Journal Description
SynBio
SynBio
is an international, peer-reviewed, open access journal on synthetic biology, biological parts, devices, and systems, published quarterly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 18.8 days after submission; acceptance to publication is undertaken in 4.8 days (median values for papers published in this journal in the second half of 2025).
- Recognition of Reviewers: APC discount vouchers, optional signed peer review, and reviewer names published annually in the journal.
- SynBio is a companion journal of IJMS.
Latest Articles
Programmable Plant Immunity: Synthetic Biology for Climate-Resilient Agriculture
SynBio 2026, 4(1), 1; https://doi.org/10.3390/synbio4010001 - 4 Jan 2026
Abstract
►
Show Figures
Agricultural systems face mounting pressures from climate change, as rising temperatures, elevated CO2, and shifting precipitation patterns intensify plant disease outbreaks worldwide. Conventional strategies, such as breeding for resistance, pesticides, and even transgenic approaches, are proving too slow or unsustainable to
[...] Read more.
Agricultural systems face mounting pressures from climate change, as rising temperatures, elevated CO2, and shifting precipitation patterns intensify plant disease outbreaks worldwide. Conventional strategies, such as breeding for resistance, pesticides, and even transgenic approaches, are proving too slow or unsustainable to meet these challenges. Synthetic biology offers a transformative paradigm for reprogramming plant immunity through genetic circuits, RNA-based defences, epigenome engineering, engineered microbiomes, and artificial intelligence (AI). We introduce the concept of synthetic immunity, a unifying framework that extends natural defence layers, PAMP-triggered immunity (PTI), and effector-triggered immunity (ETI). While pests and pathogens continue to undermine global crop productivity, synthetic immunity strategies such as CRISPR-based transcriptional activation, synthetic receptors, and RNA circuit-driven defences offer promising new avenues for enhancing plant resilience. We formalize synthetic immunity as an emerging, integrative concept that unites molecular engineering, regulatory rewiring, epigenetic programming, and microbiome modulation, with AI and computational modelling accelerating their design and climate-smart deployment. This review maps the landscape of synthetic immunity, highlights technological synergies, and outlines a translational roadmap from laboratory design to field application. Responsibly advanced, synthetic immunity represents not only a scientific frontier but also a sustainable foundation for climate-resilient agriculture.
Full article
Open AccessReview
Targeting Cancer-Associated Transcripts with Engineered RNase P Ribozymes
by
Thomas Sorrell, Ethan Ou and Fenyong Liu
SynBio 2025, 3(4), 20; https://doi.org/10.3390/synbio3040020 - 8 Dec 2025
Abstract
►▼
Show Figures
Nucleic acid-based gene interfering and editing molecules, such as antisense oligonucleotides, ribozymes, small interfering RNAs (siRNAs), and CRISPR-Cas9-associated guide RNAs, are promising gene-targeting agents for therapeutic applications. Cancer’s heterogeneous and diverse nature demands gene-silencing technologies that are both specific and adaptable. RNase P
[...] Read more.
Nucleic acid-based gene interfering and editing molecules, such as antisense oligonucleotides, ribozymes, small interfering RNAs (siRNAs), and CRISPR-Cas9-associated guide RNAs, are promising gene-targeting agents for therapeutic applications. Cancer’s heterogeneous and diverse nature demands gene-silencing technologies that are both specific and adaptable. RNase P ribozymes, called M1GS RNAs, are engineered constructs that link the catalytic M1 RNA from bacterial RNase P to a programmable guide sequence. This guide sequence directs the M1GS ribozyme to base-pair with a target RNA, inducing it to fold into a structure resembling pre-tRNA. Catalytic activity can be enhanced through in vitro selection strategies. In this review, we will discuss the application of M1GS ribozymes in targeting cancer-associated RNAs, focusing on the BCR-ABL transcript in leukemia, the internal ribosome entry site (IRES) of hepatitis C virus (HCV), and the replication and transcription activator (RTA) of Kaposi’s sarcoma-associated herpesvirus (KSHV). Together, these examples highlight the versatility of M1GS ribozymes across both viral and cellular oncogenic targets, underscoring their potential as a flexible synthetic biology platform for cancer therapy.
Full article
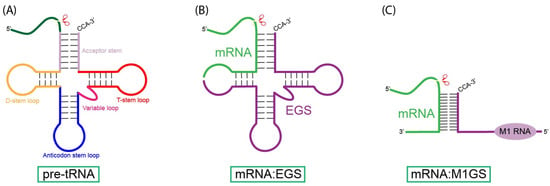
Figure 1
Open AccessReview
A Guide to Guides: An Overview of SpCas9 sgRNA Scaffold Variants and Modifications
by
Jonas De Saeger
SynBio 2025, 3(4), 19; https://doi.org/10.3390/synbio3040019 - 20 Nov 2025
Abstract
►▼
Show Figures
The CRISPR/SpCas9 system has revolutionized biology by enabling precise and programmable genome modification. While substantial effort has focused on engineering the SpCas9 protein and spacer sequences, the single-guide RNA (sgRNA) scaffold is an equally critical determinant of activity. Since the canonical scaffold was
[...] Read more.
The CRISPR/SpCas9 system has revolutionized biology by enabling precise and programmable genome modification. While substantial effort has focused on engineering the SpCas9 protein and spacer sequences, the single-guide RNA (sgRNA) scaffold is an equally critical determinant of activity. Since the canonical scaffold was introduced in 2012, numerous variants have been developed. Early designs sought to enhance editing efficiency; however, despite the first improved scaffold being reported in 2013, more than 80% of CRISPR plasmids deposited in the Addgene repository still use the original scaffold rather than an efficiency-optimized alternative, which may not provide optimal performance. Subsequent work has also addressed intra-sgRNA interactions that impair folding, as well as inter-sgRNA interactions that destabilize multiplexed arrays, yet these solutions remain largely overlooked. Beyond efficiency, scaffold engineering—and the inclusion of auxiliary RNA elements—has enabled new capabilities, including effector recruitment, conditional regulation, visualization, improved stability, and large-scale multiplexing. The main goal of this review is to (i) provide a structured overview of the diverse SpCas9 sgRNA scaffold variants and auxiliary RNA modifications developed to date, (ii) summarize their functional characteristics and contexts of use, thereby illustrating how scaffold engineering continues to expand the functional scope of CRISPR technologies, and (iii) present a curated sequence resource comprising more than 230 scaffold variants and 80 auxiliary modifications to support experimental design and benchmarking.
Full article

Figure 1
Open AccessArticle
ChronoSort: Revealing Hidden Dynamics in AlphaFold3 Structure Predictions
by
Matthew J. Argyle, William P. Heaps, Corbyn Kubalek, Spencer S. Gardiner, Bradley C. Bundy and Dennis Della Corte
SynBio 2025, 3(4), 18; https://doi.org/10.3390/synbio3040018 - 14 Nov 2025
Abstract
►▼
Show Figures
Protein function emerges from dynamic conformational changes, yet structure prediction methods provide only static snapshots. While AlphaFold3 (AF3) predicts protein structures, the potential for extracting dynamic information from its ensemble predictions has remained underexplored. Here, we demonstrate that AF3 structural ensembles contain substantial
[...] Read more.
Protein function emerges from dynamic conformational changes, yet structure prediction methods provide only static snapshots. While AlphaFold3 (AF3) predicts protein structures, the potential for extracting dynamic information from its ensemble predictions has remained underexplored. Here, we demonstrate that AF3 structural ensembles contain substantial dynamic information that correlates remarkably well with molecular dynamics simulations (MD). We developed ChronoSort, a novel algorithm that organizes static structure predictions into temporally coherent trajectories by minimizing structural differences between neighboring frames. Through systematic analysis of four diverse protein targets, we show that root-mean-square fluctuations derived from AF3 ensembles can correlate strongly with those from MD (r = 0.53 to 0.84). Principal component analysis reveals that AF3 predictions capture the same collective motion patterns observed in molecular dynamics trajectories, with eigenvector similarities significantly exceeding random distributions. ChronoSort trajectories exhibit structural evolution profiles comparable to MD. These findings suggest that modern AI-based structure prediction tools encode conformational flexibility information that can be systematically extracted without expensive MD. We provide ChronoSort as open-source software to enable broad community adoption. This work offers a novel approach to extracting functional insights from structure prediction tools in minutes, with significant implications for synthetic biology, protein engineering, drug discovery, and structure–function studies.
Full article

Figure 1
Open AccessReview
Digital to Biological Translation: How the Algorithmic Data-Driven Design Reshapes Synthetic Biology
by
Abdul Manan, Nabila Qayyum, Rajath Ramachandran, Naila Qayyum and Sidra Ilyas
SynBio 2025, 3(4), 17; https://doi.org/10.3390/synbio3040017 - 7 Nov 2025
Abstract
►▼
Show Figures
Synthetic biology, an emergent interdisciplinary field integrating principles from biology, engineering, and computer science, endeavors to rationally design and construct novel biological systems or reprogram extant ones to achieve predefined functionalities. The conventional approach relies on an iterative Design-Build-Test-Learn (DBTL) cycle, a process
[...] Read more.
Synthetic biology, an emergent interdisciplinary field integrating principles from biology, engineering, and computer science, endeavors to rationally design and construct novel biological systems or reprogram extant ones to achieve predefined functionalities. The conventional approach relies on an iterative Design-Build-Test-Learn (DBTL) cycle, a process frequently hampered by the intrinsic complexity, non-linear interactions, and vast design space inherent to biological systems. The advent of Artificial Intelligence (AI), and particularly its subfields of Machine Learning (ML) and Deep Learning (DL), is fundamentally reshaping this paradigm by offering robust computational frameworks to navigate these formidable challenges. This review elucidates the strategic integration of AI/ML/DL across the synthetic biology workflow, detailing the specific algorithms and mechanisms that enable rational design, autonomous experimentation, and pathway optimization. Their advanced applications are specifically underscored across critical facets, including de novo rational design, enhanced predictive modeling, intelligent high-throughput data analysis, and AI-driven laboratory automation. Furthermore, pivotal challenges, such as data sparsity, model interpretability, the “black box” problem, computational resource demands, and ethical considerations, have been addressed, while concurrently forecasting future trajectories for this rapidly advancing and convergent domain. The synergistic convergence of these disciplines is demonstrably accelerating biological discovery, facilitating the creation of innovative and scalable biological solutions, and fostering a more predictable and efficient paradigm for biological engineering.
Full article

Graphical abstract
Open AccessArticle
Stoichiometric Multiprotein Assembly Scaffolded by a Heterotrimeric DNA Clamp for Enzyme Colocalization and DNA Functionalization
by
Arabella Essert and Kathrin Castiglione
SynBio 2025, 3(4), 16; https://doi.org/10.3390/synbio3040016 - 6 Nov 2025
Abstract
►▼
Show Figures
Researchers strive to exploit kinetic potentials of multistep reactions by positioning enzymes in a regulated fashion. Therein, the proliferating cell nuclear antigen (PCNA) from Sulfolobus solfataricus is a promising biomolecular tool due to its extraordinary architecture. PCNA is a circular DNA sliding clamp,
[...] Read more.
Researchers strive to exploit kinetic potentials of multistep reactions by positioning enzymes in a regulated fashion. Therein, the proliferating cell nuclear antigen (PCNA) from Sulfolobus solfataricus is a promising biomolecular tool due to its extraordinary architecture. PCNA is a circular DNA sliding clamp, which can bind and move along DNA and thus, be applied for the immobilization and transport of biomolecules on versatile DNA scaffolds. Additionally, its heterotrimeric character facilitates the colocalization of enzyme cascades with defined stoichiometry. This study provides insights into the in vitro binding behavior of PCNA and its potential as protein scaffold for DNA functionalization and controlled biocatalysis: (1) PCNA was capable of binding circular DNA and wireframe DNA nanostructures. (2) DNA binding was predominantly mediated by the PCNA1 subunit. (3) PCNA assembly around DNA was compromised when cysteines were introduced at the PCNA–PCNA interfaces to stabilize the ring via disulfide bonds. (4) A two-enzyme cascade, comprising a pseudo-monomeric cytochrome P450 BM3 monooxygenase and a monomeric alcohol dehydrogenase (ADH), was successfully fused to PCNA, retaining catalytic activity. (5) When immobilized on DNA, the cascade performance was not assessable, due to nearly complete loss of ADH activity in proximity to DNA.
Full article

Figure 1
Open AccessArticle
Self-Energy-Harvesting Pacemakers: An Example of Symbiotic Synthetic Biology
by
Kuntal Kumar Das, Ashutosh Kumar Dubey, Bikramjit Basu and Yogendra Narain Srivastava
SynBio 2025, 3(4), 15; https://doi.org/10.3390/synbio3040015 - 4 Oct 2025
Abstract
►▼
Show Figures
While synthetic biology has traditionally focused on creating biological systems often through genetic engineering, emerging technologies, for example, implantable pacemakers with integrated piezo-electric and tribo-electric materials are beginning to enlarge the classical domain into what we call symbiotic synthetic biology. These devices are
[...] Read more.
While synthetic biology has traditionally focused on creating biological systems often through genetic engineering, emerging technologies, for example, implantable pacemakers with integrated piezo-electric and tribo-electric materials are beginning to enlarge the classical domain into what we call symbiotic synthetic biology. These devices are permanently attached to a body, although non-living or genetically unaltered, and closely mimic biological behavior by harvesting biomechanical energy and providing functions, such as autonomous heart pacing. They form active interfaces with human tissues and operate as hybrid systems, similar to synthetic organs. In this context, the present paper first presents a short summary of previous in vivo studies on piezo-electric composites in relation to their deployment as battery-less pacemakers. This is then followed by a summary of a recent theoretical work using a damped harmonic resonance model, which is being extended to mimic the functioning of such devices. We then extend the theoretical study further to include new solutions and obtain a sum rule for the power output per cycle in such systems. In closing, we present our quantitative understanding to explore the modulation of the quantum vacuum energy (Casimir effect) by periodic body movements to power pacemakers. Taken together, the present work provides the scientific foundation of the next generation bio-integrated intelligent implementation.
Full article

Figure 1
Open AccessFeature PaperArticle
Analysis and Application of Translation-Enhancing Peptides for Improved Production of Proteins Containing Polyproline
by
Akimichi Yoshino, Riko Shimoji, Yuma Nishikawa, Hideo Nakano and Teruyo Ojima-Kato
SynBio 2025, 3(4), 14; https://doi.org/10.3390/synbio3040014 - 3 Oct 2025
Abstract
►▼
Show Figures
Polyproline residues are well known to induce ribosomal stalling during translation. Our previous work demonstrated that inserting a short translation-enhancing peptide, Ser-Lys-Ile-Lys (SKIK), immediately upstream of such difficult-to-translate sequences can significantly alleviate ribosomal stalling in Escherichia coli. In this study, we provide
[...] Read more.
Polyproline residues are well known to induce ribosomal stalling during translation. Our previous work demonstrated that inserting a short translation-enhancing peptide, Ser-Lys-Ile-Lys (SKIK), immediately upstream of such difficult-to-translate sequences can significantly alleviate ribosomal stalling in Escherichia coli. In this study, we provide a quantitative evaluation of its translational effect by kinetically analyzing the influence of the SKIK peptide on polyproline motifs using a reconstituted E. coli in vitro translation system. Translation rates estimated under reasonable assumptions fitted well to a Hill equation within a Michaelis–Menten-like kinetic framework. We further revealed that repetition of the SKIK tag did not provide any positive effect on translation. Moreover, introduction of the SKIK tag increased the production of polyproline-containing proteins, including human interleukin 11, human G protein signaling modulator 3, and DUF58 domain–containing protein from Streptomyces sp. in E. coli cell-free protein synthesis. These findings not only provide new insight into the fundamental regulation of translation by nascent peptides but also demonstrate the potential of the SKIK peptide as a practical tool for synthetic biology, offering a strategy to improve the production of difficult-to-express proteins.
Full article

Graphical abstract
Open AccessReview
Unlocking MSC Potential: Metabolic Reprogramming via Synthetic Biology Approaches
by
Natalia Trufanova, Oleh Trufanov and Oleksandr Petrenko
SynBio 2025, 3(3), 13; https://doi.org/10.3390/synbio3030013 - 17 Sep 2025
Abstract
►▼
Show Figures
Metabolic engineering of mesenchymal stem/stromal cells (MSCs) represents a compelling frontier for advanced cellular therapies, enabling the precise tuning of their biological outputs. This feature paper examines the critical role of engineered culture microenvironments, specifically 3D platforms, hypoxic preconditioning, and other priming approaches,
[...] Read more.
Metabolic engineering of mesenchymal stem/stromal cells (MSCs) represents a compelling frontier for advanced cellular therapies, enabling the precise tuning of their biological outputs. This feature paper examines the critical role of engineered culture microenvironments, specifically 3D platforms, hypoxic preconditioning, and other priming approaches, which are synthetic biology strategies used to guide and optimize MSC metabolic states for desired functional outcomes. We show that these non-genetic approaches can significantly enhance MSC survival, immunomodulatory capacity, and regenerative potential by shifting their metabolism toward a more glycolytic phenotype. Furthermore, we propose a new paradigm of “designer” MSCs, which are programmed with synthetic circuits to sense and respond to the physiological cues of an injured microenvironment. This approach promises to transform regenerative medicine from an inconsistent field into a precise, predictable, and highly effective therapeutic discipline.
Full article

Figure 1
Open AccessPerspective
Silicon Is the Next Frontier in Plant Synthetic Biology
by
Aniruddha Acharya, Kaitlin Hopkins and Tatum Simms
SynBio 2025, 3(3), 12; https://doi.org/10.3390/synbio3030012 - 3 Aug 2025
Abstract
►▼
Show Figures
Silicon has a striking similarity to carbon and is found in plant cells. However, there is no specific role that has been assigned to silicon in the life cycle of plants. The amount of silicon in plant cells is species specific and can
[...] Read more.
Silicon has a striking similarity to carbon and is found in plant cells. However, there is no specific role that has been assigned to silicon in the life cycle of plants. The amount of silicon in plant cells is species specific and can reach levels comparable to macronutrients. Silicon is used extensively in artificial intelligence, nanotechnology, and the digital revolution, and thus can serve as an informational molecule such as nucleic acids. The diverse potential of silicon to bond with different chemical species is analogous to carbon; thus, it can serve as a structural candidate similar to proteins. The discovery of large amounts of silicon on Mars and the moon, along with the recent development of enzyme that can incorporate silicon into organic molecules, has propelled the theory of creating silicon-based life. The bacterial cytochrome has been modified through directed evolution such that it could cleave silicon–carbon bonds in organo-silicon compounds. This consolidates the idea of utilizing silicon in biomolecules. In this article, the potential of silicon-based life forms has been hypothesized, along with the reasoning that autotrophic virus-like particles could be used to investigate such potential. Such investigations in the field of synthetic biology and astrobiology will have corollary benefits for Earth in the areas of medicine, sustainable agriculture, and environmental sustainability.
Full article

Graphical abstract
Open AccessReview
A Guide in Synthetic Biology: Designing Genetic Circuits and Their Applications in Stem Cells
by
Karim S. Elnaggar, Ola Gamal, Nouran Hesham, Sama Ayman, Nouran Mohamed, Ali Moataz, Emad M. Elzayat and Nourhan Hassan
SynBio 2025, 3(3), 11; https://doi.org/10.3390/synbio3030011 - 22 Jul 2025
Cited by 2
Abstract
►▼
Show Figures
Stem cells, unspecialized cells with regenerative and differentiation capabilities, hold immense potential in regenerative medicine, exemplified by hematopoietic stem cell transplantation. However, their clinical application faces significant limitations, including their tumorigenic risk due to uncontrolled proliferation and cellular heterogeneity. This review explores how
[...] Read more.
Stem cells, unspecialized cells with regenerative and differentiation capabilities, hold immense potential in regenerative medicine, exemplified by hematopoietic stem cell transplantation. However, their clinical application faces significant limitations, including their tumorigenic risk due to uncontrolled proliferation and cellular heterogeneity. This review explores how synthetic biology, an interdisciplinary approach combining engineering and biology, offers promising solutions to these challenges. It discusses the concepts, toolkit, and advantages of synthetic biology, focusing on the design and integration of genetic circuits to program stem cell differentiation and engineer safety mechanisms like inducible suicide switches. This review comprehensively examines recent advancements in synthetic biology applications for stem cell engineering, including programmable differentiation circuits, cell reprogramming strategies, and therapeutic cell engineering approaches. We highlight specific examples of genetic circuits that have been successfully implemented in various stem cell types, from embryonic stem cells to induced pluripotent stem cells, demonstrating their potential for clinical translation. Despite these advancements, the integration of synthetic biology with mammalian cells remains complex, necessitating further research, standardized datasets, open access repositories, and interdisciplinary collaborations to build a robust framework for predicting and managing this complexity.
Full article

Figure 1
Open AccessReview
Cell-Free Protein Synthesis Reactor Formats: A Brief History and Analysis
by
Dallin M. Chipman, Anna C. Woolley, Davu N. Chau, William A. Lance, Joseph P. Talley, Tyler P. Green, Benjamin C. Robbins and Bradley C. Bundy
SynBio 2025, 3(3), 10; https://doi.org/10.3390/synbio3030010 - 1 Jul 2025
Abstract
►▼
Show Figures
Cell-free protein synthesis (CFPS) has transformed protein production capabilities by eliminating cellular constraints, enabling the rapid expression of difficult-to-produce proteins in an open, customizable environment. As CFPS applications expand from fundamental research to industrial production, therapeutic manufacturing, and point-of-care diagnostics, the diverse array
[...] Read more.
Cell-free protein synthesis (CFPS) has transformed protein production capabilities by eliminating cellular constraints, enabling the rapid expression of difficult-to-produce proteins in an open, customizable environment. As CFPS applications expand from fundamental research to industrial production, therapeutic manufacturing, and point-of-care diagnostics, the diverse array of reactor formats has become increasingly important yet challenging to navigate. This review examines the evolution and characteristics of thirteen major CFPS reactor formats, from traditional batch systems to advanced platforms. The historical development of CFPS reactors from the 1960s to present day is presented. Additionally, for each format, operational principles, advantages, limitations, and notable applications are evaluated. The review concludes with a comparative assessment of reactor performance across critical parameters, including productivity, scalability, technical complexity, environmental stability, and application suitability. To our knowledge this structured analysis is the first to focus predominantly on the various reactor formats of cell-free systems and to provide a guide to assist researchers in choosing the reactor type that best fits their specific applications.
Full article

Figure 1
Open AccessReview
Plant Transformation and Genome Editing for Precise Synthetic Biology Applications
by
Sharathchandra Kambampati, Pankaj K. Verma and Madhusudhana R. Janga
SynBio 2025, 3(3), 9; https://doi.org/10.3390/synbio3030009 - 27 Jun 2025
Abstract
►▼
Show Figures
Synthetic biology (SynBio) is an emerging interdisciplinary field that applies engineering principles to the design and construction of novel biological systems or the redesign of existing natural systems for new functions. As autotrophs with complex cellular architectures, plants possess inherent capabilities to serve
[...] Read more.
Synthetic biology (SynBio) is an emerging interdisciplinary field that applies engineering principles to the design and construction of novel biological systems or the redesign of existing natural systems for new functions. As autotrophs with complex cellular architectures, plants possess inherent capabilities to serve as “living factories” for SynBio applications. Recent advancements in genetic engineering, genome editing, and transformation techniques are improving the precision and programmability of plant systems. Innovations, such as CRISPR systems, prime editing strategies, and in planta and nanoparticle-mediated delivery, are expanding the SynBio toolkit for plants. However, the efficient delivery of genetic constructs remains a barrier due to plant systems’ complexity. To address these limitations, SynBio is increasingly integrating iterative Design–Build–Test–Learn (DBTL) cycles, standardization, modular DNA assembly systems, and plant-optimized toolkits to enable predictable trait engineering. This review explores the technological foundations of plant SynBio, including genome editing and transformation methods, and examines their integration into engineered systems. Applications, such as biofuel production, pharmaceutical biosynthesis, and agricultural innovation, are highlighted, along with their ethical, technical, and regulatory challenges. Ultimately, SynBio could offer a transformative path toward sustainable solutions, provided it continues to align technological advances with public interest and global sustainability goals.
Full article

Figure 1
Open AccessReview
Polyamine-Mediated Growth Regulation in Microalgae: Integrating Redox Balance and Amino Acids Pathway into Metabolic Engineering
by
Leandro Luis Lavandosque and Flavia Vischi Winck
SynBio 2025, 3(2), 8; https://doi.org/10.3390/synbio3020008 - 28 May 2025
Cited by 3
Abstract
►▼
Show Figures
Polyamines play a pivotal role in regulating the growth and metabolic adaptation of microalgae, yet their integrative regulatory roles remain underexplored. This review advances a comprehensive perspective of microalgae growth, integrating polyamine dynamics, amino acid metabolism, and redox balance. Polyamines (putrescine, spermidine, and
[...] Read more.
Polyamines play a pivotal role in regulating the growth and metabolic adaptation of microalgae, yet their integrative regulatory roles remain underexplored. This review advances a comprehensive perspective of microalgae growth, integrating polyamine dynamics, amino acid metabolism, and redox balance. Polyamines (putrescine, spermidine, and spermine) biology in microalgae, particularly Chlamydomonas reinhardtii, is reviewed, exploring their critical function in modulating cell cycle progression, enzymatic activity, and stress responses through nucleic acid stabilization, protein synthesis regulation, and post-translational modifications. This review explores how the exogenous supplementation of polyamines modifies their intracellular dynamics, affecting growth phases and metabolic transitions, highlighting the complex regulation of internal pools of these molecules. Comparative analyses with Chlorella ohadii and Scenedesmus obliquus indicated species-specific responses to polyamine fluctuations, linking putrescine and spermine levels to important tunable metabolic shifts and fast growth phenotypes in phototrophic conditions. The integration of multi-omic approaches and computational modeling has already provided novel insights into polyamine-mediated growth regulation, highlighting their potential in optimizing microalgae biomass production for biotechnological applications. In addition, genomic-based modeling approaches have revealed target genes and cellular compartments as bottlenecks for the enhancement of microalgae growth, including mitochondria and transporters. System-based analyses have evidenced the overlap of the polyamines biosynthetic pathway with amino acids (especially arginine) metabolism and Nitric Oxide (NO) generation. Further association of the H2O2 production with polyamines metabolism reveals novel insights into microalgae growth, combining the role of the H2O2/NO rate regulation with the appropriate balance of the mitochondria and chloroplast functionality. System-level analysis of cell growth metabolism would, therefore, be beneficial to the understanding of the regulatory networks governing this phenotype, fostering metabolic engineering strategies to enhance growth, stress resilience, and lipid accumulation in microalgae. This review consolidates current knowledge and proposes future research directions to unravel the complex interplay of polyamines in microalgal physiology, opening new paths for the optimization of biomass production and biotechnological applications.
Full article

Figure 1
Open AccessArticle
Functional and Evolutionary Characterization of the NSP6 Protein in SARS-CoV-2 Omicron Variants
by
Joyhare Barbosa Souza and Samir Mansour Moraes Casseb
SynBio 2025, 3(2), 7; https://doi.org/10.3390/synbio3020007 - 27 Apr 2025
Abstract
►▼
Show Figures
The SARS-CoV-2 virus, which causes COVID-19, has rapidly evolved, producing highly transmissible variants like Omicron. Non-structural protein 6 (NSP6) is essential for viral replication and immune evasion. This study analyzed the NSP6 protein of the Omicron variant, focusing on conserved motifs, mutations, and
[...] Read more.
The SARS-CoV-2 virus, which causes COVID-19, has rapidly evolved, producing highly transmissible variants like Omicron. Non-structural protein 6 (NSP6) is essential for viral replication and immune evasion. This study analyzed the NSP6 protein of the Omicron variant, focusing on conserved motifs, mutations, and residual properties to better understand its structure, function, and potential for immune evasion. Sequences from humans in South America were obtained from GISAID and aligned using Clustal Omega 1.2.4, with mutations identified by a Python 3 algorithm and conserved motifs detected using the MEME tool. Sequence diversity was assessed with Shannon’s entropy, while hydrophilicity, flexibility, accessibility, and antigenicity were analyzed using EMBOSS PEPSTATS and Expasy’s ProtScale tools. Phylogenetic analysis was performed with IQ-TREE software. Analysis of 161 NSP6 protein sequences revealed significant divergence from the reference sequence, with mutations proximal to conserved regions indicating potential functional and structural changes. The analysis also identified distinct hydrophobic and hydrophilic regions, with specific amino acid positions showing high flexibility and antigenicity. Phylogenetic analysis identified three clades with varying degrees of similarity to the reference sequence. This comprehensive study of the NSP6 protein in the Omicron variant provides insights into its role in viral replication and immune evasion, contributing to the development of targeted interventions against COVID-19.
Full article

Figure 1
Open AccessReview
The Role of the Sda Carbohydrate Antigen and That of Its Cognate Glycosyltransferase B4GALNT2 in Health and Disease
by
Martina Duca, Nadia Malagolini and Fabio Dall’Olio
SynBio 2025, 3(1), 6; https://doi.org/10.3390/synbio3010006 - 3 Mar 2025
Abstract
►▼
Show Figures
The carbohydrate antigen Sda is expressed on the cells and secretions of the vast majority of Caucasians. The epitope is formed by a terminal GalNAc residue β4-linked to an α3-sialylated galactose. Different carbohydrate chains N- or O-linked to glycoproteins can
[...] Read more.
The carbohydrate antigen Sda is expressed on the cells and secretions of the vast majority of Caucasians. The epitope is formed by a terminal GalNAc residue β4-linked to an α3-sialylated galactose. Different carbohydrate chains N- or O-linked to glycoproteins can be terminated by this epitope. The final step of Sda biosynthesis is catalyzed by the GalNAc transferase B4GALNT2. In this review, we discuss the multifaceted aspects of B4GALNT2/Sda in fertility and pregnancy, susceptibility to infectious diseases, cancer, chronic kidney diseases, and Duchenne muscular dystrophy. We show how multiple synthetic biology approaches have been adopted to investigate its role.
Full article

Figure 1
Open AccessArticle
Streamlined Production, Protection, and Purification of Enzyme Biocatalysts Using Virus-like Particles and a Cell-Free Protein Synthesis System
by
Seung O. Yang, Joseph P. Talley, Gregory H. Nielsen, Kristen M. Wilding and Bradley C. Bundy
SynBio 2025, 3(1), 5; https://doi.org/10.3390/synbio3010005 - 5 Feb 2025
Cited by 2
Abstract
►▼
Show Figures
Enzymes play an essential role in many different industries; however, their operating conditions are limited due to the loss of enzyme activity in the presence of proteases and at temperatures significantly above physiological conditions. One way to improve the stability of these enzymes
[...] Read more.
Enzymes play an essential role in many different industries; however, their operating conditions are limited due to the loss of enzyme activity in the presence of proteases and at temperatures significantly above physiological conditions. One way to improve the stability of these enzymes against high temperatures and proteases is to encapsulate them in protective shells or virus-like particles. This work presents a streamlined, three-step, cell-free protein synthesis (CFPS) procedure that enables rapid in vitro enzyme production, targeted encapsulation in protective virus-like particles (VLPs), and facile purification using a 6× His-tag fused to the VLP coat protein. This process is performed in under 12 h and overcomes several limitations of enzyme encapsulation, such as the control of packing density, speed, and complexity of the process. Here, we encapsulate the enzyme Candida antarctica lipase B in the VLP from the bacteriophage Qβ, while in the presence of a linking RNA aptamer. The encapsulated enzymes largely retained their activity in comparison to the free enzymes. Additionally, when subjected to 90 °C temperatures or 5 h incubation with proteases, the encapsulated enzymes maintained their activity, whereas the free enzymes lost their activity. In this work, we also demonstrate control over packing density by achieving packing densities of 4.7 and 6.5 enzymes per VLP based off the concentration of enzyme added to the encapsulation step.
Full article

Figure 1
Open AccessReview
Synthetic Biology-Based Approaches to Investigate Host–Pathogen Interactions
by
Rajdeep Banerjee
SynBio 2025, 3(1), 4; https://doi.org/10.3390/synbio3010004 - 3 Feb 2025
Cited by 4
Abstract
►▼
Show Figures
The increasing prevalence of multi-drug-resistant (MDR) bacterial pathogens presents a critical global health threat, highlighting the urgent need for innovative approaches to understanding bacterial pathogenesis and developing effective therapies. This review underscores the potential of synthetic biology in elucidating host–pathogen interactions and facilitating
[...] Read more.
The increasing prevalence of multi-drug-resistant (MDR) bacterial pathogens presents a critical global health threat, highlighting the urgent need for innovative approaches to understanding bacterial pathogenesis and developing effective therapies. This review underscores the potential of synthetic biology in elucidating host–pathogen interactions and facilitating the creation of advanced diagnostic tools and targeted therapies to combat MDR infections. We first explore CRISPR-based strategies that modulate essential gene expression, providing insights into the molecular mechanisms underlying host–pathogen interactions. Next, we discuss engineered microbial synthetic circuits for rapid pathogen detection by identifying molecular signatures involved in interspecies communication and facilitating swift pathogen elimination. Additionally, we explore phage therapy (PT), which leverages bacteriophages to selectively target and eliminate specific bacterial pathogens, presenting a targeted and promising approach to combat MDR infections. Finally, we review the application of organ-on-a-chip (OOAC) technology, which overcomes the limitations of animal models in predicting human immune responses by using microfluidic devices that simulate organ-level physiology and pathophysiology, thereby enabling more accurate disease modeling, drug testing, and the development of personalized medicine. Collectively, these synthetic biology tools provide transformative insights into the molecular mechanisms of host–pathogen interactions, advancing the development of precise diagnostic and therapeutic strategies against MDR infections.
Full article

Graphical abstract
Open AccessArticle
Design of an Effective sgRNA for CRISPR/Cas9 Knock-Ins and Full Mutant Segregation in Polyploid Synechocystis sp. PCC 6803
by
Maria Isabel Nares-Rodriguez and Esther Karunakaran
SynBio 2025, 3(1), 3; https://doi.org/10.3390/synbio3010003 - 27 Jan 2025
Abstract
►▼
Show Figures
Synechocystis sp. PCC 6803 is a highly promising organism for the production of diverse recombinant compounds, including biofuels. However, conventional genetic engineering in Synechocystis presents challenges due to its highly polyploid genome, which not only results in low product yields but also compromises
[...] Read more.
Synechocystis sp. PCC 6803 is a highly promising organism for the production of diverse recombinant compounds, including biofuels. However, conventional genetic engineering in Synechocystis presents challenges due to its highly polyploid genome, which not only results in low product yields but also compromises the reliability of recombinant strains for biomanufacturing applications. The CRISPR/Cas9 system, renowned for its precision, efficiency, and versatility across a wide range of chassis, offers significant potential to address the limitations posed by polyploid genomes. In this study, we developed and optimized an effective sgRNA for the targeted knock-in of nucleotide sequences of varying lengths into the neutral locus slr0168 of polyploid Synechocystis using CRISPR/Cas9. The gene encoding di-geranylgeranylglycerophospholipid reductase from Sulfolobus acidocaldarius and the methyl ketone operon from Solanum habrochaites were chosen as the exemplar nucleotide sequences for incorporation into the chromosome of Synechocystis. Our results demonstrate that the designed sgRNA effectively facilitated both knock-in events and that CRISPR/Cas9 enabled complete mutant segregation in a single round of selection and induction.
Full article

Figure 1
Open AccessReview
Artificial Intelligence-Based Target for Personalized Interventions of Atherosclerosis from Gut Microbiota Signature
by
Suravi Majumder, Koushik Sen and Rabimba Karanjai
SynBio 2025, 3(1), 2; https://doi.org/10.3390/synbio3010002 - 9 Jan 2025
Cited by 2
Abstract
Atherosclerosis remains a major driver for cardiovascular disease (CVD), despite advancements in traditional risk factor management therapies. Recent evidence emphasizes the crucial role of the gut microbiome in the progression of atherosclerosis and plaque rupture, highlighting a promising therapeutic avenue. This review focuses
[...] Read more.
Atherosclerosis remains a major driver for cardiovascular disease (CVD), despite advancements in traditional risk factor management therapies. Recent evidence emphasizes the crucial role of the gut microbiome in the progression of atherosclerosis and plaque rupture, highlighting a promising therapeutic avenue. This review focuses on the intertwined relationship between the gut microbiome, its metabolites, and atherosclerosis and CVD, also highlighting the potential therapeutic role of probiotics and prebiotics. Given the diverse and unique gut microbiota signatures among individuals, a one-size-fits-all therapeutic approach is unlikely to be effective. Personalized treatment strategies are therefore necessary. Here, we discussed how Artificial Intelligence (AI) can be leveraged to analyze individual gut microbiome profiles from microbiome sequencing, predict treatment response, and optimize therapeutic strategies based on individual patients, which would significantly improve outcomes of the treatment for atherosclerosis patients.
Full article
(This article belongs to the Special Issue AI-Powered Synthetic Biology: Emerging Technologies and Biosecurity)
►▼
Show Figures

Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
SynBio
Synthetic Biology and Metabolic Engineering in Fungi
Guest Editor: Shengmin ZhouDeadline: 20 June 2026








