie, C.H.; Patra, B.; Yuan, L. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 2010, 231, 1061–1076. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2010, 39, 366–380. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2010, 46, 768–779. [Google Scholar] [CrossRef]
- An, J.P.; An, X.H.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiol. 2018, 38, 1578–1587. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.L.; Xie, L.F.; Ma, Y.Y.; Ren, C.H.; Xing, M.Y.; Fu, Z.S.; Wu, X.Y.; Yin, X.R.; Xu, C.J.; Li, X. PpMYB15 and PpMYBF1 transcription factors are involved in regulating flavonol biosynthesis in peach fruit. J. Agric. Food Chem. 2019, 67, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hao, R.J.; Xu, Z.D.; Yang, W.R.; Wang, J.; Cheng, T.R.; Pan, H.T.; Zhang, Q.X. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell. Tiss. Org. Cult. 2017, 131, 417–429. [Google Scholar] [CrossRef]
- Wang, N.; Xu, H.F.; Jiang, S.H.; Zhang, Z.Y.; Lu, N.L.; Qiu, H.R.; Qu, C.Z.; Wang, Y.C.; Wu, S.J.; Chen, X.S. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017, 90, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Long, J.M.; Zhu, K.J.; Liu, L.L.; Yang, W.; Zhang, H.Y.; Li, L.; Xu, Q.; Deng, X.X. Characterization of a Citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 2016, 6, 25352. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Drummond, B.J.; Bowen, B.; Peterson, T. The myb -homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 1994, 76, 543–553. [Google Scholar] [CrossRef]
- Petroni, K.; Cominelli, E.; Consonni, G.; Gusmaroli, G.; Gavazzi, G.; Tonelli, C. The developmental expression of the maize regulatory gene Hopi determines germination-dependent anthocyanin accumulation. Genetics 2000, 155, 323–336. [Google Scholar] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.; Emiliani, J.; Pourcel, L.; Feller, A.; Morohashi, K.; Casati, P.; Grotewold, E. Cloning and characterization of a UV-B-inducible maize flavonol synthase. Plant J. 2010, 62, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero, D.L.I.; Van Houwelingen, A.M.; Vos, R.C.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [PubMed]
- An, X.H.; Tian, Y.; Chen, K.Q.; Wang, X.F.; Hao, Y.J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 2012, 169, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.P.; Mérillon, J.M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006, 140, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [PubMed]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verriès, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, H.; Liu, Z.; Cui, X.; Zhang, T.; Li, Y.; Zhang, L. Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 2016, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, 1182–1187. [Google Scholar] [CrossRef]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, X.; Chen, W.; Zheng, Y.; Yang, Z. Comparative transcriptomic analysis of white and red Chinese bayberry (Myrica rubra) fruits reveals flavonoid biosynthesis regulation. Sci. Hortic. 2018, 235, 9–20. [Google Scholar] [CrossRef]

| Name | Deduced Polypeptide | ||||
|---|---|---|---|---|---|
| Length (aa) | PI | MW (Da) | Pfam | Pfam ID | |
| MYBL1 | 265 | 8.57 | 30057.30 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL2 | 157 | 9.71 | 17780.38 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL3 | 348 | 5.37 | 39436.40 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL4 | 451 | 5.58 | 49663.84 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL5 | 315 | 5.22 | 35681.21 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL6 | 129 | 9.00 | 14908.11 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL7 | 375 | 7.12 | 42961.38 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL8 | 200 | 9.26 | 22052.74 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL9 | 360 | 6.26 | 39085.01 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL10 | 204 | 9.97 | 22644.72 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL11 | 471 | 5.82 | 52808.93 | MYB_DNA-binding | PF00249.30, PF13921.5 |
| MYBL12 | 123 | 9.36 | 13594.37 | ||
| MYBL13 | 209 | 5.33 | 22759.35 | ||
| bHLHL1 | 729 | 5.49 | 80280.20 | bHLH-MYC_N | PF14215.5 |
| bHLHL2 | 589 | 6.70 | 64930.88 | bHLH-MYC_N | PF14215.5 |
| bHLHL3 | 491 | 6.16 | 54127.08 | bHLH-MYC_N | PF14215.5 |
| bHLHL4 | 491 | 6.32 | 54049.06 | bHLH-MYC_N | PF14215.5 |
| bHLHL5 | 589 | 6.92 | 65060.05 | bHLH-MYC_N | PF14215.5 |
| bHLHL6 | 468 | 9.21 | 51764.32 | bHLH-MYC_N | PF14215.5 |
| bHLHL7 | 498 | 9.34 | 55037.05 | bHLH-MYC_N | PF14215.5 |
| bHLHL8 | 371 | 9.44 | 40555.56 | ||
| WDL1 | 694 | 8.77 | 76244.98 | WD40 | PF00400.31 |
| WDL2 | 313 | 4.74 | 34289.96 | WD40 | PF00400.31 |
| WDL3 | 347 | 6.41 | 38427.24 | WD40 | PF00400.31 |
| WDL4 | 438 | 8.73 | 50248.11 | WD40 | PF00400.31 |
| WDL5 | 898 | 5.67 | 100036.07 | WD40 | PF00400.31 |
| WDL6 | 898 | 5.67 | 100036.07 | WD40 | PF00400.31 |
| WDL7 | 254 | 4.80 | 28932.00 | ||
| WDL8 | 252 | 4.85 | 28703.75 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development. Genes 2019, 10, 496. https://doi.org/10.3390/genes10070496
Zhao M, Li J, Zhu L, Chang P, Li L, Zhang L. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development. Genes. 2019; 10(7):496. https://doi.org/10.3390/genes10070496
Chicago/Turabian StyleZhao, Mengran, Jian Li, Ling Zhu, Pan Chang, Lingli Li, and Lingyun Zhang. 2019. "Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development" Genes 10, no. 7: 496. https://doi.org/10.3390/genes10070496
APA StyleZhao, M., Li, J., Zhu, L., Chang, P., Li, L., & Zhang, L. (2019). Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development. Genes, 10(7), 496. https://doi.org/10.3390/genes10070496




