Abstract
The previously reported 1-(2,4-dihydroxy-5-methylphenyl)ethan-1-one (1), (1’Z)-2-(1’,5’-dimethylhexa-1’,4’-dieny1)-5-methylbenzene-1,4-diol (2), and 1,8-epoxy-1(6),2,4,7,10-bisaborapentaen-4-ol (5) together with four new structures of aromatic bisabolane-related compounds (3, 4, 6, 7) were isolated from the marine sponge Myrmekioderma sp. Compounds 1, 2, and 5 were identified based on spectral data available in the literature. The structures of the four new compounds were experimentally established by 1D and 2D-NMR and (−)-HRESIMS spectral analysis. Cytotoxic and lipid-reducing activities of the isolated compounds were evaluated. None of the isolated compounds were active against the tested cancer cell lines; however, lipid-reducing activity was found for compounds 2–5 and 7 in the zebrafish Nile red fat metabolism assay. This class of compounds should be further explored for their suitability as possible agents for the treatment of lipid metabolic disorders and obesity.
1. Introduction
Marine organisms are exposed to continuous and strong selection pressures due to the huge variations in predation, temperature, pressure, and light. For these reasons, they are known to produce secondary metabolites as a mechanism of defense [1]. These secondary metabolites represent an impressive source of structurally diverse molecules with biological activities which can lead to major advances in the field of medicinal chemistry [2,3].
Among marine organisms, sponges represent a prolific source of a vast number of diverse molecules with potential applications for human health. The numbers of compounds isolated from sponges have been increasing every year [4]. Among these compounds, marine sesquiterpenes are recognized as an important class with great structural diversity and a wide range of bioactivities such as anti-HIV, antitumor, antibiotic, antiviral, cytotoxic, insecticidal, antifeedant, and antifungal activities [5,6]. Bisabolane compounds constitute a class of sesquiterpene bioactive metabolites that have been identified from both terrestrial plants and marine invertebrates [7,8]. Several bioactivities are associated with this class of compounds, such as cytotoxicity [9,10] and antifungal [10] properties. Furthermore, their suitability for use as biodiesel is also under investigation [11].
Obesity is increasing at epidemic rates and new therapeutics are needed in order to prevent and control this disorder [12]. Scientists have been working hard to find new compounds from different natural sources, both terrestrial and marine, that show anti-obesity activity [13,14,15]. Several marine secondary metabolites with anti-obesity properties have already been reported, such as the 5-alkylpyrrole-2-carboxaldehyde derivatives, isolated from the sponge Mycale lissochela, which have protein-tyrosine phosphatase 1B (a recognized target for obesity) inhibitory activity [6]. Also, citreorosein and questinol, isolated from the marine sponge-associated fungus Talaromyces stipitatus KUFA 0207, decreased the neutral lipids in the zebrafish Nile red fat metabolism assay [16].
As a part of our on-going screening program for the discovery of new secondary metabolites from marine sponges, the study of an organic extract of Myrmekioderma sp. resulted in the isolation of seven natural compounds: three known compounds 1-(2,4-dihydroxy-5-methylphenyl)ethan-1-one (1), (1’Z)-2-(1’,5’-dimethylhexa-1’,4’-dieny1)-5-methylbenzene-1,4-diol (2), 1,8-epoxy-1(6),2,4,7,10-bisaborapentaen-4-ol (5), and four new bisabolane derivatives (3, 4, 6 and 7). Their planar structures were fully elucidated using spectroscopic and spectrometric techniques. All compounds were tested for their cytotoxic and lipid-reducing activities. Compounds 2, 5, and 7 were highly active in the zebrafish Nile red fat metabolism assay and compounds 3 and 4 showed moderate activity in the same bioassay. Cytotoxic activity in the four cancer cell lines tested was not observed for any of the isolated compounds.
2. Results and Discussion
Isolation and Structure Elucidation
The sponge Myrmekioderma sp. was collected by hand while scuba diving in Boano (Indonesia). The specimen was repeatedly extracted using dichloromethane:methanol (1:1 v/v). The crude organic extract was subsequently partitioned between n-hexane, ethyl acetate, n-butanol, and water. The n-hexane and ethyl acetate fractions, after vacuum liquid chromatography (VLC) and semi-preparative reverse-phase HPLC separations, led to the isolation of the seven pure compounds shown in Figure 1.
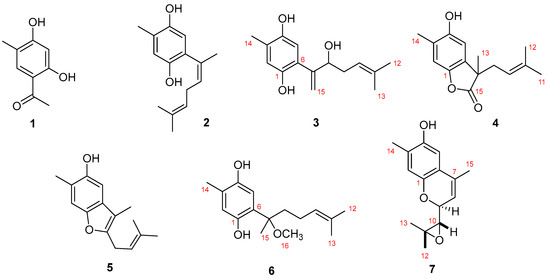
Figure 1.
Chemical structures of the compounds 1–7 isolated from Myrmekioderma sp.
Compound 1 was isolated as a dark-brown oil. It was identified as 1-(2,4-dihydroxy-5-methylphenyl)ethan-1-one, as shown in Figure 1, based on spectral data available in the literature [17].
Also based on spectral data available in the literature, compound 2 was identified as (1’Z)-2-(1’,5’-dimethylhexa-1’,4’-dieny1)-5-methylbenzene-1,4-diol [18].
Compound 3 was isolated as a yellow amorphous solid. The molecular formula C15H20O3 was established based on the (−)-HRESIMS molecular ion m/z 247.1344 [M − H]− (calculated 247.1334), which imposed six degrees of unsaturation. The 13C-NMR spectrum of 3, compiled in Table 1, confirmed the presence of fifteen carbon signals which were assigned, by DEPT and HMQC spectral analysis, to two tertiary (δC 26.1, 18.1) and one secondary (δC 15.8) methyls, two methylenes (δC 120.3, 34.3) of which one was double bonded (δC 120.3), two aromatic (δC 118.8, 117.2), one double-bonded (δC 118.8), and one hydroxylated (δC 76.7) methine and six non-protonated carbons (δC 148.0, 147.5, 147.0, 136.9, 125.4, 124.2). From the listed non-protonated carbons, two were hydroxylated (δC 148.0, 147.5). In accordance, the 1H-NMR spectrum exhibited three methyl singlets (δH 2.20, 1.71, 1.53), two splitting methylenes (δH 5.43 and 5.24, 2.30, and 2.15), the first two suggesting a double bond, and four methines (δH 6.68, 6.50, 5.06, 4.40). Based on COSY and HMBC spectral data, as shown in Figure 2, a simple sesquiterpene structure was proposed for this compound. 1H and 13C data, together with the H-2 HMBC correlations with C-1 and C-5 revealed the presence of a tetrasubstituted benzene ring. C-1 and C-4 deshielded carbon resonances (δC 147.5, 147.0) indicated the presence of a benzene-1,4-diol. Me-14 was assigned based on the HMBC correlation Me-14/C-4 and C-6 substitution based on the correlations H-5/C-7 and H2-15/C-6. The double bond, suggested by H2-15 resonances (δC 120.3, δH 5.43, 5.24), was elucidated based on the previously described HMBC correlation of H2-15/C-6 with the hydroxylated C-8 (δC 76.7). The COSY correlations H-8/H2-9 and H2-9/H-10 allowed assignment of the methylene and Δ10(11) double bond. The HMBC cross signals of the methyl groups Me-12 and Me-13 with each other and of both of them with C-10 and C-11 completed the structure. Unfortunately, a paucity of material prevented the assignment of the C-8 absolute stereochemistry. Thus, the structure of 3 was elucidated as the curcuhydroquinone derivative shown in Figure 1: 6-(3-hydroxy-6-methyl-1,5-heptadien-2-yl)-3-methylbenzene-1,4-diol.

Table 1.
1H and 13C-NMR (400 and 100 MHz, respectively) for compounds 3 and 4. The experiments were performed in CDCl3.
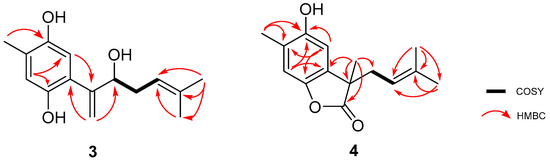
Figure 2.
Key 1H-1H COSY and HMBC correlations of compounds 3 and 4.
Compound 4 was isolated as a yellow amorphous powder. The molecular formula C15H18O3 was calculated based on the (−)-HRESIMS m/z 245.1126 [M − H]− (calculated 245.1177) molecular ion peak indicating the existence of seven degrees of unsaturation. Compound 4 1H and 13C-NMR spectral data, compiled in Table 1, resembled those of compounds 2 and 3. The 13C-NMR spectrum confirmed the presence of fifteen carbon signals which were assigned, by DEPT and HMQC spectral analysis, to four methyls (δC 25.8, 23.5, 18.0, 16.2), one methylene (δC 37.4), two aromatic (δC 112.5, 110.0), and one olefinic (δC 117.2) methines and seven non-protonated carbons (δC 180.8, 150.4, 146.4, 136.5, 130.4, 123.9, 48.1), of which two were hydroxylated (δC 150.4, 146.4) and one an ester (δC 180.8). In accordance, the 1H-NMR spectrum showed four singlet methyls (δH 2.26, 1.60, 1.56, 1.44), one splitting methylene (δH 2.58, 2.42), two aromatic (δH 6.87, 6.64), and one olefinic (δH 4.85) methine. The same tetrasubstituted benzene ring found in compounds 2 and 3 was also proposed for compound 4 due to the similarity of the 1H and 13C-NMR data. The HMBC correlations H-2/C-4, H-2/C-6, H-5/C-1, H-5/C-3, and H-5/C-4 confirmed the proposed sub-structure. Further HMBC correlations Me-14/C-3 and Me-14/C-4 corroborated the assignment of this methyl group. The most notable new features of compound 4 were the carbonyl resonance (δC 180.8) and a non-protonated alkane carbon (δC 48.1). Allocation of these was accomplished based on the HMBC correlations of Me-13 with C-6, C-7, C-8, and C-15, confirming a lactone sub-structure. The COSY correlation H2-8/H-9 allowed assignment of the methylene and the Δ9(10) double bond, which was linked to the non-protonated C-10 based on the HMBC correlations of Me-11 and Me-12 with both C-9 and C-10. As a result, the structure of compound 4 was elucidated as the sesquiterpene shown in Figure 1: 4-hydroxy-3,7-dimethyl-7-(3-methylbut-2-en-1-yl)benzofuran-15-one.
Compound 5 was isolated as a yellow amorphous powder. Spectral data available in the literature allowed its identification as 1,8-epoxy-1(6),2,4,7,10-bisaborapentaen-4-ol [19].
Compound 6 was isolated as dark-brown oil. The molecular formula C16H24O3 was established based on the (−)-HRESIMS m/z molecular ion peak 263.1610 [M − H]− (calculated 263.1647), indicating five degrees of unsaturation. Both 1H and 13C-NMR indicated structural similarities with compounds 2–5 (Table 2). The same tetrasubstituted hydroquinone ring found in compounds 2 and 3 was suggested for compound 6. The HMBC correlations H-5/C-1, H-5/C-3, and H-5/C-4, represented in Figure 3, confirmed the proposed hydroquinone ring. The methyl-14 substitution was assigned based on HMBC correlations of this group with C-2, C-3, and C-4. The C-6 substitution was confirmed based on the HMBC correlations H-5/C-7, Me-15/C-6, and Me-15/C-7. The last correlation, together with Me-16/C-7, provided the key to methyl groups -15 and -16. The 13C-NMR and DEPT data suggested the presence of two methylenes (δC 39.8, 22.8), consistent with a side chain one carbon longer than found previously. Me-12 and Me-13 were assigned based on their HMBC correlations between each other and with C-10 and C-11. The COSY correlation H-10/H-9 allowed completion of this second sub-structure.

Table 2.
1H and 13C-NMR (400 and 100 MHz, respectively) for compounds 6 and 7. Experiments with compound 6 were done in CD3OD and with compound 7 in CDCl3.
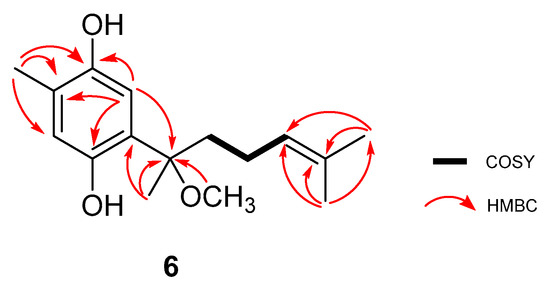
Figure 3.
Key 1H-1H COSY, HMBC, and TOCSY correlations of compound 6.
The configuration of the chiral center present in compound 6 could not be clearly elucidated with the material available and the physio-chemical information obtained for this compound. Thus, the structure of 6 was elucidated as the curcuhydroquinone derivative 6-(2-methoxy-6-methylhept-5-en-2-yl)-3-methylbenzene-1,4-diol (Figure 1).
Compound 7 was isolated as a green crystal. The (−)-HRESIMS showed the molecular ion peak m/z 245.1126 [M − H]− (calculated 245.1177), very similar to the one reported for compound 4. As for compound 4, C15H18O3 was the calculated molecular formula, indicating the existence of seven degrees of unsaturation. Analysis of the 1H and 13C-NMR spectral data, compiled in Table 2, and a comparison with of the data for the previously elucidated compounds revealed the presence of the phenolic part of the structure, but with considerable modifications in the side chain. As seen in Figure 4a, the HMBC correlations Me-15/C-6 and Me-15/C-7, together with the deshielded resonance of C-7 (δC 122.0) allowed assignment of this methyl group and the Δ7(8) double bond. Based on the COSY correlations between H-8, H-9, and H-10, a spin system was defined. Chemical shifts of the positions 9 (δ 4.51/55.6) and 10 (δ 3.06/63.8) indicated they were bearing an oxygen atom. The HMBC correlations between Me-15 with C-7/C-8/C-9 and H-10 C-11/Me-12/Me-13 allowed us to establish the position of the side chain. Furthermore, the chemical shift of the quaternary C-11 (δC 57.7) indicated that it was also oxygenated. This fact, along with the molecular formula, suggests cyclization of the phenol OH to the C-9 position and an epoxide between C-10 and C-11. Additionally, the chemical shift of the epoxide positions supported the proposed structure when compared to other related epoxide fragments described in the literature [20,21]. Finally, the MS fragmentation pattern showing the m/z fragments 230.1421, 165.0497, and 122.0332 (see Supplementary Materials Figure S45) reinforced this proposal.
Figure 4.
Key correlations for the elucidation of compound 7. (a) 1H-1H COSY and HMBC. (b) NOESY (partial structure).
A comparison of the resonances, together with a large coupling constant between H-9 and H-10 (8.3 Hz), allowed the relative configuration at C-9/C-10 to be determined as anti [22]. Furthermore, the NOE correlations H-12/H-10 and H-13/H-9, represented in Figure 4b, confirmed this configuration. Thus, the structure of 7 was elucidated as the curcuphenol derivative shown in Figure 1: 9-(3,3-dimethyloxiran-2-yl)-1,7-dimethyl-7-chromen-4-ol.
Several bisabolane-type sesquiterpenoids have been reported from different marine organisms, such as the marine sponge Halichondria sp. [18], the gorgonians Pseudopterogorgia spp. [7] or the red algae Laurencia scoparia [23]. The isolation of bisabolane-related compounds from microorganisms, such as the marine-derived fungus Aspergillus sp. [24], has been used to suggest that these compounds are produced by microbial-associated organisms and not directly by the host. In this work, we were able to isolate four new bisabolane-related compounds. For these new compounds from this work, no assumptions can be made as to whether the producer is the sponge or possible associated microorganisms since the metabolites were extracted indistinctly.
Bisabolane-like compounds have previously been isolated from marine sponges [9,10]. However, to the best of our knowledge, this work represents the first report of this class of compounds from Myrmekioderma sp. Besides belonging to a known class of compounds, the four new isolated bisabolane-related metabolites show novel structural features. Cyclic bisabolane and metabolites bearing oxo functionality are both uncommon among this group of compounds, highlighting the importance of these discoveries.
From the biosynthetic point of view, bisabolane-related compounds have already been described as a result of the combination of the shikimic and mevalonic acid pathways [25,26] and the same routes are proposed for the described compounds. Compound 4, however, has a migrated carbon relative to the curcuphenol skeleton, a feature that can be found in other related-compounds [27]. Compound 4 is, therefore, proposed to be obtained from tetraketide 3-methyl-orsellinic acid [27,28]. As such, there is strong evidence that the compounds originate from a fungi-associated strain. All seven isolated compounds were tested for their cytotoxic activity against A-549 human lung carcinoma cells, MDA-MB-231 human breast adenocarcinoma cells, HT-29 human colorectal carcinoma, and PSN1 human pancreatic adenocarcinoma cells. Compounds 1–7 were inactive (IC50 > 20 μM) in all the cancer cell lines tested.
The lipid-reducing activity of compounds 1–7 was also tested using the zebrafish Nile red fat metabolism assay (Figure 5 and Figure 6). This whole small animal model was already successfully used in the discovery of lipid-reducing compounds from fungus [16], chemically modified polyphenols [29] and cyanobacteria [30], and offers higher physiological relevance compared to commonly used cellular in vitro models. Furthermore, it was previously shown that zebrafish larvae responded similarly to humans if challenged with known lipid regulator drugs [31]. The results showed that compounds 2, 5, and 7 have potent lipid-reducing activity (IC50 = 1.78, 0.84, and 1.22 µM, respectively), reducing significantly the zebrafish Nile red fluorescence intensity, which is indicative of the total amount of neutral lipids. Compounds 3 and 4 also showed moderate lipid-reducing activity (IC50 = 7.89, 12.61 µM, respectively). None of the compounds 1–7 had any general toxicity on zebrafish larvae and additionally did not cause any malformations. It is interesting to observe that all the active compounds are bisabolane-related, while compound 1 did not show activity. The structural differences between compounds 2 or 3 compared to compound 6 caused the inactivation of the compound, but cyclizing the side chain (compound 7) did not. Therefore, more studies are needed in order to understand the relationship between the chemical structure and its lipid-reducing activity.
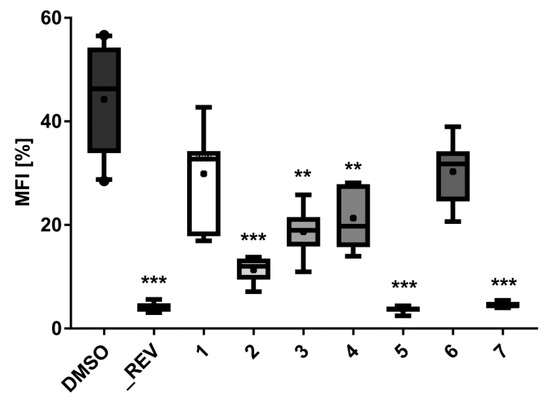
Figure 5.
Lipid-reducing activity of compounds 1–7 in zebrafish Nile red fat metabolism assay. MFI represents the mean fluorescence intensity, indicative of neutral lipids. A solvent control with 0.1% DMSO was included in the bioassay, together with the positive control 50 µM resveratrol (REV). Per treatment, 6–8 individual zebrafish larvae were used. ** p < 0.01, *** p < 0.001.
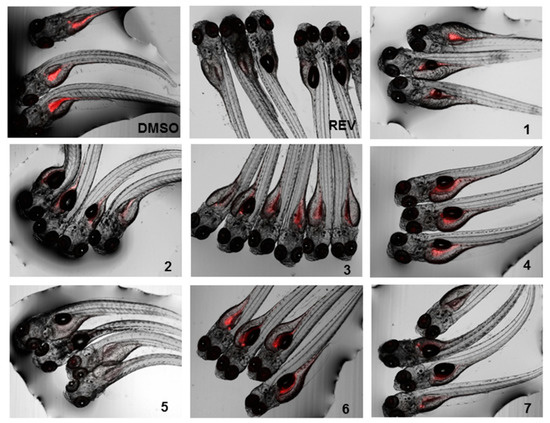
Figure 6.
Representative images of the zebrafish Nile red assay. Images show the overlay of the fluorescence and bright field images; 0.1% DMSO was used as solvent control and 50 µM resveratrol (REV) as positive control.
3. Materials and Methods
3.1. General Experiments
Optical rotations were measured on a Jasco P-1020 polarimeter. The UV spectra were measured using an Agilent 8453 UV-vis spectrometer. The IR spectra were recorded on a Nicolet iZ10 (ThermoFisher Scientific) FTIR spectrophotometer. The NMR experiments were performed on a Bruker 400 spectrometer at 400/100 MHz (1H/13C). Chemical shifts were reported in ppm using residual CD3OD (δ 3.31 for 1H and 49.0 for 13C) and CDCl3 (δ 7.26 for 1H and 77.2 for 13C) as internal references. The HRESIMS was performed on a Waters Synapt G1 UPLC-QTOF mass spectrometer in negative ionization mode.
3.2. Biological Sample
The Myrmekioderma sp. sponge was collected by hand while scuba diving in Boano (Indonesia). The sponge was immediately frozen and kept under these conditions until extraction. The specimen was identified by María Jesús Uriz at CEAB, Blanes, Spain. A voucher specimen (ORMA135834) is deposited at PharmaMar facilities (Madrid, Spain).
3.3. Extraction and Isolation
The frozen sponge specimen (320 g) was repeatedly extracted using dichloromethane: methanol (CH2:Cl2:MeOH 1:1 v/v). The extract was concentrated under vacuum to yield 25.91 g of crude extract. This crude extract was dissolved in 300 mL of water and subsequently extracted with n-hexane (3 × 300 mL), ethyl acetate (EtOAc) (3 × 300 mL), and butanol (n-BuOH) (2 × 250 mL). The n-hexane extract (6.11 g) was subjected to reversed phase VLC over RP-18 silica gel with a stepped gradient from H2O:MeOH (3:1 v/v) to dichloromethane (CH2Cl2). Fraction 1 (95.6 mg), eluted with H2O:MeOH (3:1 v/v), was subjected to semi-preparative HPLC (Gemini-NX C18 110A, Phenomenex, 5µ, 10.0 × 250 mm, gradient H2O:MeCN 60:40 v/v to 50:50 v/v, in 15 min, 3 mL/min) to yield compound 1 (6.4 mg) at 10 min. Fraction 3 (1640.7 mg), eluted with pure MeOH, was initially separated by preparative HPLC (Luna C18 100A, Phenomenex, 5 µ, 21.20 × 250 mm, gradient H2O:MeCN (25:75 v/v) to 0:100, in 30 min, 6 mL/min), yielding HPLC Fraction 2 at 14 minutes (444.2 mg). This fraction was again separated by preparative HPLC (Luna C18 100A, Phenomenex, 5 µ, 21.20 × 250 mm, gradient H2O:MeCN 50:50 v/v to 40:60 v/v, in 25 min, 10 mL/min), yielding compound 2 (98.6 mg) at 21 minutes and HPLC Fraction 4 at 24 minutes (146.6 mg). The HPLC Fraction 4 was submitted to a final semi-preparative HPLC separation (Gemini-NX C18 110A column, 5 µ, Phenomenex, 10.0 × 250 mm, gradient H2O:MeCN 50:50 v/v to 30:70 v/v, in 35 min, 2.3 mL/min) to yield compounds 3 (1.3 mg) at 11 min, 4 (4.9 mg) at 21 min and 5 (9.4 mg) at 34 min. The EtOAc extract from the original liquid/liquid extraction was also subjected to reversed phase VLC over RP-18 silica gel with a stepped gradient from H2O:MeOH (3:1 v/v) to CH2Cl2. Fraction 2 (1021.7 mg) eluted with H2O:MeOH (1:3 v/v) and was further separated by preparative HPLC (Luna C18 100A, Phenomenex, 5 µ, 21.20 × 250 mm, gradient H2O:MeCN (50:50 v/v) to 20:80 v/v, in 30 min, 8 mL/min), to yield compounds 6 (46.5 mg) at 28 min and 7 (23.3 mg) at 19 min.
1-(2,4-dihydroxy-5-methylphenyl)ethan-1-one (1): Dark-brown oil; IR (neat) ʋmax, 3314 (br), 2971, 2853, 1652, 1406, 1038 cm−1; UV/Vis (MeOH) λmax 194, 210, 232, 265, 360 nm. HRESIMS: m/z 165.0552 [M − H]− (calcd for C9H9O3, 165.0552).
(1’Z)-2-(1’,5’-dimethylhexa-1’,4’-dieny1)-5-methylbenzene-1,4-diol (2): Dark-brown oil; IR (neat) ʋmax 3413 (br), 2970, 2913, 1416, 1187 cm−1; UV/Vis (MeOH) λmax 229, 299 nm. HRESIMS: m/z 231.1496 [M − H]− (calcd for C15H19O2, 231.1385).
6-(3-hydroxy-6-methyl-1,5-heptadien-2-yl)-3-methylbenzene-1,4-diol (3): Yellow amorphous solid; (α) +0.72 (c 0.484, CH3OH); IR (MeOH) ʋmax 3314 (br), 2943, 2831, 1033 cm−1; UV/Vis (CH3OH) λmax 195, 299 nm. 1H-NMR (400 MHz, CDCl3) and 13C-NMR (100 MHz, CDCl3) see Table 1; HRESIMS: m/z 247.1344 [M − H]− (calcd for C15H19O3, 247.1334), m/z 149.0575 (M − C6H11O)− (calcd for C9H9O2, 149.0602).
4-hydroxy-3,7-dimethyl-7-(3-methylbut-2-en-1-yl)benzofuran-15-one (4): Yellow amorphous solid; (α) +2.2 (c 0.115, MeOH); IR (MeOH) ʋmax 3313 (br), 2944, 2832, 1656, 1451, 1035 cm−1; UV/Vis (MeOH) λmax 196, 294 nm. 1H-NMR (400 MHz, CDCl3) and 13C-NMR (100 MHz, CDCl3) see Table 1; HRESIMS: m/z 245.1126 [M − H]− (calcd for C15H17O3, 245.1177).
1,8-epoxy-1(6),2,4,7,10-bisaborapentaen-4-ol (5): Yellow amorphous solid; IR (neat) ʋmax 3266 (br), 2915, 1437, 1168, 805, 434 cm−1; UV/Vis (MeOH) λmax 203, 257, 297 nm. HRESIMS: m/z 229.1234 [M − H]− (calcd for C15H17O2, 229.1229).
6-(2-methoxy-5-methylhept-4-en-2-yl)-3-methylbenzene-1,4-diol (6): Dark-brown oil; (α) +5.0 (c 0.0337, CH3OH); IR (MeOH) ʋmax 3339 (br), 2926, 1453, 1374, 1183, 1051 cm−1; UV/Vis (CH3OH) λmax 196, 297 nm. 1H-NMR (400 MHz, CDCl3) and 13C-NMR (100 MHz, CDCl3) see Table 2; HRESIMS: m/z 263.1610 [M − H]− (calcd for C16H23O3, 263.1647).
9-(3,3-dimethyloxiran-2-yl)-1,7-dimethyl-7-chromen-4-ol (7): Green crystals; (α) −10.4 (c 0.0322, CH3OH); IR (neat) ʋmax 3388 (br), 2926, 1412, 1178, 994, 829, 597 cm−1; UV/Vis (CH3OH) λmax 194, 217, 330 nm. 1H-NMR (400 MHz, CDCl3) and 13C-NMR (100 MHz, CDCl3) see Table 2; HRESIMS: m/z 245.1126 [M − H]− (calcd for C15H17O3, 245.1177).
3.4. Biological Activities
3.4.1. Cytotoxicity
The cytotoxic activity of compounds 1–7 was tested against A-549 human lung carcinoma cells, MDA-MB-231 human breast adenocarcinoma cells, HT-29 human colorectal carcinoma cells, and PSN1 human pancreatic adenocarcinoma cells. The four cell lines were provided by the American Type Culture Collection (ATCC): A549 from ATCC CCL-185, MDA-MB-231 from ATCC HTB-26, HT-29 from ATCC HTB-38 and PSN-1 from ATCC CRM-CRL-3211. The concentration giving half maximum inhibitory concentration (IC50) was calculated according to the procedure described in the literature [32]. Cell survival was estimated using the National Cancer Institute (NCI) algorithm [33]. Dose-response parameters were determined at three different concentrations of each one of the compounds.
3.4.2. Zebrafish Nile Red Fat Metabolism Assay
The lipid reducing activity of the compounds was analyzed using the zebrafish Nile red fat metabolism assay as previously described [16,25]. Approval by an ethics committee was not necessary for the presented work since the procedures used are not considered animal experimentation according to EC Directive 86/609/EEC for animal experiments. In brief, zebrafish embryos were raised from 1 DPF (days post fertilization) in egg water (60 µg/mL marine sea salt dissolved in distilled H2O) with 200 µM PTU (1-phenyl-2-thiourea) to inhibit pigmentation. From 3 DPF to 5 DPF, zebrafish larvae were exposed to compounds at a final concentration of 10 µM with the daily renewal of water and compounds in a 48 well plate with a density of 6–8 larvae/well (n = 6–8). A solvent control (0.1% DMSO) and positive control (REV, resveratrol, final concentration of 50 µM) were included in the assay. Lipids were stained with Nile red overnight at the final concentration of 10 ng/mL. For imaging, the larvae were anesthetized with tricaine (MS-222, 0.03%) for 5 minutes and fluorescence analyzed with a fluorescence microscope (Olympus BX43, Hamburg, Germany). Fluorescence intensity was quantified in individual zebrafish larvae by ImageJ [34]. Effective concentrations 50% (EC50) values were determined in further assays by dose-response curves by using a 1:2 v/v dilution series from 20 µM to 312.5 nM (final concentrations) in 7 dilution steps.
4. Conclusions
This work represents the first isolation and structural elucidation of novel compounds 3, 4, 6, and 7. It is also the first report of the isolation of compounds 1, 2, and 5 from marine sources. Besides being a known and wide-spread class of compounds, the structures of the new compounds isolated present unique structural features. The isolation of these novel compounds, as well as related analogs previously found in marine-derived organisms, raises the question of who is the real metabolite producer, the host or the associated-microorganisms. Further studies are needed in order to answer that question. All of the isolated compounds except for 1 and 6, showed significant lipid-reducing activity when tested in the zebrafish Nile red fat metabolism assay, but no general toxicity, reinforcing their biotechnological potential. More studies are needed in order to relate the bioactivity with structural features.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/17/6/375/s1. Figure S1: Picture of the fresh sponge; Figures S2–S44: HRESIMS and NMR spectra of compounds 1–7; Figure S45: MS fragmentation pattern of compound 7.
Author Contributions
M.C. performed the isolation and structural elucidation of the compounds and wrote the paper. L.C. performed the organic extractions. R.U. conducted the zebrafish Nile red fat metabolism assay. M.P. and M.T. designed and guided the experiments. All the authors read, reviewed, and agreed with the structure and content of the manuscript.
Funding
The research leading to these results received funding from the Marie Curie Actions of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA grant agreement No. 607786, BluePharmTrain, and by the European ERA-NET Marine Biotechnology project CYANOBESITY (ERA-MBT/0001/2015), financed by national funds through FCT (Foundation for Science and Technology, Portugal), RANNIS (Icelandic Center of Research, Iceland), and FCT strategic fund UID/Multi/04423/2019. Ralph Urbatzka was supported by FCT grant SFRH/BPD/112287/2015.
Acknowledgments
The authors gratefully acknowledge the help of their PharmaMar colleagues and all the assistance given, including R. Fernández for revising the spectroscopic data and the manuscript and J.M. Dominguez for the cytotoxicity assays. The authors would also like to thank Andalas University (Indonesia) for helping with the sponge collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Haefner, B. Drugs from the deep: Marine natural products as drug candidates. Drug Discov. Today 2003, 8, 536–544. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Jansen, B.J.M.; de Groot, A. Occurrence, biological activity and synthesis of drimane sesquiterpenoids. Nat. Prod. Rep. 2004, 21, 449–477. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.-Q.; Liu, H.-L.; Chen, S.-H.; Mollo, E.; Gavagnin, M.; Li, J.; Li, X.-W.; Guo, Y.-W. 5-Alkylpyrrole-2-carboxaldehyde derivatives from the Chinese sponge Mycale lissochela and their PTP1B inhibitory activities. Chin. Chem. Lett. 2017, 28, 1190–1193. [Google Scholar] [CrossRef]
- Miller, S.L.; Tinto, W.F.; McLean, S.; Reynolds, W.F.; Yu, M. Bisabolane Sesquiterpenes from Barbadian Pseudopterogorgia spp. J. Nat. Prod. 1995, 58, 1116–1119. [Google Scholar] [CrossRef]
- Jin, A.; Wu, W.-M.; Yu, H.-Y.; Zhou, M.; Liu, Y.; Tian, T.; Ruan, H.-L. Bisabolane-Type Sesquiterpenoids from the Whole Plant of Parasenecio rubescens. J. Nat. Prod. 2015, 78, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Yegdaneh, A.; Putchakarn, S.; Yuenyongsawad, S.; Ghannadi, A.; Plubrukarn, A. 3-Oxoabolene and 1-Oxocurcuphenol, Aromatic Bisabolanes from the Sponge Myrmekioderma sp. Nat. Prod. Commun. 2013, 8, 1355–1357. [Google Scholar] [CrossRef]
- Wright, A.E.; Pomponi, S.A.; McConnell, O.J.; Kohmoto, S.; McCarthy, P.J. (+)-Curcuphenol and (+)-Curcudiol, Sesquiterpene Phenols from Shallow and Deep Water Collections of the Marine Sponge Didiscus flavus. J. Nat. Prod. 1987, 50, 976–978. [Google Scholar] [CrossRef]
- Peralta-Yahya, P.P.; Ouellet, M.; Chan, R.; Mukhopadhyay, A.; Keasling, J.D.; Lee, T.S. Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2011, 2, 483. [Google Scholar] [CrossRef] [PubMed]
- Hurt, R.T.; Kulisek, C.; Buchanan, L.A.; McClave, S.A. The Obesity Epidemic: Challenges, Health Initiatives, and Implications for Gastroenterologists. Gastroenterol. Hepatol. 2010, 6, 780–792. [Google Scholar]
- Fu, C.; Jiang, Y.; Guo, J.; Su, Z. Natural Products with Anti-obesity Effects and Different Mechanisms of Action. J. Agric. Food Chem. 2016, 64, 9571–9585. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Tao, N.; Wang, X.; Xiao, J.; Wang, M. Marine-derived bioactive compounds with anti-obesity effect: A review. J. Funct. Foods 2016, 21, 372–387. [Google Scholar] [CrossRef]
- Castro, M.; Preto, M.; Vasconcelos, V.; Urbatzka, R. Obesity: The Metabolic Disease, Advances on Drug Discovery and Natural Product Research. Curr. Top. Med. Chem. 2016, 16, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Noinart, J.; Buttachon, S.; Dethoup, T.; Gales, L.; Pereira, J.A.; Urbatzka, R.; Freitas, S.; Lee, M.; Silva, A.M.S.; Pinto, M.M.M.; et al. A New Ergosterol Analog, a New Bis-Anthraquinone and Anti-Obesity Activity of Anthraquinones from the Marine Sponge-Associated Fungus Talaromyces stipitatus KUFA 0207. Mar. Drugs 2017, 15, 139. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.G.; Pavkov-Keller, T.; Richter, N.; Wiltschi, B.; Gruber, K.; Kroutil, W. Biocatalytic Friedel–Crafts Acylation and Fries Reaction. Angew. Chem. Int. Ed. 2017, 56, 7615–7619. [Google Scholar] [CrossRef] [PubMed]
- Capon, R.; Ghisalberti, E.; Jefferies, P. New aromatic sesquiterpenes from a Halichondria sp. Aust. J. Chem. 1982, 35, 2583–2587. [Google Scholar] [CrossRef]
- Arihara, S.; Umeyama, A.; Bando, S.; Imoto, S.; Ono, M.; Yoshikawa, K. Three New Sesquiterpenes from the Black Heartwood of Cryptomeria japonica. Chem. Pharm. Bull. 2004, 52, 463–465. [Google Scholar] [CrossRef]
- Raju, B.; Subbaraju, G.; Rao, C.; Trimurtulu, G. Two New Oxigenated Lobanes from a Soft Coral of Lobophytum species of the Andaman and Nicobar Coasts. J. Nat. Prod. 1993, 56, 961–966. [Google Scholar] [CrossRef]
- Duh, C.; El-Gamal, A.; Chiang, C.; Chu, C.; Wang, S.; Dai, C. New Cytotoxic Xenia Diterpenoids from the Formosan Soft Coral Xenia umbellata. J. Nat. Prod. 2002, 65, 1882–1885. [Google Scholar] [CrossRef] [PubMed]
- Bishara, A.; Rudi, A.; Goldberg, I.; Benayahu, Y.; Kashman, Y. Novaxenicins A–D and xeniolides I–K, seven new diterpenes from the soft coral Xenia novaebrittanniae. Tetrahedron 2006, 62, 12092–12097. [Google Scholar] [CrossRef]
- Davyt, D.; Fernandez, R.; Suescun, L.; Mombru, A.W.; Saldana, J.; Dominguez, L.; Fujii, M.T.; Manta, E. Bisabolanes from the red alga Laurencia scoparia. J. Nat. Prod. 2006, 69, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.Y.; Wang, C.Y.; Liu, Q.A.; Shao, C.L.; She, Z.G.; Lin, Y.C. Five Sesquiterpenoids from a Marine-Derived Fungus Aspergillus sp Isolated from a Gorgonian Dichotella gemmacea. Mar. Drugs 2010, 8, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Hansson, D.; Menkis, A.; Olson, Å.; Stenlid, J.; Broberg, A.; Karlsson, M. Biosynthesis of fomannoxin in the root rotting pathogen Heterobasidion occidentale. Phytochemistry 2012, 84, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Prompanya, C.; Dethoup, T.; Gales, L.; Lee, M.; Pereira, J.A.C.; Silva, A.M.S.; Pinto, M.M.M.; Kijjoa, A. New Polyketides and New Benzoic Acid Derivatives from the Marine Sponge-Associated Fungus Neosartorya quadricincta KUFA 0081. Mar Drugs 2016, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Kehraus, S.; Prudêncio, M.; König, M.K. Marilones A–C, phthalides from the sponge-derived fungus Stachylidium sp. Beilstein J. Org. Chem. 2011, 7, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- El Maddah, F.; Eguereva, E.; Kehraus, S.; König, G.M. Biosynthetic studies of novel polyketides from the marine sponge-derived fungus Stachylidium sp. 293K04. Org. Biomol. Chem. 2019, 17, 2747–2752. [Google Scholar] [CrossRef] [PubMed]
- Urbatzka, R.; Freitas, S.; Palmeira, A.; Almeida, T.; Moreira, J.; Azevedo, C.; Afonso, C.; Correia-da-Silva, M.; Sousa, E.; Pinto, M.; et al. Lipid reducing activity and toxicity profiles of a library of polyphenol derivatives. Eur. J. Med. Chem. 2018, 151, 272–284. [Google Scholar] [CrossRef]
- Costa, M.; Rosa, F.; Ribeiro, T.; Hernandez-Bautista, R.; Bonaldo, M.; Goncalves Silva, N.; Eiriksson, F.; Thorsteinsdottir, M.; Ussar, S.; Urbatzka, R. Identification of Cyanobacterial Strains with Potential for the Treatment of Obesity-Related Co-Morbidities by Bioactivity, Toxicity Evaluation and Metabolite Profiling. Mar. Drugs 2019, 17, 280. [Google Scholar] [CrossRef]
- Jones, K.S.; Alimov, A.P.; Rilo, H.L.; Jandacek, R.J.; Woollett, L.A.; Penberthy, W.T. A high throughput live transparent animal bioassay to identify non-toxic small molecules or genes that regulate vertebrate fat metabolism for obesity drug development. Nutr. Metab. 2008, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. JNCI 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Available online: https://imagej.nih.gov/ij/index.html (accessed on 7 September 2018).
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).