ChronoSort: Revealing Hidden Dynamics in AlphaFold3 Structure Predictions
Abstract
1. Introduction
2. Results and Discussion
2.1. Conceptual Framework: Extracting Dynamics from Static Predictions
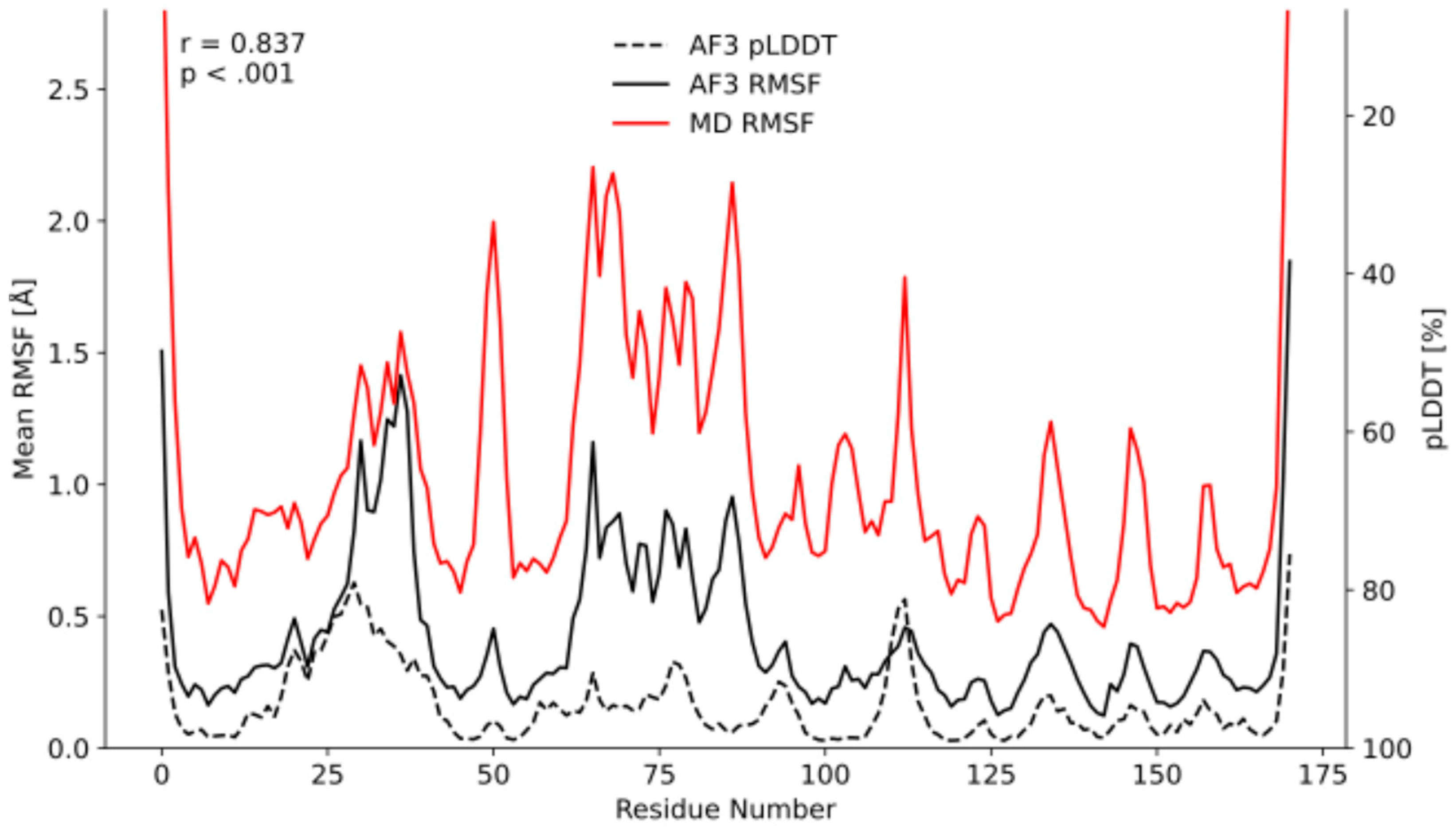
2.2. Initial Discovery: Dynamic Signatures in NanoLuc Luciferase
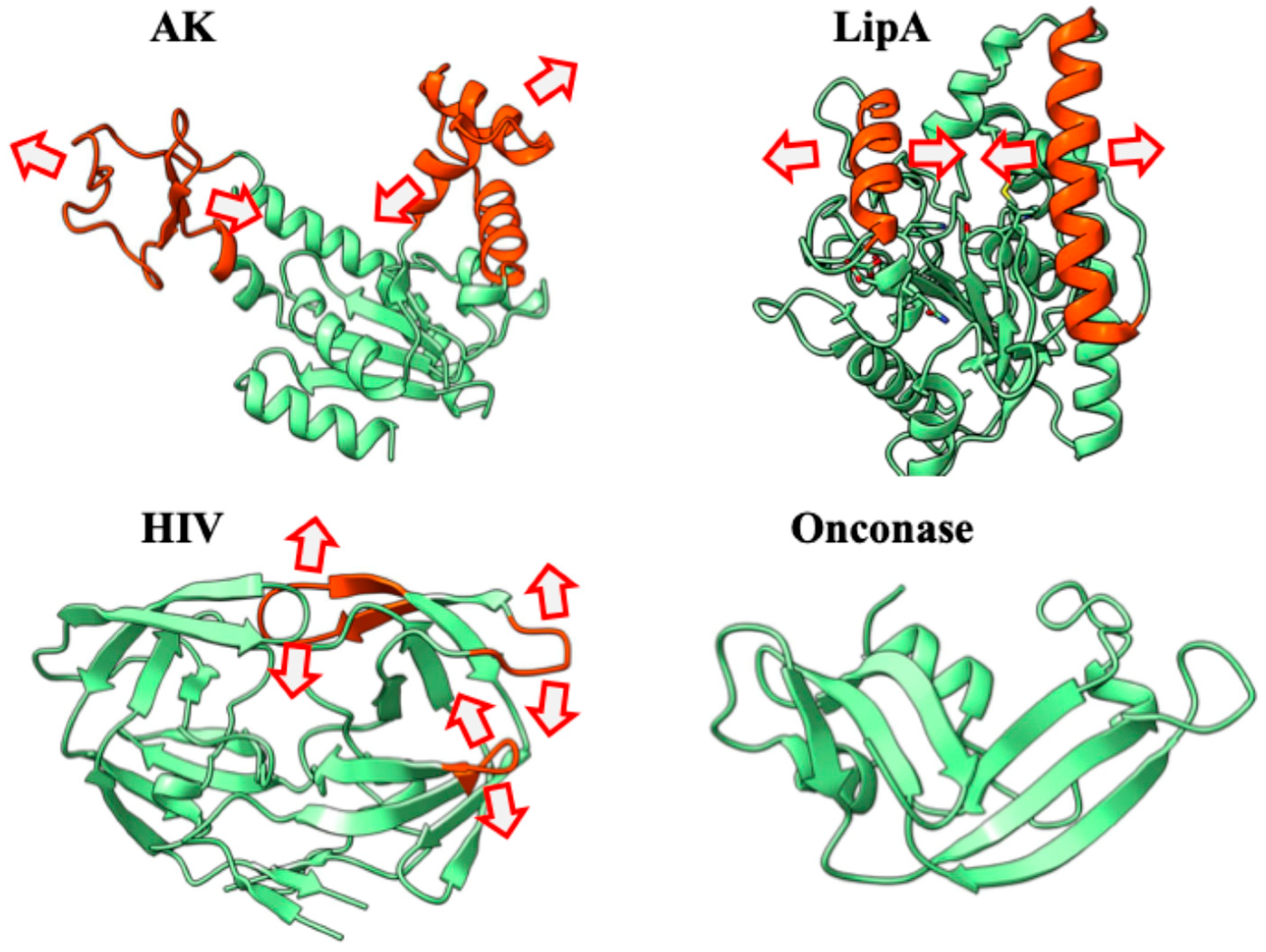
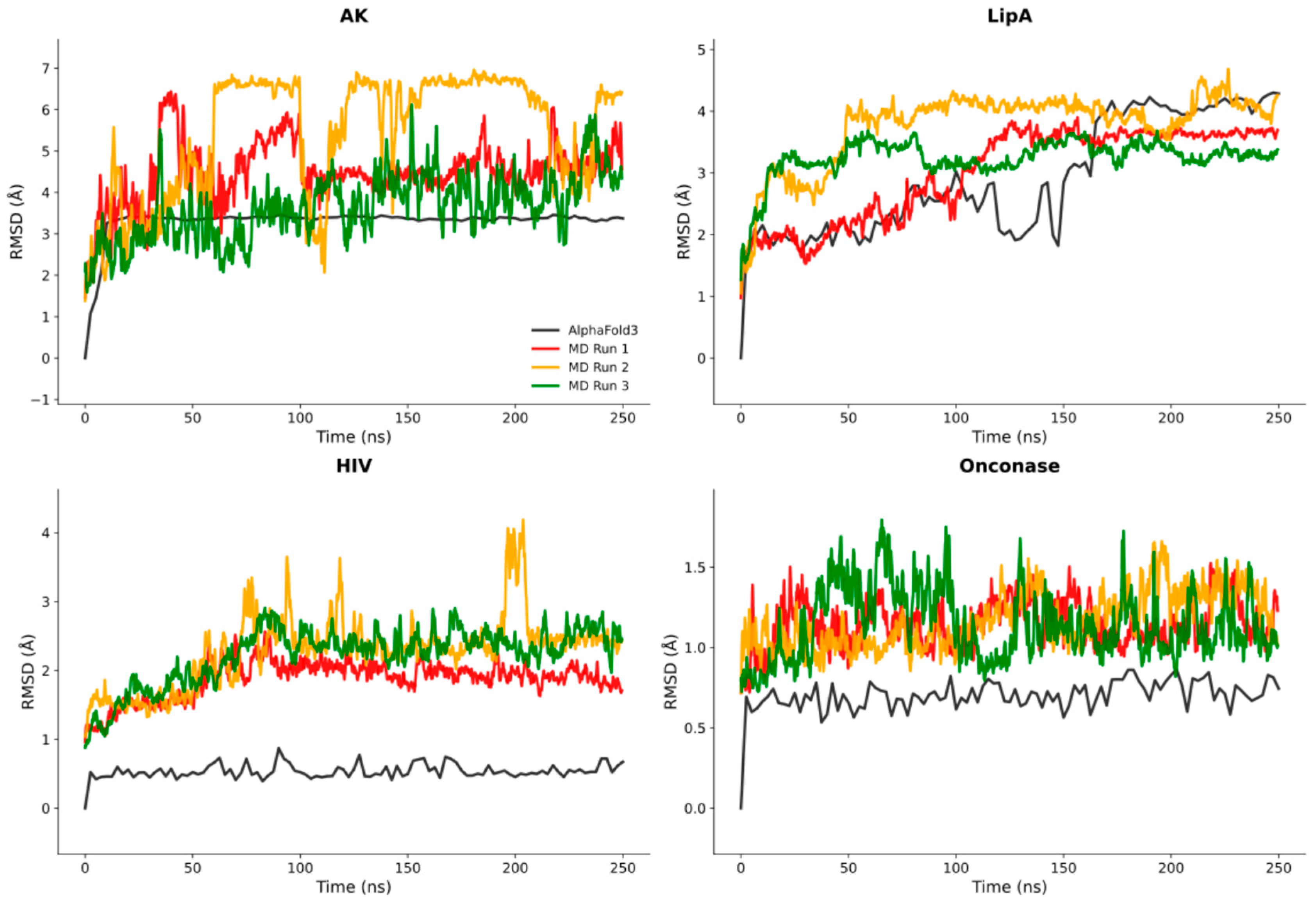
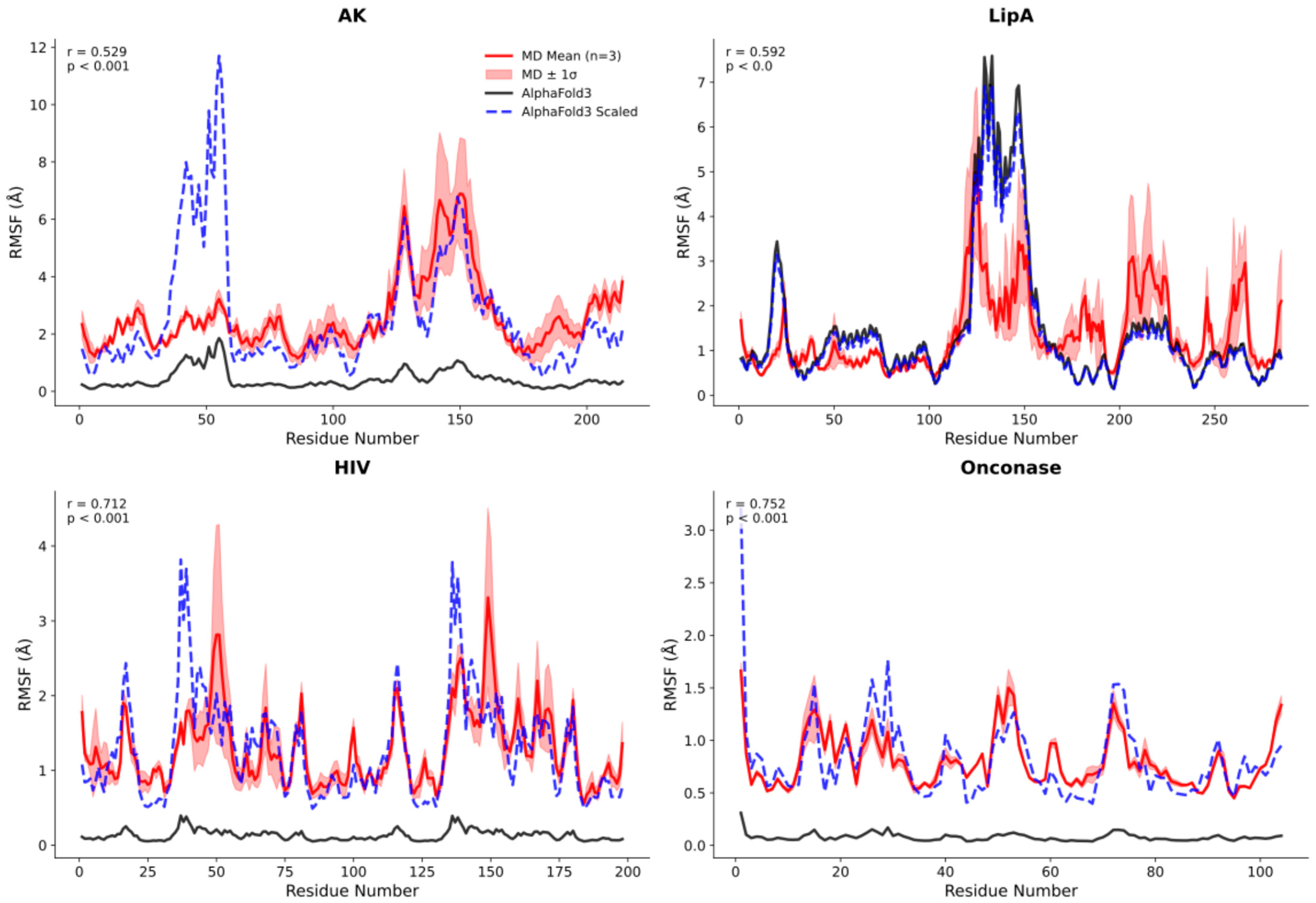
2.3. Systematic Validation Across Diverse Protein Targets
2.4. ChronoSort Algorithm Validates Temporal Organization
2.5. Quantitative Validation of Flexibility Patterns
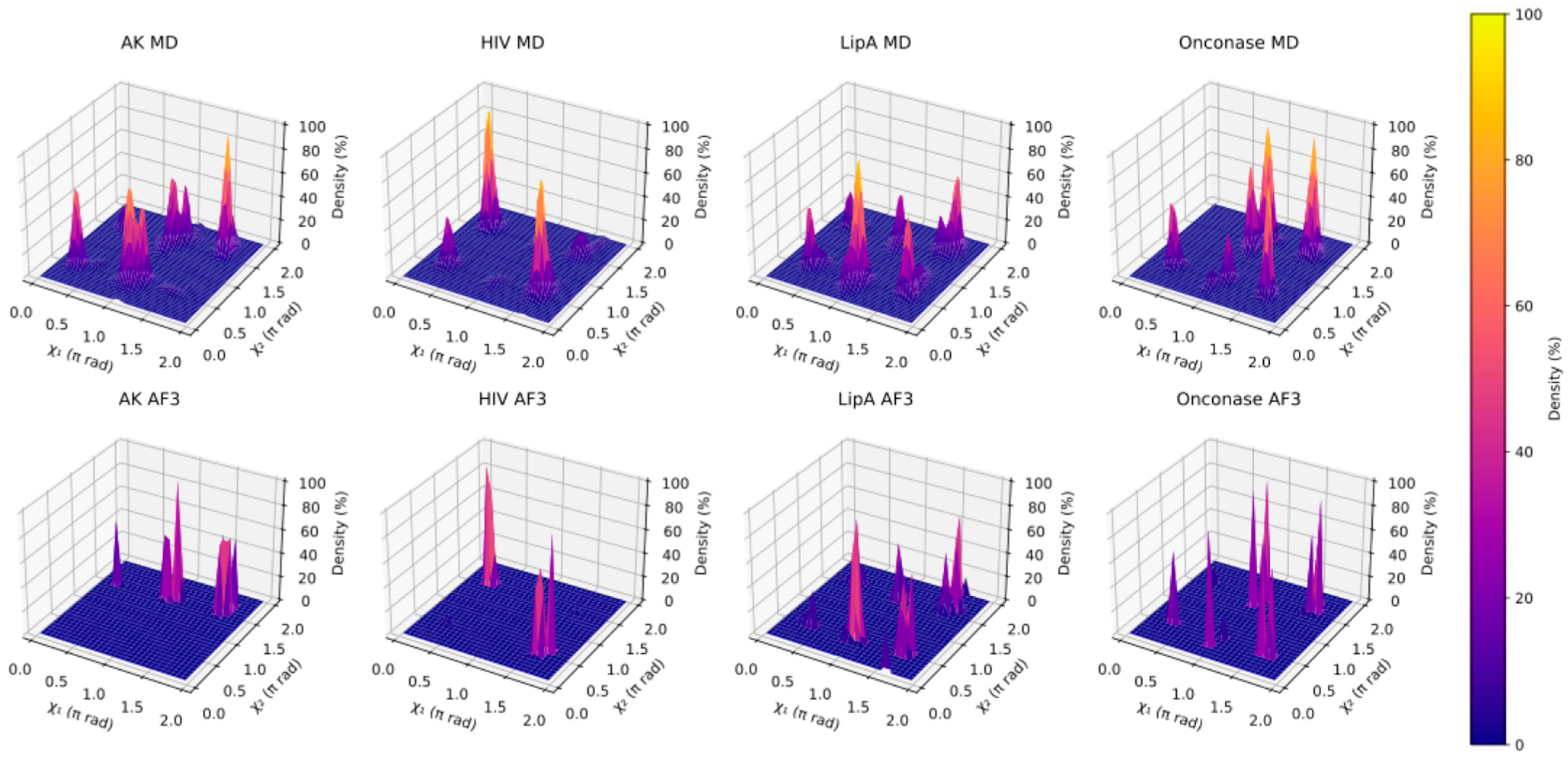
2.6. Analysis of Side Chain Conformation
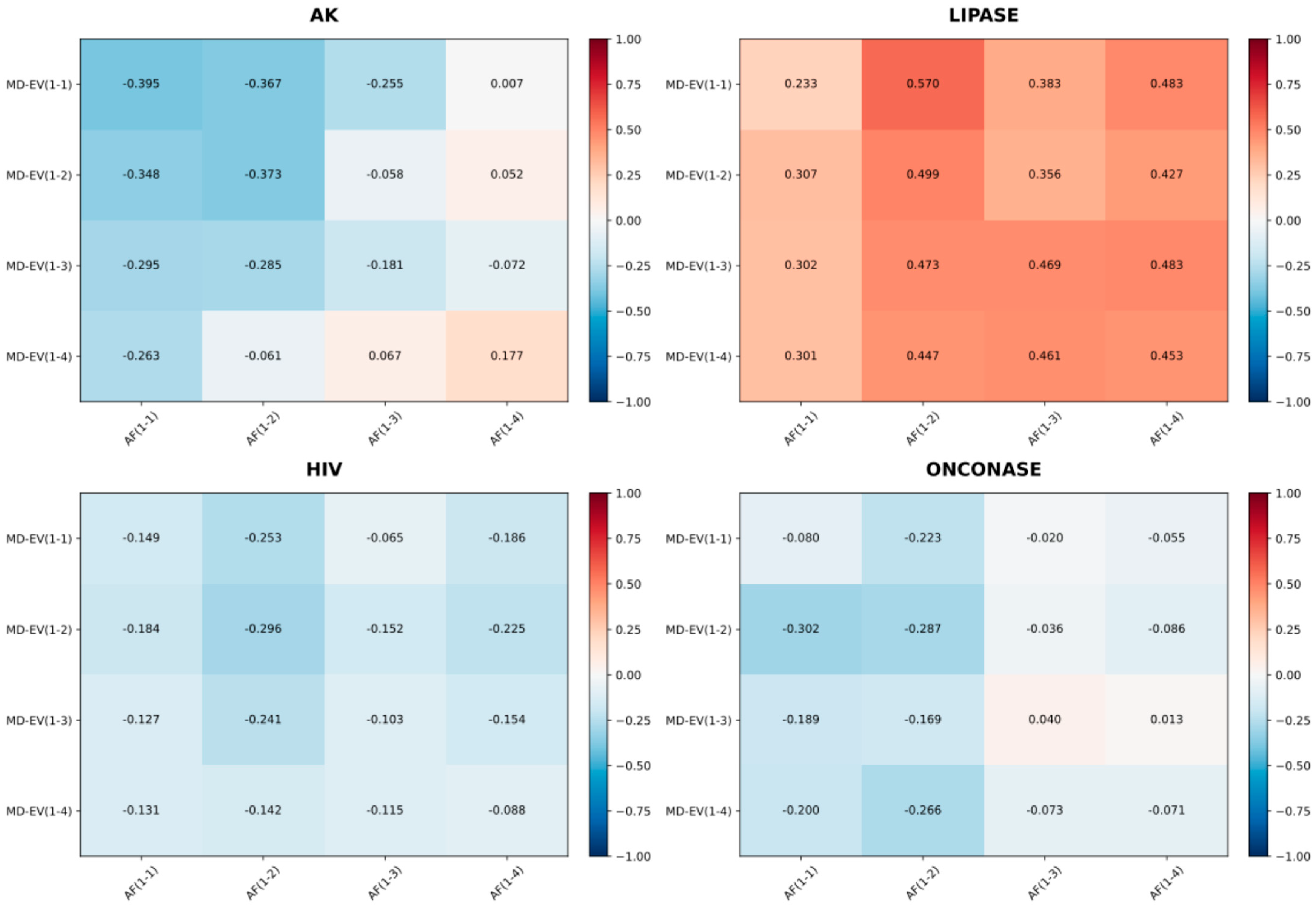
2.7. Principal Component Analysis Reveals Shared Motion Patterns
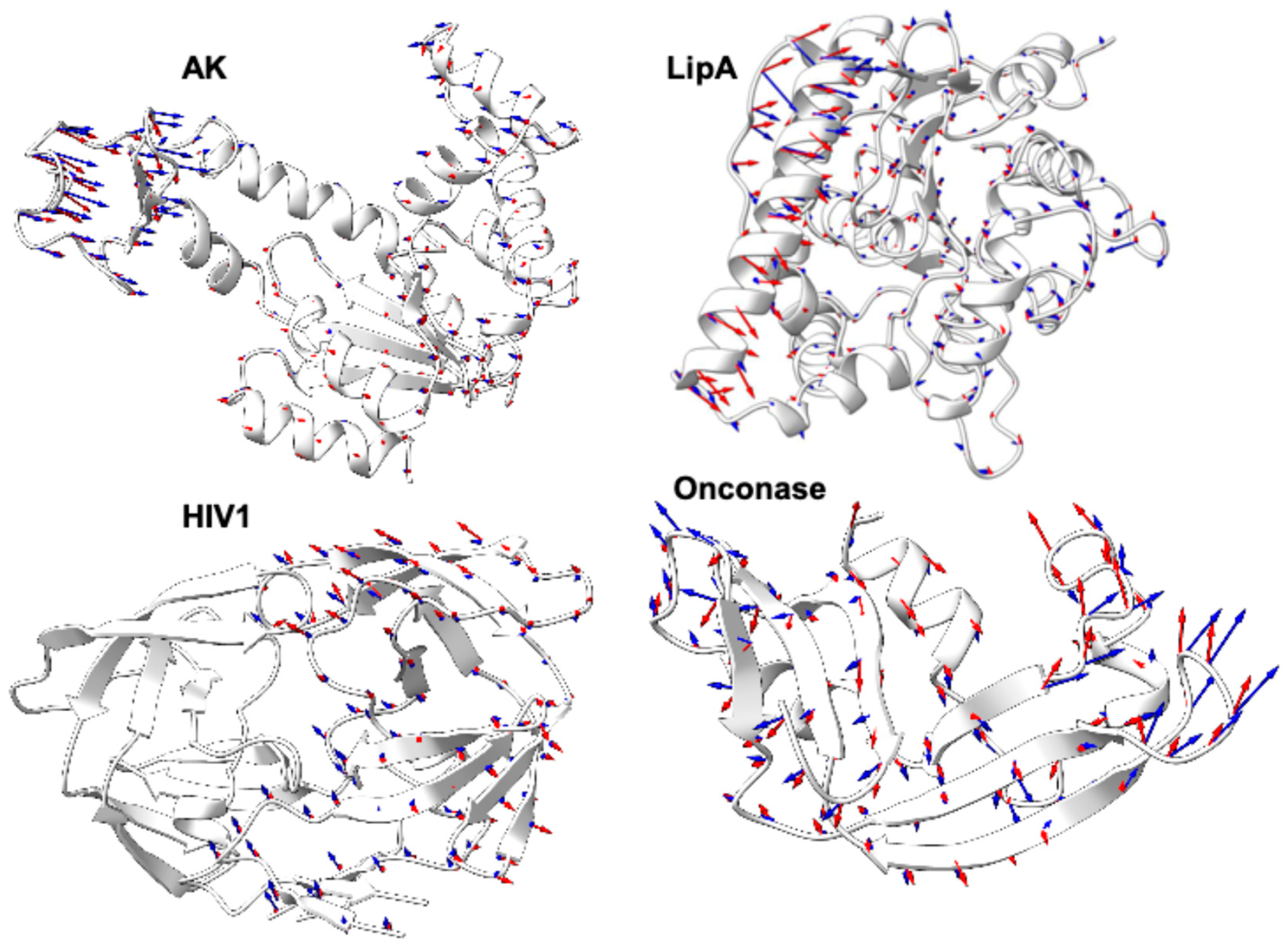
2.8. Visual Validation Through Porcupine Plot Analysis
2.9. Implications for Structure–Function Relationships
2.10. Methodological Considerations and Limitations
3. Methods
3.1. Protein Target Preparation
3.2. AlphaFold3 Ensemble Generation
3.3. Molecular Dynamics Simulations
3.4. ChronoSort Algorithm
3.5. Root-Mean-Squared Deviation Analysis
3.6. Root-Mean-Square Fluctuation Calculations
3.7. Side Chain Analysis
3.8. Principal Component Analysis
3.9. Eigenvector Visualization
3.10. Eigenvector Similarity Analysis
3.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AF3 | AlphaFold3 |
| MD | Molecular dynamics simulations |
| NMR | Nuclear magnetic resonance spectroscopy |
| PCA | Principal component analysis |
| RMSF | Root-mean-squared fluctuation |
| RMSD | Root-mean-squared deviation |
| pLDDT | per-residue predicted Local Distance Difference Test |
References
- Guo, J.; Zhou, H.-X. Protein allostery and conformational dynamics. Chem. Rev. 2016, 116, 6503–6515. [Google Scholar] [CrossRef]
- Wei, G.; Xi, W.; Nussinov, R.; Ma, B. Protein ensembles: How does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem. Rev. 2016, 116, 6516–6551. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.J.; Corte, D.D. Using molecular docking and molecular dynamics to investigate protein-ligand interactions. Mod. Phys. Lett. B 2021, 35, 2130002. [Google Scholar] [CrossRef]
- Della Corte, D.; van Beek, H.L.; Syberg, F.; Schallmey, M.; Tobola, F.; Cormann, K.U.; Schlicker, C.; Baumann, P.T.; Krumbach, K.; Sokolowsky, S. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020, 11, 4851. [Google Scholar] [CrossRef] [PubMed]
- Möckel, C.; Kubiak, J.; Schillinger, O.; Kühnemuth, R.; Della Corte, D.; Schröder, G.F.; Willbold, D.; Strodel, B.; Seidel, C.A.; Neudecker, P. Integrated NMR, fluorescence, and molecular dynamics benchmark study of protein mechanics and hydrodynamics. J. Phys. Chem. B 2018, 123, 1453–1480. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Billings, W.M.; Morris, C.J.; Della Corte, D. The whole is greater than its parts: Ensembling improves protein contact prediction. Sci. Rep. 2021, 11, 8039. [Google Scholar] [CrossRef]
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796, Erratum in Nat. Chem. Biol. 2009, 5, 954. [Google Scholar] [CrossRef] [PubMed]
- Motlagh, H.N.; Wrabl, J.O.; Li, J.; Hilser, V.J. The ensemble nature of allostery. Nature 2014, 508, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular dynamics simulation for all. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.E.; Grossman, J.; Bank, J.A.; Batson, B.; Butts, J.A.; Chao, J.C.; Deneroff, M.M.; Dror, R.O.; Even, A.; Fenton, C.H. Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. In Proceedings of the SC’14: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, New Orleans, LA, USA, 16–21 November 2014; pp. 41–53. [Google Scholar]
- Palmer, A.G., III. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004, 104, 3623–3640. [Google Scholar] [CrossRef]
- Kay, L.E. NMR studies of protein structure and dynamics. J. Magn. Reson. 2011, 213, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Mittermaier, A.K.; Kay, L.E. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 2009, 34, 601–611. [Google Scholar] [CrossRef]
- Della Corte, D.; Wildberg, A.; Schröder, G.F. Protein structure refinement with adaptively restrained homologous replicas. Proteins Struct. Funct. Bioinform. 2016, 84, 302–313. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Monteiro da Silva, G.; Cui, J.Y.; Dalgarno, D.C.; Lisi, G.P.; Rubenstein, B.M. High-throughput prediction of protein conformational distributions with subsampled AlphaFold2. Nat. Commun. 2024, 15, 2464, Correction in Nat Commun. 2024, 15, 3089. https://doi.org/10.1038/s41467-024-47504-0. [Google Scholar] [CrossRef]
- Audagnotto, M.; Czechtizky, W.; De Maria, L.; Käck, H.; Papoian, G.; Tornberg, L.; Tyrchan, C.; Ulander, J. Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble. Sci. Rep. 2022, 12, 10018. [Google Scholar] [CrossRef] [PubMed]
- Ruff, K.M.; Pappu, R.V. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Brotzakis, Z.; Zhang, S.; Vendruscolo, M. AlphaFold prediction of structural ensembles of disordered proteins. Nat. Commun. 2025, 16, 1632. [Google Scholar] [CrossRef]
- Wallerstein, J.; Han, X.; Levkovets, M.; Lesovoy, D.; Malmodin, D.; Mirabello, C.; Wallner, B.; Sun, R.; Sandalova, T.; Agback, P. Insights into mechanisms of MALT1 allostery from NMR and AlphaFold dynamic analyses. Commun. Biol. 2024, 7, 868. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy protein structure prediction in CASP14. Proteins Struct. Funct. Bioinform. 2021, 89, 1687–1699. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.; Berendsen, H.J. Essential dynamics of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 412–425. [Google Scholar] [CrossRef] [PubMed]
- García, A.E. Large-amplitude nonlinear motions in proteins. Phys. Rev. Lett. 1992, 68, 2696. [Google Scholar] [CrossRef] [PubMed]
- Ichiye, T.; Karplus, M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins Struct. Funct. Bioinform. 1991, 11, 205–217. [Google Scholar] [CrossRef]
- Bahar, I.; Lezon, T.R.; Yang, L.-W.; Eyal, E. Global dynamics of proteins: Bridging between structure and function. Annu. Rev. Biophys. 2010, 39, 23–42. [Google Scholar] [CrossRef]
- Zheng, W.; Brooks, B. Identification of dynamical correlations within the myosin motor domain by the normal mode analysis of an elastic network model. J. Mol. Biol. 2005, 346, 745–759. [Google Scholar] [CrossRef]
- Dror, R.O.; Dirks, R.M.; Grossman, J.; Xu, H.; Shaw, D.E. Biomolecular simulation: A computational microscope for molecular biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef] [PubMed]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef] [PubMed]
- Coates, T.L.; Young, N.; Jarrett, A.J.; Morris, C.J.; Moody, J.D.; Corte, D.D. Current computational methods for enzyme design. Mod. Phys. Lett. B 2021, 35, 2150155. [Google Scholar] [CrossRef]
- Stern, J.A.; Free, T.J.; Stern, K.L.; Gardiner, S.; Dalley, N.A.; Bundy, B.C.; Price, J.L.; Wingate, D.; Della Corte, D. A probabilistic view of protein stability, conformational specificity, and design. Sci. Rep. 2023, 13, 15493. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.R.; Pande, V.S.; Noé, F. An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; Volume 797. [Google Scholar]
- Plattner, N.; Noé, F. Protein conformational plasticity and complex ligand-binding kinetics explored by atomistic simulations and Markov models. Nat. Commun. 2015, 6, 7653. [Google Scholar] [CrossRef]
- Husic, B.E.; Pande, V.S. Markov state models: From an art to a science. J. Am. Chem. Soc. 2018, 140, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.K.; Trendelkamp-Schroer, B.; Paul, F.; Pérez-Hernández, G.; Hoffmann, M.; Plattner, N.; Wehmeyer, C.; Prinz, J.-H.; Noé, F. PyEMMA 2: A software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput. 2015, 11, 5525–5542. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.-B.; Perminov, A.; Bekele, S.; Kedziora, G.; Farajollahi, S.; Varaljay, V.; Hinkle, K.; Molinero, V.; Meister, K.; Hung, C. AlphaFold2 models indicate that protein sequence determines both structure and dynamics. Sci. Rep. 2022, 12, 10696. [Google Scholar] [CrossRef]
- Ma, P.; Li, D.W.; Brüschweiler, R. Predicting protein flexibility with AlphaFold. Proteins Struct. Funct. Bioinform. 2023, 91, 847–855. [Google Scholar] [CrossRef]
- Brown, B.P.; Stein, R.A.; Meiler, J.; Mchaourab, H.S. Approximating projections of conformational Boltzmann distributions with AlphaFold2 predictions: Opportunities and limitations. J. Chem. Theory Comput. 2024, 20, 1434–1447. [Google Scholar] [CrossRef] [PubMed]
- Alderson, T.R.; Pritišanac, I.; Kolarić, Đ.; Moses, A.M.; Forman-Kay, J.D. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2. Proc. Natl. Acad. Sci. USA 2023, 120, e2304302120. [Google Scholar] [CrossRef] [PubMed]
- Coskuner-Weber, O. Structures prediction and replica exchange molecular dynamics simulations of α-synuclein: A case study for intrinsically disordered proteins. Int. J. Biol. Macromol. 2024, 276, 133813. [Google Scholar] [CrossRef]
- Gardiner, S.; Talley, J.; Haynie, C.; Ebbert, J.; Argyle, M.; Allen, D.; Heaps, W.; Green, T.; Chipman, D.; Bundy, B.C. Advancing Luciferase Activity and Stability beyond Directed Evolution and Rational Design through Expert Guided Deep Learning. bioRxiv 2025. [Google Scholar] [CrossRef]
- Kitao, A. Principal component analysis and related methods for investigating the dynamics of biological macromolecules. J 2022, 5, 298–317. [Google Scholar] [CrossRef]
- Richard, J.P. Protein Flexibility and Stiffness Enable Efficient Enzymatic Catalysis. J. Am. Chem. Soc. 2019, 141, 3320–3331. [Google Scholar] [CrossRef] [PubMed]
- Aranda-García, D.; Stepniewski, T.M.; Torrens-Fontanals, M.; García-Recio, A.; Lopez-Balastegui, M.; Medel-Lacruz, B.; Morales-Pastor, A.; Peralta-García, A.; Dieguez-Eceolaza, M.; Sotillo-Nuñez, D.; et al. Large scale investigation of GPCR molecular dynamics data uncovers allosteric sites and lateral gateways. Nat. Commun. 2025, 16, 2020. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J. Protein complex prediction with AlphaFold-Multimer. Biorxiv 2021. [Google Scholar] [CrossRef]
- Sharma, N.; Sonavane, U.; Joshi, R. Comparative MD simulations and advanced analytics based studies on wild-type and hot-spot mutant A59G HRas. PLoS ONE 2020, 15, e0234836. [Google Scholar] [CrossRef] [PubMed]
- Wankowicz, S.A.; de Oliveira, S.H.; Hogan, D.W.; van den Bedem, H.; Fraser, J.S. Ligand binding remodels protein side-chain conformational heterogeneity. eLife 2022, 11, e74114. [Google Scholar] [CrossRef]
- Bonardi, A.; Turelli, M.; Moro, G.; Greco, C.; Cosentino, U.; Adamo, C. Behind the glow: Unveiling the nature of NanoLuc reactants and products. Phys. Chem. Chem. Phys. 2024, 26, 27447–27458. [Google Scholar] [CrossRef]
- Gardiner, S.; Dollinger, P.; Kovacic, F.; Pietruszka, J.; Ess, D.H.; Jaeger, K.-E.; Schröder, G.F.; Della Corte, D. Resolution of physics and deep learning-based protein engineering filters: A case study with a lipase for industrial substrate hydrolysis. PLoS ONE 2025, 20, e0332409. [Google Scholar] [CrossRef] [PubMed]
- Ping, J.; Hao, P.; Li, Y.-X.; Wang, J.-F. Molecular dynamics studies on the conformational transitions of adenylate kinase: A computational evidence for the conformational selection mechanism. BioMed Res. Int. 2013, 2013, 628536. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, S.; Momoh, R.; Lin, L.; Mallareddy, J.R.; Krstenansky, J.L. Identification of potential binding pocket on viral oncoprotein HPV16 E6: A promising anti-cancer target for small molecule drug discovery. BMC Mol. Cell Biol. 2019, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Holloway, D.E.; Singh, U.P.; Shogen, K.; Acharya, K.R. Crystal structure of Onconase at 1.1 Å resolution–insights into substrate binding and collective motion. FEBS J. 2011, 278, 4136–4149. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Sapundzhi, F.; Popstoilov, M.; Lazarova, M. RMSD Calculations for Comparing Protein Three-Dimensional Structures. In Proceedings of the International Conference on Numerical Methods and Applications, Borovets, Bulgaria, 22–26 August 2022; pp. 279–288. [Google Scholar]
- Cohen, F.E.; Sternberg, M.J. On the prediction of protein structure: The significance of the root-mean-square deviation. J. Mol. Biol. 1980, 138, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Li, S. Concise Formulas for the Area and Volume of a Hyperspherical Cap. 2011. Available online: https://stacks.cdc.gov/view/cdc/191858/cdc_191858_DS1.pdf (accessed on 30 September 2025).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272, Correction in Nat. Methods 2020, 17, 352. https://doi.org/10.1038/s41592-020-0772-5.. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Argyle, M.J.; Heaps, W.P.; Kubalek, C.; Gardiner, S.S.; Bundy, B.C.; Della Corte, D. ChronoSort: Revealing Hidden Dynamics in AlphaFold3 Structure Predictions. SynBio 2025, 3, 18. https://doi.org/10.3390/synbio3040018
Argyle MJ, Heaps WP, Kubalek C, Gardiner SS, Bundy BC, Della Corte D. ChronoSort: Revealing Hidden Dynamics in AlphaFold3 Structure Predictions. SynBio. 2025; 3(4):18. https://doi.org/10.3390/synbio3040018
Chicago/Turabian StyleArgyle, Matthew J., William P. Heaps, Corbyn Kubalek, Spencer S. Gardiner, Bradley C. Bundy, and Dennis Della Corte. 2025. "ChronoSort: Revealing Hidden Dynamics in AlphaFold3 Structure Predictions" SynBio 3, no. 4: 18. https://doi.org/10.3390/synbio3040018
APA StyleArgyle, M. J., Heaps, W. P., Kubalek, C., Gardiner, S. S., Bundy, B. C., & Della Corte, D. (2025). ChronoSort: Revealing Hidden Dynamics in AlphaFold3 Structure Predictions. SynBio, 3(4), 18. https://doi.org/10.3390/synbio3040018




