Lysozyme Amyloid Fibril Structural Variability Dependence on Initial Protein Folding State
Abstract
:1. Introduction
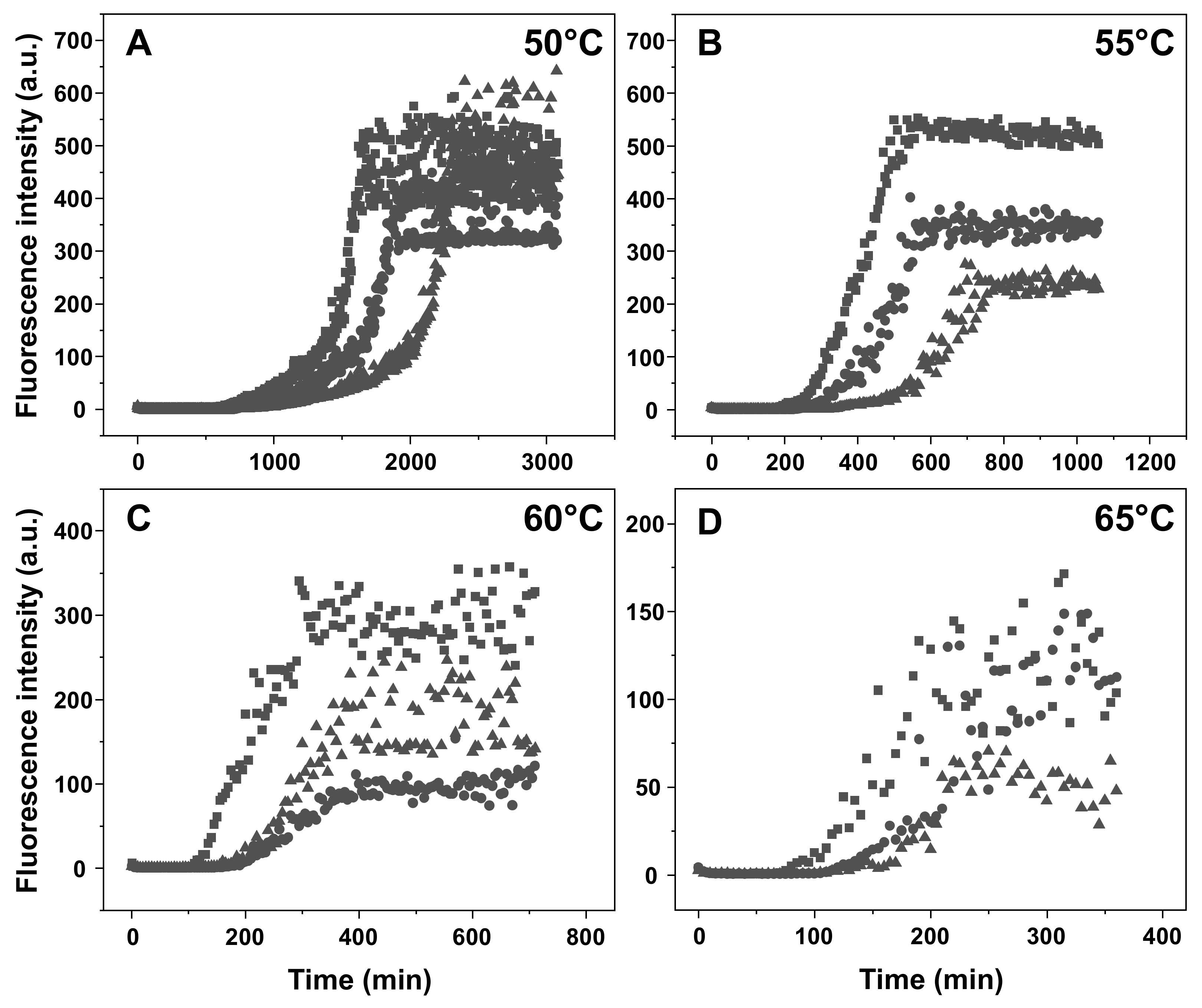
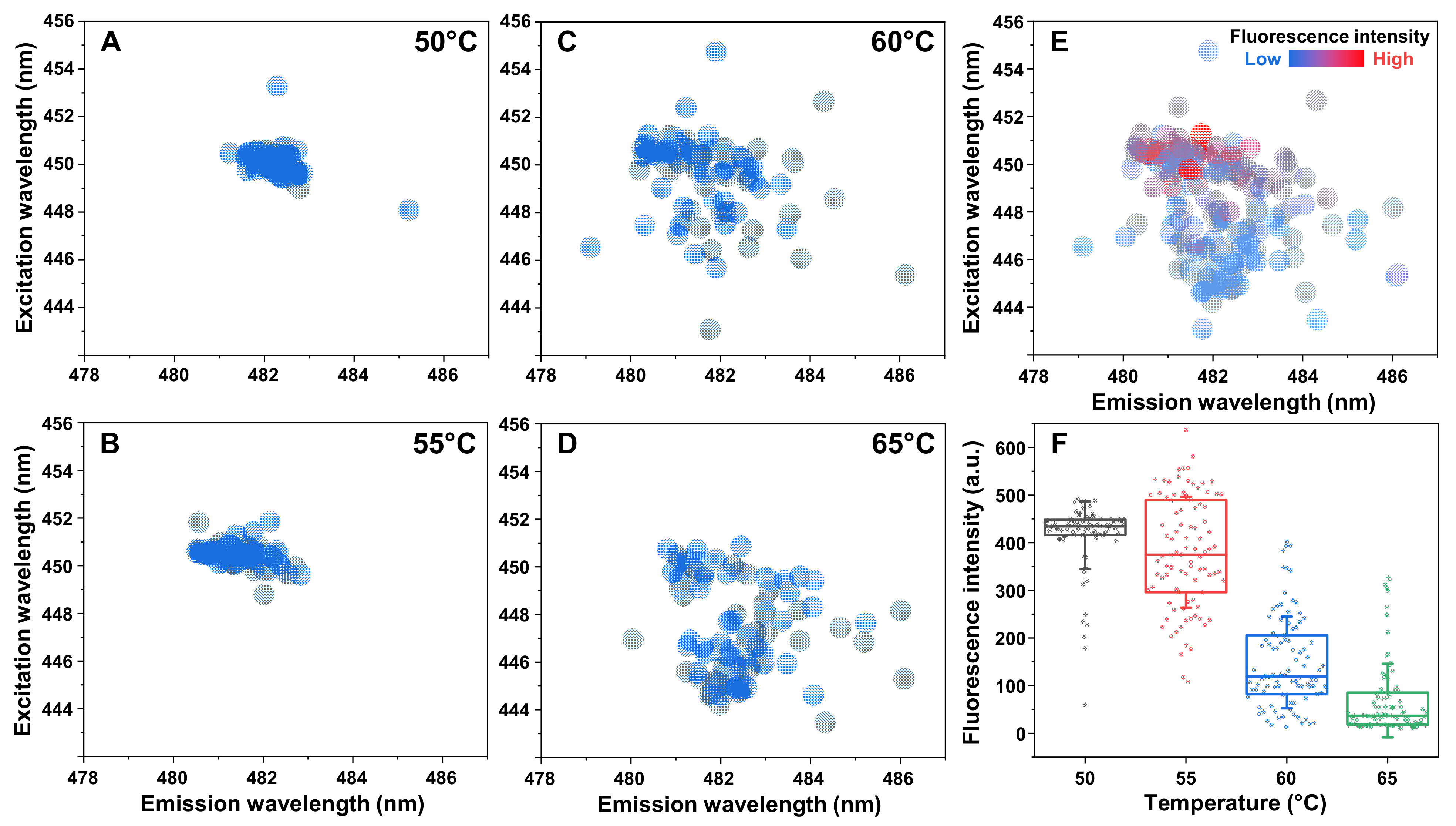
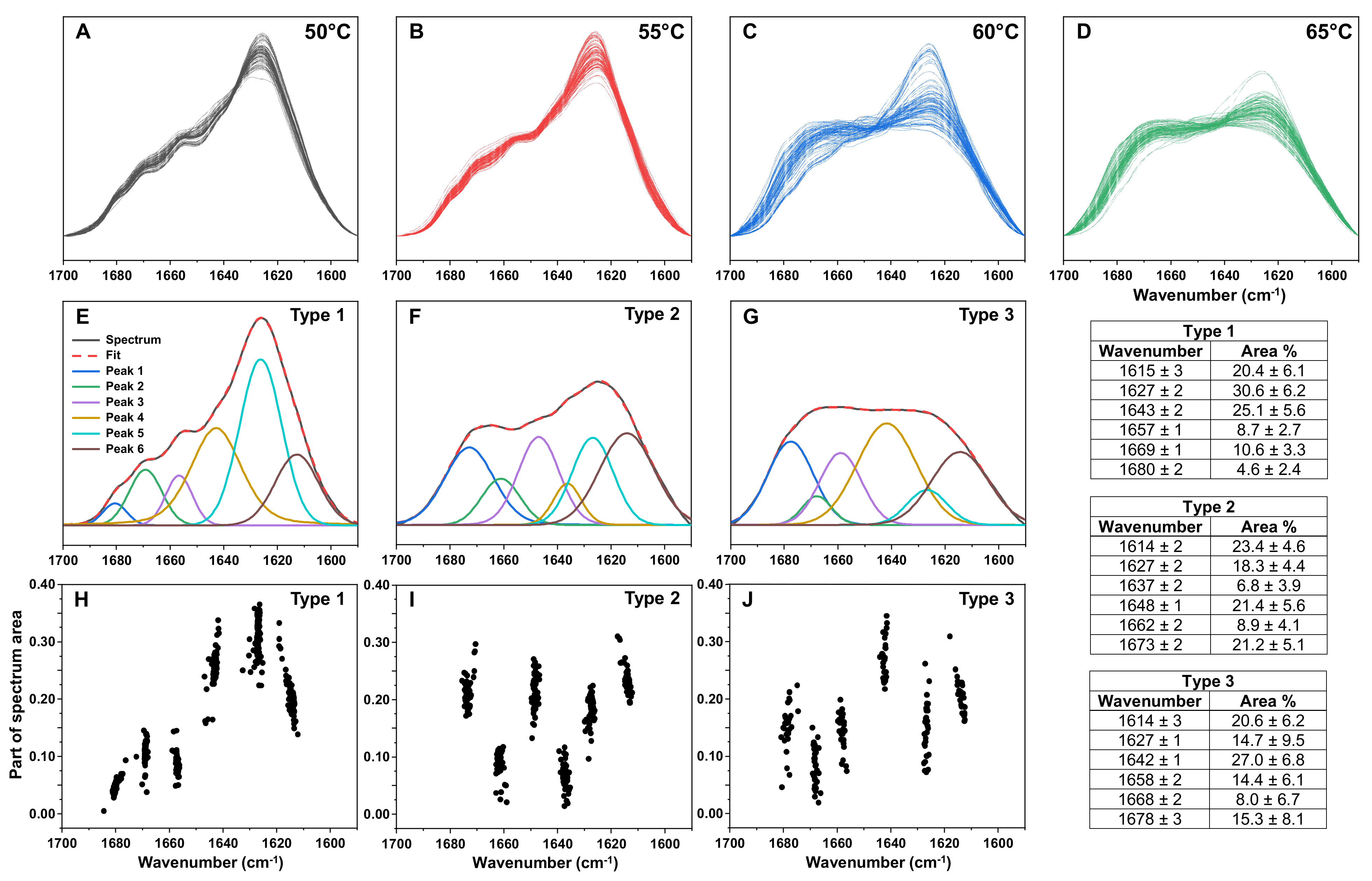
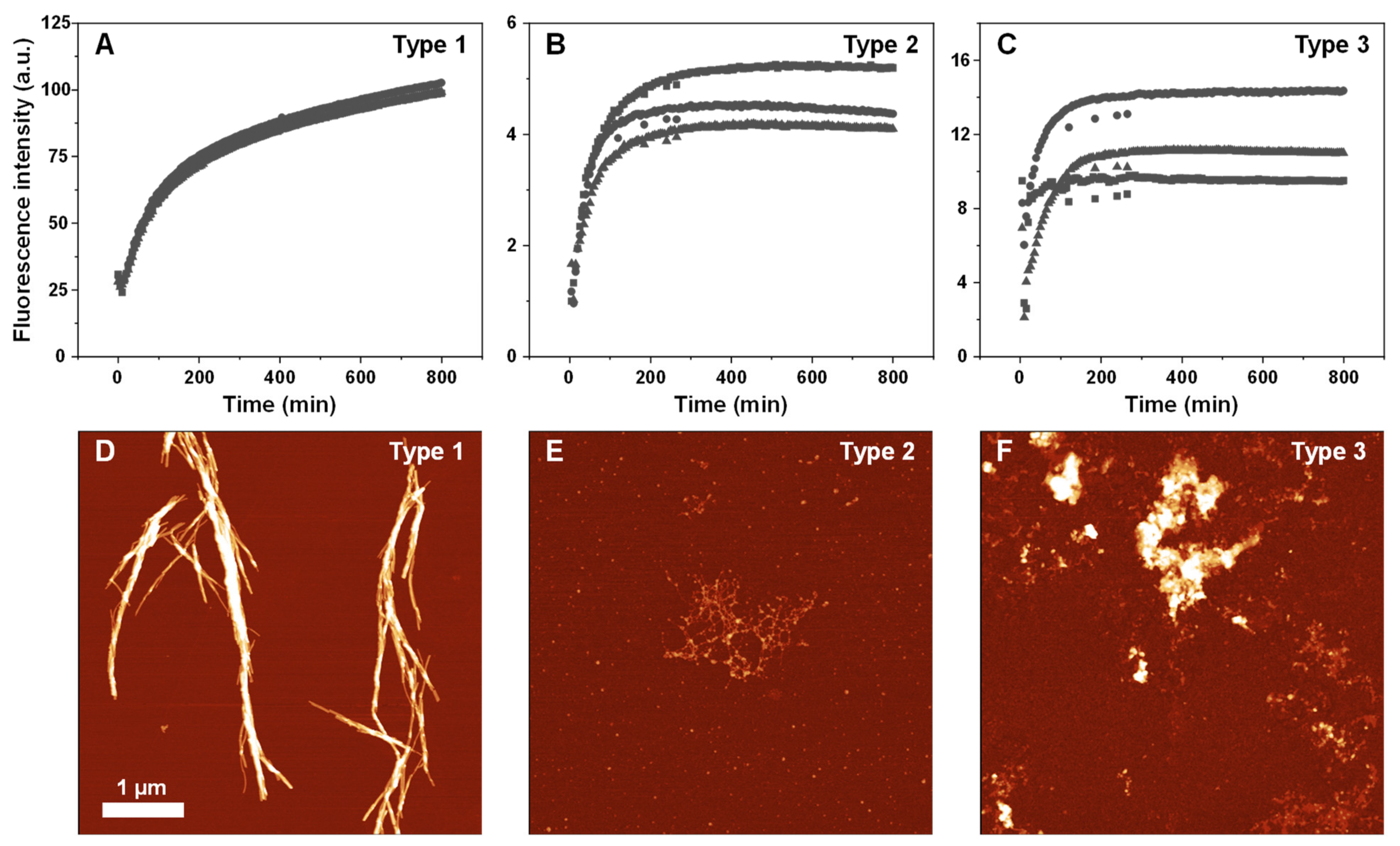
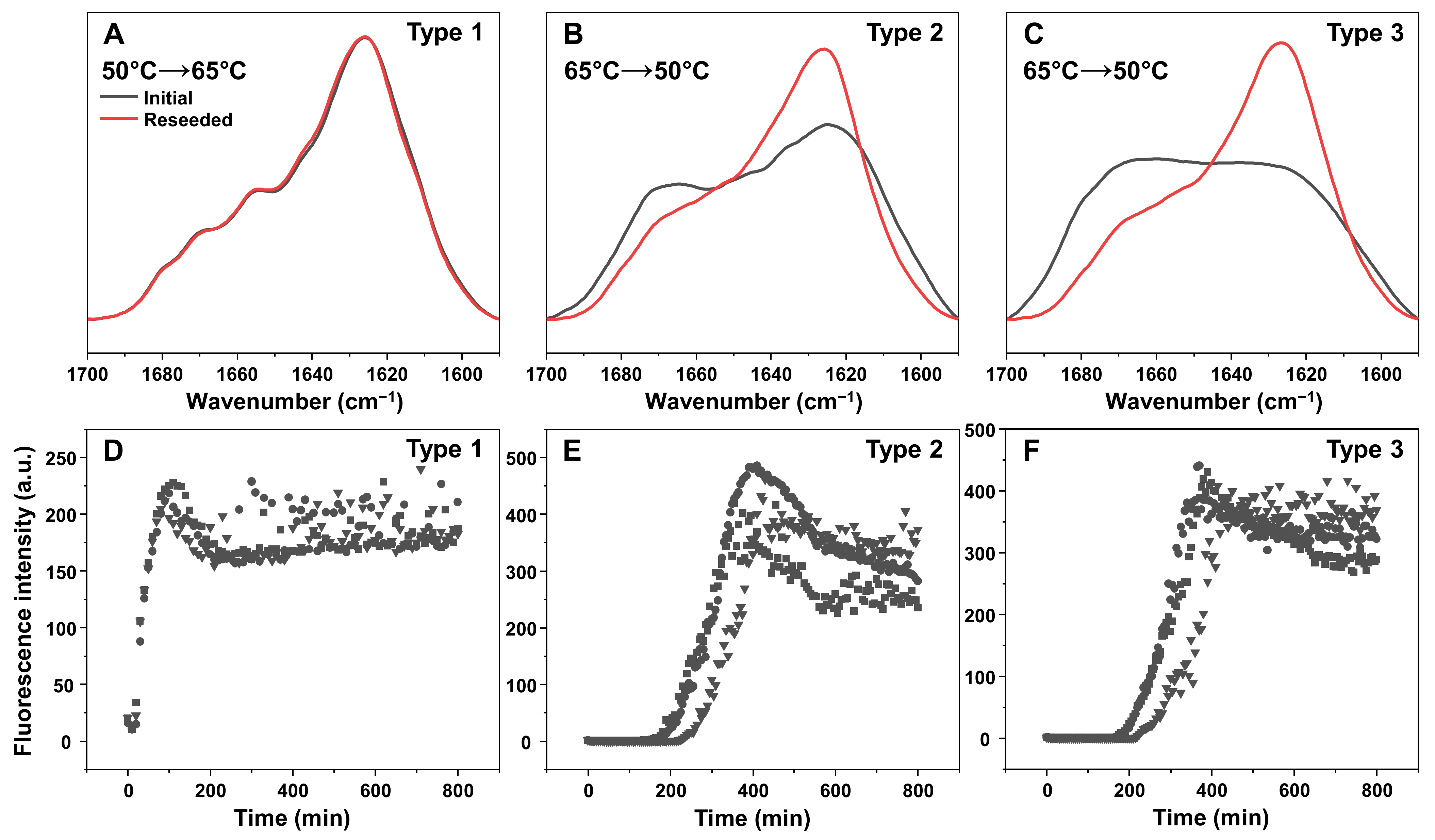
2. Results
3. Discussion
4. Materials and Methods
4.1. Lysozyme Aggregation Kinetics
4.2. Lysozyme Melt Assay
4.3. Excitation–Emission Matrices
4.4. Fourier-Transform Infrared Spectroscopy
4.5. Aggregate Reseeding
4.6. Atomic Force Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Baker, K.R.; Rice, L. The Amyloidoses: Clinical Features, Diagnosis and Treatment. Methodist Debakey Cardiovasc. J. 2012, 8, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Lee, G.; Zhong, K.; Fonseca, J.; Taghva, K. Alzheimer’s disease drug development pipeline: 2021. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2021, 7, e12179. [Google Scholar] [CrossRef] [PubMed]
- Chatani, E.; Yamamoto, N. Recent progress on understanding the mechanisms of amyloid nucleation. Biophys. Rev. 2018, 10, 527–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, D.; Jackson, R.; Paul, G.; Shi, J.; Sabbagh, M. Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010–2015. Expert Opin. Investig. Drugs 2017, 26, 735–739. [Google Scholar] [CrossRef]
- Doig, A.J.; del Castillo-Frias, M.P.; Berthoumieu, O.; Tarus, B.; Nasica-Labouze, J.; Sterpone, F.; Nguyen, P.H.; Hooper, N.M.; Faller, P.; Derreumaux, P. Why Is Research on Amyloid-β Failing to Give New Drugs for Alzheimer’s Disease? ACS Chem. Neurosci. 2017, 8, 1435–1437. [Google Scholar] [CrossRef] [Green Version]
- Arthur, K.C.; Calvo, A.; Price, T.R.; Geiger, J.T.; Chiò, A.; Traynor, B.J. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat. Commun. 2016, 7, 12408. [Google Scholar] [CrossRef] [Green Version]
- Brookmeyer, R.; Gray, S.; Kawas, C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am. J. Public Health 1998, 88, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Meisl, G.; Yang, X.; Hellstrand, E.; Frohm, B.; Kirkegaard, J.B.; Cohen, S.I.A.; Dobson, C.M.; Linse, S.; Knowles, T.P.J. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl. Acad. Sci. USA 2014, 111, 9384–9389. [Google Scholar] [CrossRef] [Green Version]
- Buell, A.K.; Galvagnion, C.; Gaspar, R.; Sparr, E.; Vendruscolo, M.; Knowles, T.P.J.; Linse, S.; Dobson, C.M. Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc. Natl. Acad. Sci. USA 2014, 111, 7671–7676. [Google Scholar] [CrossRef] [Green Version]
- Gurry, T.; Stultz, C.M. Mechanism of amyloid-β fibril elongation. Biochemistry 2014, 53, 6981–6991. [Google Scholar] [CrossRef]
- Milto, K.; Botyriute, A.; Smirnovas, V. Amyloid-Like Fibril Elongation Follows Michaelis-Menten Kinetics. PLoS ONE 2013, 8, e68684. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.A.; Chen, L.Y.; Plascencia-Villa, G.; Perry, G. Thermodynamics of Amyloid-β Fibril Elongation: Atomistic Details of the Transition State. ACS Chem. Neurosci. 2018, 9, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Nicoud, L.; Lazzari, S.; Balderas Barragán, D.; Morbidelli, M. Fragmentation of Amyloid Fibrils Occurs in Preferential Positions Depending on the Environmental Conditions. J. Phys. Chem. B 2015, 119, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Törnquist, M.; Michaels, T.C.T.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.A.; Knowles, T.P.J.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [Green Version]
- Foderà, V.; Librizzi, F.; Groenning, M.; Van De Weert, M.; Leone, M. Secondary nucleation and accessible surface in insulin amyloid fibril formation. J. Phys. Chem. B 2008, 112, 3853–3858. [Google Scholar] [CrossRef]
- Gaspar, R.; Meisl, G.; Buell, A.K.; Young, L.; Kaminski, C.F.; Knowles, T.P.J.; Sparr, E.; Linse, S. Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Q. Rev. Biophys. 2017, 50, e6. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, A.; Segers-Nolten, I.; Raussens, V.; Claessens, M.M.A.E.; Subramaniam, V. Distinct Mechanisms Determine α-Synuclein Fibril Morphology during Growth and Maturation. ACS Chem. Neurosci. 2017, 8, 538–547. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, S.; Matsumoto, F.; Yonezawa, Y. Effects of salt concentration on association of the amyloid protofilaments of Hen egg white lysozyme studied by time-resolved neutron scattering. J. Mol. Biol. 2003, 331, 21–28. [Google Scholar] [CrossRef]
- Loquet, A.; El Mammeri, N.; Stanek, J.; Berbon, M.; Bardiaux, B.; Pintacuda, G.; Habenstein, B. 3D structure determination of amyloid fibrils using solid-state NMR spectroscopy. Methods 2018, 138, 26–38. [Google Scholar] [CrossRef] [Green Version]
- Lattanzi, V.; André, I.; Gasser, U.; Dubackic, M.; Olsson, U.; Linse, S. Amyloid β 42 fibril structure based on small-angle scattering. Proc. Natl. Acad. Sci. USA 2021, 118, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, T.K.; Kalverda, A.P.; Thompson, G.S.; Radford, S.E. Mechanisms of amyloid formation revealed by solution NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 88, 86–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, S.D.; Zanni, M.T. How to Get Insight into Amyloid Structure and Formation from Infrared Spectroscopy. J. Phys. Chem. Lett. 2014, 5, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Sneideris, T.; Darguzis, D.; Botyriute, A.; Grigaliunas, M.; Winter, R.; Smirnovas, V. pH-Driven Polymorphism of Insulin Amyloid-Like Fibrils. PLoS ONE 2015, 10, e0136602. [Google Scholar] [CrossRef]
- Wawer, J.; Szociński, M.; Olszewski, M.; Piątek, R.; Naczk, M.; Krakowiak, J. Influence of the ionic strength on the amyloid fibrillogenesis of hen egg white lysozyme. Int. J. Biol. Macromol. 2019, 121, 63–70. [Google Scholar] [CrossRef]
- Gaspar, R.; Lund, M.; Sparr, E.; Linse, S. Anomalous Salt Dependence Reveals an Interplay of Attractive and Repulsive Electrostatic Interactions in α-synuclein Fibril Formation. QRB Discov. 2020, 1, e2. [Google Scholar] [CrossRef]
- Noormägi, A.; Valmsen, K.; Tõugu, V.; Palumaa, P. Insulin Fibrillization at Acidic and Physiological pH Values is Controlled by Different Molecular Mechanisms. Protein J. 2015, 34, 398–403. [Google Scholar] [CrossRef]
- Sakalauskas, A.; Ziaunys, M.; Smirnovas, V. Concentration-dependent polymorphism of insulin amyloid fibrils. PeerJ 2019, 7, e8208. [Google Scholar] [CrossRef]
- Fändrich, M.; Meinhardt, J.; Grigorieff, N. Structural polymorphism of Alzheimer Aβ and other amyloid fibrils. Prion 2009, 3, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Ziaunys, M.; Sakalauskas, A.; Mikalauskaite, K.; Snieckute, R.; Smirnovas, V. Temperature-dependent structural variability of prion protein amyloid fibrils. Int. J. Mol. Sci. 2021, 22, 5075. [Google Scholar] [CrossRef]
- Zhang, W.; Falcon, B.; Murzin, A.G.; Fan, J.; Crowther, R.A.; Goedert, M.; Scheres, S.H.W. Heparin-induced tau filaments are polymorphic and differ from those in alzheimer’s and pick’s diseases. eLife 2019, 8, e43584. [Google Scholar] [CrossRef]
- Collinge, J.; Clarke, A.R. A General Model of Prion Strains and Their Pathogenicity. Science 2007, 318, 930–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Li, Y.; Bai, Y. Elevated temperatures accelerate the formation of toxic amyloid fibrils of hen egg-white lysozyme. Vet. Med. Sci. 2021, 7, 1938–1947. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, R.; Ravi, V.K.; Kumar, S.; Kumar, M.V.S.; Chandra, N. Lysozyme: A model protein for amyloid research. In Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 84, pp. 63–111. ISBN 9780123864833. [Google Scholar]
- Ow, S.-Y.; Dunstan, D.E. The effect of concentration, temperature and stirring on hen egg white lysozyme amyloid formation. Soft Matter 2013, 9, 9692. [Google Scholar] [CrossRef] [PubMed]
- Arnaudov, L.N.; De Vries, R. Thermally induced fibrillar aggregation of hen egg white lysozyme. Biophys. J. 2005, 88, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Sulatskaya, A.I.; Rodina, N.P.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K. Different conditions of fibrillogenesis cause polymorphism of lysozyme amyloid fibrils. J. Mol. Struct. 2017, 1140, 52–58. [Google Scholar] [CrossRef]
- Rahimi Araghi, L.; Dee, D.R. Cross-Species and Cross-Polymorph Seeding of Lysozyme Amyloid Reveals a Dominant Polymorph. Front. Mol. Biosci. 2020, 7, 206. [Google Scholar] [CrossRef]
- Ziaunys, M.; Sakalauskas, A.; Mikalauskaite, K.; Smirnovas, V. Polymorphism of Alpha-Synuclein Amyloid Fibrils Depends on Ionic Strength and Protein Concentration. Int. J. Mol. Sci. 2021, 22, 12382. [Google Scholar] [CrossRef]
- Ziaunys, M.; Sakalauskas, A.; Smirnovas, V. Identifying Insulin Fibril Conformational Differences by Thioflavin-T Binding Characteristics. Biomacromolecules 2020, 21, 4989–4997. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikalauskaite, K.; Ziaunys, M.; Smirnovas, V. Lysozyme Amyloid Fibril Structural Variability Dependence on Initial Protein Folding State. Int. J. Mol. Sci. 2022, 23, 5421. https://doi.org/10.3390/ijms23105421
Mikalauskaite K, Ziaunys M, Smirnovas V. Lysozyme Amyloid Fibril Structural Variability Dependence on Initial Protein Folding State. International Journal of Molecular Sciences. 2022; 23(10):5421. https://doi.org/10.3390/ijms23105421
Chicago/Turabian StyleMikalauskaite, Kamile, Mantas Ziaunys, and Vytautas Smirnovas. 2022. "Lysozyme Amyloid Fibril Structural Variability Dependence on Initial Protein Folding State" International Journal of Molecular Sciences 23, no. 10: 5421. https://doi.org/10.3390/ijms23105421
APA StyleMikalauskaite, K., Ziaunys, M., & Smirnovas, V. (2022). Lysozyme Amyloid Fibril Structural Variability Dependence on Initial Protein Folding State. International Journal of Molecular Sciences, 23(10), 5421. https://doi.org/10.3390/ijms23105421






