Digital to Biological Translation: How the Algorithmic Data-Driven Design Reshapes Synthetic Biology
Abstract
1. Introduction
2. Strategic Integration: ML/DL as Catalysts in Synthetic Biology
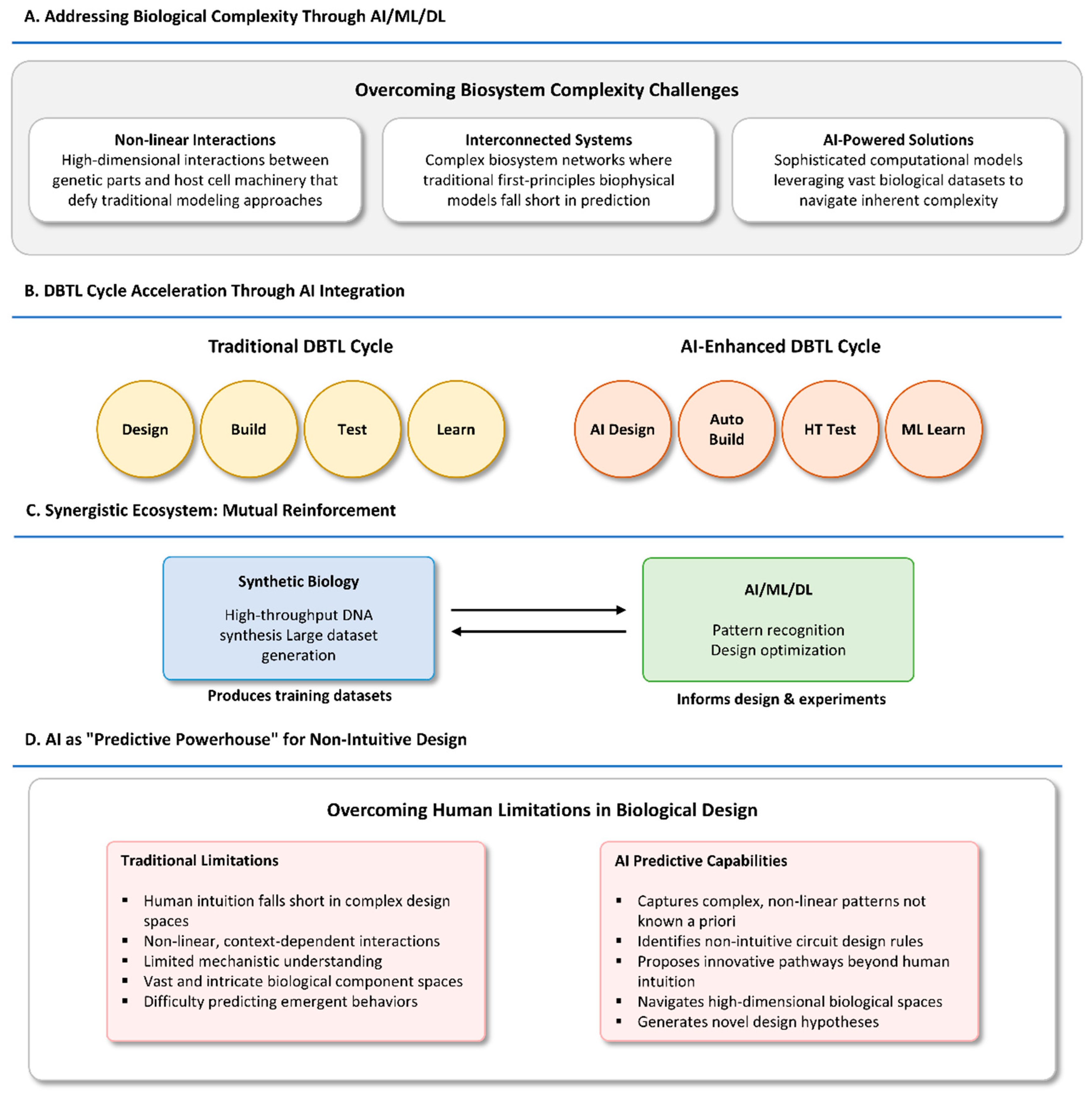
2.1. Addressing Biological Complexity and Non-Linearity with ML/DL
2.2. Accelerating the Synthetic Biology DBTL Cycle Through Data-Driven Approaches
2.3. Synergistic Relationship: Data Generation from Synthetic Biology for ML/DL Training
2.4. AI as a “Predictive Powerhouse” for Non-Intuitive Design
2.5. The Virtuous Cycle of Data and Design
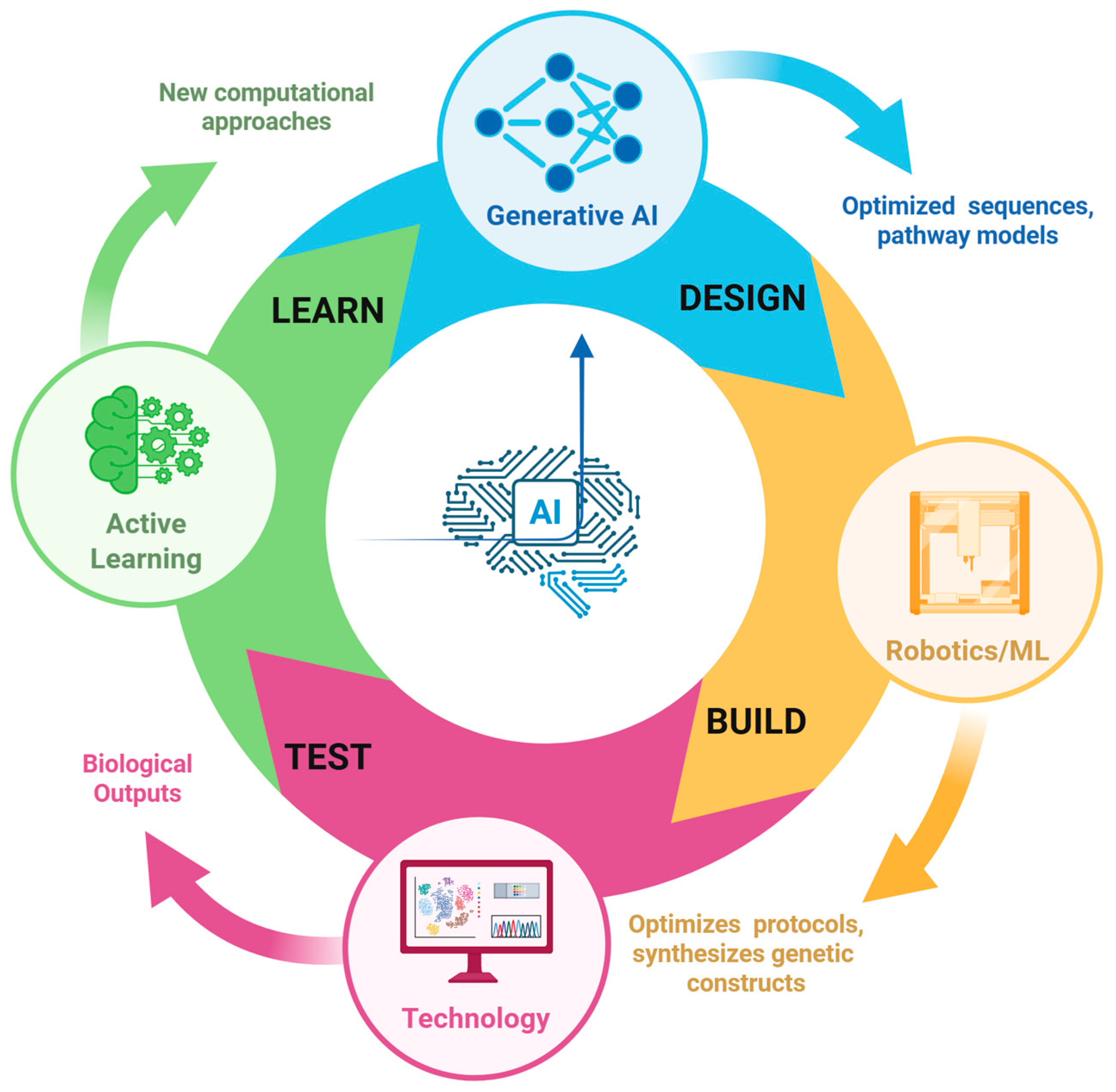
2.6. Integration Process of AI/ML/DL in the Synthetic Biology Workflow
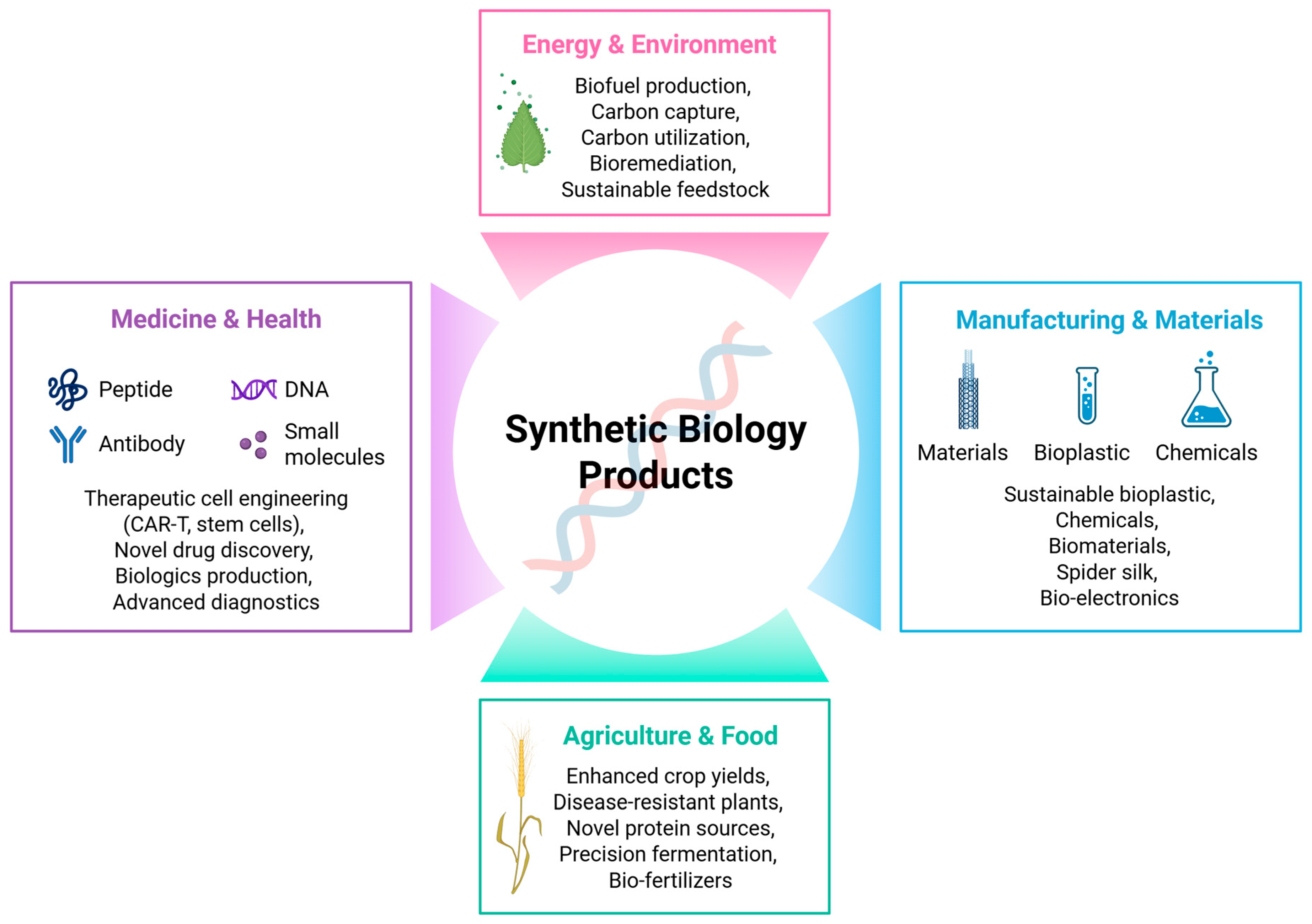
2.7. Real-World Applications of AI in Synthetic Biology
3. Advanced Applications of ML/DL in Synthetic Biology
3.1. Rational Design and Engineering of Biological Systems
3.1.1. Genetic Circuit Design and Optimization
3.1.2. De Novo Design of Novel Biological Components and Parts
3.1.3. Protein Engineering and Enzyme Optimization
3.1.4. Metabolic Pathway Design and Optimization
3.1.5. DNA Sequence Optimization and Gene Editing
3.2. Predictive Modeling and Simulation of Biological Behavior
3.3. High-Throughput Data Analysis and Knowledge Extraction
3.4. Automation of Laboratory Processes and Experimental Design
3.5. The Role of Foundation Models and Generative AI
3.6. From Prediction to Generation: The Evolution of AI’s Impact
3.7. AI-Driven Automation as a Force Multiplier for Discovery
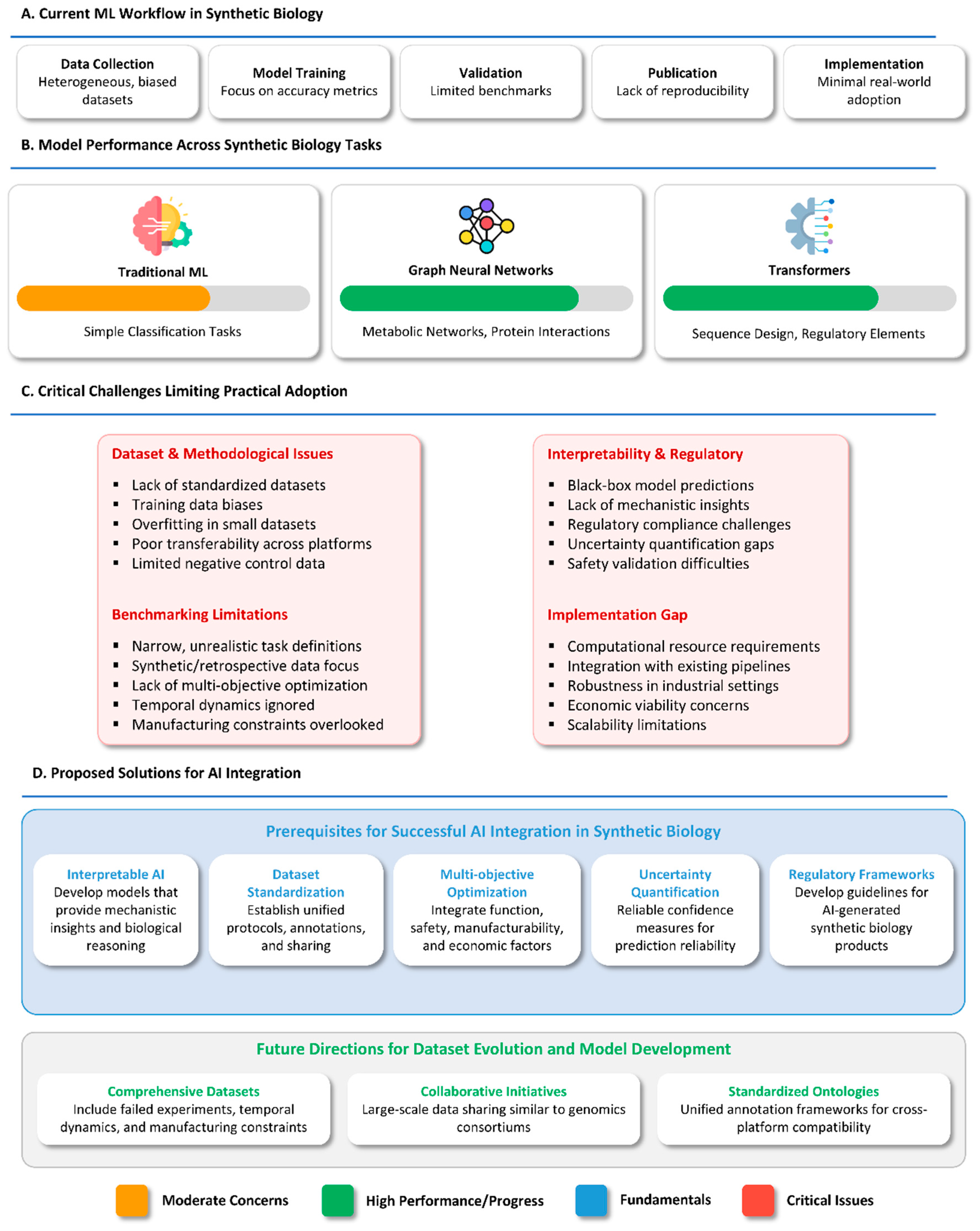
4. Critical Analysis of ML/DL in Synthetic Biology
4.1. Performance Disparities in ML/DL Approaches
4.2. Conditional Failures and Success of DL Models
4.3. Limitations in Current Benchmarking Practices
4.4. The Accuracy–Interpretability Trade-Off
4.5. Dataset Standardization and Reproducibility Concerns
4.6. Fundamental Limitations in Current DL Approaches
4.7. The Implementation Gap in Real-World Applications
4.8. Are DL Models Truly Learning Biological Principles?
4.9. Feasibility of AI-Generated Synthetic Biology Products
4.10. Prerequisites for AI Integration in Synthetic Biology
4.11. Evolution of Datasets for Multi-Objective Optimization
4.12. Outlook: Hybrid Models, Advanced Automation and Societal Impact
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CNNs | Convolutional Neural Networks |
| DBTL | Design-Build-Test-Learn |
| DL | Deep Learning |
| D-MPNN | Directed Message Passing Neural Network |
| FMs | Foundation Models |
| GANs | Generative Adversarial Networks |
| GEMs | Genome-Scale Metabolic Models |
| GNNs | Graph Neural Networks |
| Graph Attention | G-Attn |
| LLMs | Large Language Models |
| MINN | Multi-omics Integrated Neural Network |
| ML | Machine Learning |
| NLP | Natural Language processing |
| R&D | Research and Development |
| RBS | Ribosomal Binding Site |
| RNNs | Recurrent Neural Networks |
References
- Krohs, U.; Bedau, M.A. Interdisciplinary Interconnections in Synthetic Biology. Biol. Theory 2013, 8, 313–317. [Google Scholar] [CrossRef][Green Version]
- Calvert, J. Synthetic Biology: Constructing Nature? Sociol. Rev. 2010, 58, 95–112. [Google Scholar] [CrossRef]
- Bensaude-Vincent, B. Building Multidisciplinary Research Fields: The Cases of Materials Science. In Nanotechnology and Synthetic Biology; Springer: Cham, Switzerland, 2016; pp. 45–60. [Google Scholar]
- Ilyas, S.; Lee, J.; Hwang, Y.; Choi, Y.; Lee, D. Deciphering Cathepsin K Inhibitors: A Combined QSAR, Docking and MD Simulation Based Machine Learning Approaches for Drug Design. SAR QSAR Environ. Res. 2024, 35, 771–793. [Google Scholar] [CrossRef]
- Ogliore, T. Heme Is Not Just for Impossible Burgers-The Source-WashU. Available online: https://source.washu.edu/2021/05/heme-is-not-just-for-impossible-burgers/ (accessed on 28 July 2025).
- Le Feuvre, R.A.; Scrutton, N.S. A Living Foundry for Synthetic Biological Materials: A Synthetic Biology Roadmap to New Advanced Materials. Synth. Syst. Biotechnol. 2018, 3, 105–112. [Google Scholar] [CrossRef]
- Shapira, P.; Kwon, S.; Youtie, J. Tracking the Emergence of Synthetic Biology. Scientometrics 2017, 112, 1439–1469. [Google Scholar] [CrossRef]
- Mao, N.; Aggarwal, N.; Poh, C.L.; Cho, B.K.; Kondo, A.; Liu, C.; Yew, W.S.; Chang, M.W. Future Trends in Synthetic Biology in Asia. Adv. Genet. 2021, 2, e10038. [Google Scholar] [CrossRef]
- Rai, K.; Wang, Y.; O’Connell, R.W.; Patel, A.B.; Bashor, C.J. Using Machine Learning to Enhance and Accelerate Synthetic Biology. Curr. Opin. Biomed. Eng. 2024, 31, 100553. [Google Scholar] [CrossRef]
- Jeon, S.; Sohn, Y.J.; Lee, H.; Park, J.Y.; Kim, D.; Lee, E.S.; Park, S.J. Recent Advances in the Design-Build-Test-Learn (DBTL) Cycle for Systems Metabolic Engineering of Corynebacterium glutamicum. J. Microbiol. 2025, 63, e2501021. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Dewan, A.; Sahu, A.K.; Taye, M.M. Understanding of Machine Learning with Deep Learning: Architectures, Workflow, Applications and Future Directions. Computers 2023, 12, 91. [Google Scholar] [CrossRef]
- Goshisht, M.K. Machine Learning and Deep Learning in Synthetic Biology: Key Architectures, Applications, and Challenges. ACS Omega 2024, 9, 9921–9945. [Google Scholar] [CrossRef]
- Freemont, P.S. Synthetic Biology Industry: Data-Driven Design Is Creating New Opportunities in Biotechnology. Emerg. Top. Life Sci. 2019, 3, 651. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Wang, X.; Chen, G.; Liang, C.; Li, Q.; Jiang, H.; Xiong, W. Combining Diffusion and Transformer Models for Enhanced Promoter Synthesis and Strength Prediction in Deep Learning. mSystems 2025, 10, e0018325. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Chu, W.; Sheng, Y.; Tang, K.; Wang, S.; Liu, Y.; Wong, W.-F. Integrating Deep Learning and Synthetic Biology: A Co-Design Approach for Enhancing Gene Expression via N-Terminal Coding Sequences. ACS Synth. Biol. 2024, 13, 2960–2968. [Google Scholar] [CrossRef]
- Lutz, I.D.; Wang, S.; Norn, C.; Courbet, A.; Borst, A.J.; Zhao, Y.T.; Dosey, A.; Cao, L.; Xu, J.; Leaf, E.M.; et al. Top-down Design of Protein Architectures with Reinforcement Learning. Science 2023, 380, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Fu, L.; Guo, E.; Zhang, Z.; Wang, Z.; Ma, C.; Zhang, Z.; Zhang, J.; Huang, J.; Si, T. Automation in Synthetic Biology Using Biological Foundries. Chin. Sci. Bull. 2021, 66, 300–309. [Google Scholar] [CrossRef]
- Radivojević, T.; Costello, Z.; Workman, K.; Garcia Martin, H. A Machine Learning Automated Recommendation Tool for Synthetic Biology. Nat. Commun. 2020, 11, 4879. [Google Scholar] [CrossRef]
- Pandi, A.; Diehl, C.; Yazdizadeh Kharrazi, A.; Scholz, S.A.; Bobkova, E.; Faure, L.; Nattermann, M.; Adam, D.; Chapin, N.; Foroughijabbari, Y.; et al. A Versatile Active Learning Workflow for Optimization of Genetic and Metabolic Networks. Nat. Commun. 2022, 13, 3876. [Google Scholar] [CrossRef]
- Arboleda-Garcia, A.; Stiebritz, M.; Boada, Y.; Picó, J.; Vignoni, A. DBTL Bioengineering Cycle for Part Characterization and Refactoring. IFAC-PapersOnLine 2024, 58, 7–12. [Google Scholar] [CrossRef]
- Helmy, M.; Smith, D.; Selvarajoo, K. Systems Biology Approaches Integrated with Artificial Intelligence for Optimized Metabolic Engineering. Metab. Eng. Commun. 2020, 11, e00149, Erratum in Metab. Eng. Commun. 2021, 13, e00186. [Google Scholar] [CrossRef]
- Groff-Vindman, C.S.; Trump, B.D.; Cummings, C.L.; Smith, M.; Titus, A.J.; Oye, K.; Prado, V.; Turmus, E.; Linkov, I. The Convergence of AI and Synthetic Biology: The Looming Deluge. npj Biomed. Innov. 2025, 2, 20. [Google Scholar] [CrossRef]
- Sieow, B.F.-L.; De Sotto, R.; Seet, Z.R.D.; Hwang, I.Y.; Chang, M.W. Synthetic Biology Meets Machine Learning. Methods Mol. Biol. 2023, 2553, 21–39. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Nguyen-Tran, T.; Bennett, S.A.L. Machine Learning and Hybrid Methods for Metabolic Pathway Modeling. Methods Mol. Biol. 2023, 2553, 417–439. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M. Machine Learning Methods for Analysis of Metabolic Data and Metabolic Pathway Modeling. Metabolites 2018, 8, 4. [Google Scholar] [CrossRef]
- Azrag, M.A.K.; Kadir, T.A.A.; Kabir, M.N.; Jaber, A.S. Large-Scale Kinetic Parameters Estimation of Metabolic Model of Escherichia Coli. Int. J. Mach. Learn. Comput. 2019, 9, 160–167. [Google Scholar] [CrossRef]
- Li, Y.; Wu, F.-X.; Ngom, A. A Review on Machine Learning Principles for Multi-View Biological Data Integration. Brief. Bioinform. 2018, 19, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Johnston, K.E.; Arnold, F.H.; Yang, K.K. Protein Sequence Design with Deep Generative Models. Curr. Opin. Chem. Biol. 2021, 65, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, G.S.; Bakken, T.I.; Fages-Lartaud, M.; Lale, R. From Context to Code: Rational De Novo DNA Design and Predicting Cross-Species DNA Functionality Using Deep Learning Transformer Models. bioRxiv 2023. [Google Scholar] [CrossRef]
- Tazza, G.; Moro, F.; Ruggeri, D.; Teusink, B.; Vidács, L. MINN: A Metabolic-Informed Neural Network for Integrating Omics Data into Genome-Scale Metabolic Modeling. Comput. Struct. Biotechnol. J. 2025, 27, 3609–3617. [Google Scholar] [CrossRef] [PubMed]
- Hasibi, R.; Michoel, T.; Oyarzún, D.A. Integration of Graph Neural Networks and Genome-Scale Metabolic Models for Predicting Gene Essentiality. npj Syst. Biol. Appl. 2024, 10, 24. [Google Scholar] [CrossRef]
- Han, X.; Jia, M.; Chang, Y.; Li, Y.; Wu, S. Directed Message Passing Neural Network (D-MPNN) with Graph Edge Attention (GEA) for Property Prediction of Biofuel-Relevant Species. Energy AI 2022, 10, 100201. [Google Scholar] [CrossRef]
- Treloar, N.J.; Fedorec, A.J.H.; Ingalls, B.; Barnes, C.P. Deep Reinforcement Learning for the Control of Microbial Co-Cultures in Bioreactors. PLoS Comput. Biol. 2020, 16, e1007783. [Google Scholar] [CrossRef] [PubMed]
- Patron, N.J. Beyond Natural: Synthetic Expansions of Botanical Form and Function. New Phytol. 2020, 227, 295–310. [Google Scholar] [CrossRef]
- Palacios, S.; Collins, J.J.; Del Vecchio, D. Machine Learning for Synthetic Gene Circuit Engineering. Curr. Opin. Biotechnol. 2025, 92, 103263. [Google Scholar] [CrossRef]
- Müller, M.M.; Arndt, K.M.; Hoffmann, S.A. Genetic Circuits in Synthetic Biology: Broadening the Toolbox of Regulatory Devices. Front. Synth. Biol. 2025, 3, 1548572. [Google Scholar] [CrossRef]
- Prasad, K.; Cross, R.S.; Jenkins, M.R. Synthetic Biology, Genetic Circuits and Machine Learning: A New Age of Cancer Therapy. Mol. Oncol. 2023, 17, 946–949. [Google Scholar] [CrossRef]
- Chang, J.; Ye, J.C. Bidirectional Generation of Structure and Properties Through a Single Molecular Foundation Model. Nat. Commun. 2023, 15, 2323. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, H.H.; He, J.; Tibo, A.; Janet, J.P.; Voronov, A.; Mervin, L.H.; Engkvist, O. Reinvent 4: Modern AI–Driven Generative Molecule Design. J. Cheminform. 2024, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wu, L.; Zhao, B.; Huff, C.; Zhang, J.; Wu, J.; Lin, L.; Wei, P.; Wu, C. Benchmarking DNA Foundation Models for Genomic Sequence Classification. bioRxiv 2024, bioRxiv:2024.08.16.608288. [Google Scholar] [CrossRef]
- Thomas, N.; Belanger, D.; Xu, C.; Lee, H.; Hirano, K.; Iwai, K.; Polic, V.; Nyberg, K.D.; Hoff, K.G.; Frenz, L.; et al. Engineering Highly Active Nuclease Enzymes with Machine Learning and High-Throughput Screening. Cell Syst. 2025, 16, 101236. [Google Scholar] [CrossRef]
- Kouba, P.; Kohout, P.; Haddadi, F.; Bushuiev, A.; Samusevich, R.; Sedlar, J.; Damborsky, J.; Pluskal, T.; Sivic, J.; Mazurenko, S. Machine Learning-Guided Protein Engineering. ACS Catal. 2023, 13, 13863–13895. [Google Scholar] [CrossRef]
- Liu, S.-H.; Bai, L.; Wang, X.-D.; Wang, Q.-Q.; Wang, D.-X.; Bornscheuer, U.T.; Ao, Y.-F. Machine Learning-Guided Protein Engineering to Improve the Catalytic Activity of Transaminases under Neutral PH Conditions. Org. Chem. Front. 2025, 12, 4788–4793. [Google Scholar] [CrossRef]
- van Lent, P.; Schmitz, J.; Abeel, T. Simulated Design-Build-Test-Learn Cycles for Consistent Comparison of Machine Learning Methods in Metabolic Engineering. ACS Synth. Biol. 2023, 12, 2588–2599. [Google Scholar] [CrossRef]
- Zhou, K.; Ng, W.; Cortés-Peña, Y.; Wang, X. Increasing Metabolic Pathway Flux by Using Machine Learning Models. Curr. Opin. Biotechnol. 2020, 66, 179–185. [Google Scholar] [CrossRef]
- Cheng, Y.; Bi, X.; Xu, Y.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Machine Learning for Metabolic Pathway Optimization: A Review. Comput. Struct. Biotechnol. J. 2023, 21, 2381–2393. [Google Scholar] [CrossRef]
- Tabane, E.; Mnkandla, E.; Wang, Z. Optimizing DNA Sequence Classification via a Deep Learning Hybrid of LSTM and CNN Architecture. Appl. Sci. 2025, 15, 8225. [Google Scholar] [CrossRef]
- Li, J.; Wu, P.; Cao, Z.; Huang, G.; Lu, Z.; Yan, J.; Zhang, H.; Zhou, Y.; Liu, R.; Chen, H.; et al. Machine Learning-Based Prediction Models to Guide the Selection of Cas9 Variants for Efficient Gene Editing. Cell Rep. 2024, 43, 113765. [Google Scholar] [CrossRef] [PubMed]
- Einarson, D.; Frisk, F.; Klonowska, K.; Sennersten, C. A Machine Learning Approach to Simulation of Mallard Movements. Appl. Sci. 2024, 14, 1280. [Google Scholar] [CrossRef]
- Okoro, O.V.; Hippolyte, D.E.C.; Nie, L.; Karimi, K.; Denayer, J.F.M.; Shavandi, A. Machine Learning-Based Predictive Modeling and Optimization: Artificial Neural Network-Genetic Algorithm vs. Response Surface Methodology for Black Soldier Fly (Hermetia Illucens) Farm Waste Fermentation. Biochem. Eng. J. 2025, 218, 109685. [Google Scholar] [CrossRef]
- Smith, G.D.; Ching, W.H.; Cornejo-Páramo, P.; Wong, E.S. Decoding Enhancer Complexity with Machine Learning and High-Throughput Discovery. Genome Biol. 2023, 24, 116. [Google Scholar] [CrossRef]
- Alcantar, M.A.; English, M.A.; Valeri, J.A.; Collins, J.J. A High-Throughput Synthetic Biology Approach for Studying Combinatorial Chromatin-Based Transcriptional Regulation. Mol. Cell 2024, 84, 2382–2396.e9. [Google Scholar] [CrossRef]
- Rapp, J.T.; Bremer, B.J.; Romero, P.A. Self-Driving Laboratories to Autonomously Navigate the Protein Fitness Landscape. Nat. Chem. Eng. 2024, 1, 97–107. [Google Scholar] [CrossRef]
- Martin, H.G.; Radivojevic, T.; Zucker, J.; Bouchard, K.; Sustarich, J.; Peisert, S.; Arnold, D.; Hillson, N.; Babnigg, G.; Marti, J.M.; et al. Perspectives for Self-Driving Labs in Synthetic Biology. Curr. Opin. Biotechnol. 2023, 79, 102881. [Google Scholar] [CrossRef]
- Nguyen, E.; Poli, M.; Durrant, M.G.; Kang, B.; Katrekar, D.; Li, D.B.; Bartie, L.J.; Thomas, A.W.; King, S.H.; Brixi, G.; et al. Sequence Modeling and Design from Molecular to Genome Scale with Evo. Science 2024, 386, eado9336. [Google Scholar] [CrossRef]
- Tang, B.; Ewalt, J.; Ng, H.-L. Generative AI Models for Drug Discovery. In Topics in Medicinal Chemistry; Springer: Berlin/Heidelberg, Germany, 2021; Volume 37, pp. 221–243. [Google Scholar]
- Seo, E.; Choi, Y.-N.; Shin, Y.R.; Kim, D.; Lee, J.W. Design of Synthetic Promoters for Cyanobacteria with Generative Deep-Learning Model. Nucleic Acids Res. 2023, 51, 7071–7082. [Google Scholar] [CrossRef]
- Kitano, S.; Lin, C.; Foo, J.L.; Chang, M.W. Synthetic Biology: Learning the Way toward High-Precision Biological Design. PLoS Biol. 2023, 21, e3002116. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Han, R.; Jang, J.; Kim, J.; Paik, J.; Heo, J.; Lee, Y.; Park, J. MetaboGNN: Predicting Liver Metabolic Stability with Graph Neural Networks and Cross-Species Data. J. Cheminform. 2025, 17, 140. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph Neural Networks: A Review of Methods and Applications. AI Open 2020, 1, 57–81, Erratum in AI Open 2024. [Google Scholar] [CrossRef]
- Zhang, X.M.; Liang, L.; Liu, L.; Tang, M.J. Graph Neural Networks and Their Current Applications in Bioinformatics. Front. Genet. 2021, 12, 690049. [Google Scholar] [CrossRef]
- Yasmeen, E.; Wang, J.; Riaz, M.; Zhang, L.; Zuo, K. Designing Artificial Synthetic Promoters for Accurate, Smart, and Versatile Gene Expression in Plants. Plant Commun. 2023, 4, 100558. [Google Scholar] [CrossRef]
- Orzechowski, P.; Moore, J.H. Generative and Reproducible Benchmarks for Comprehensive Evaluation of Machine Learning Classifiers. Sci. Adv. 2022, 8, 4747. [Google Scholar] [CrossRef] [PubMed]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Scardapane, S.; Spinelli, I.; Mahmud, M.; Hussain, A. Interpreting Black-Box Models: A Review on Explainable Artificial Intelligence. Cogn. Comput. 2024, 16, 45–74. [Google Scholar] [CrossRef]
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Semmelrock, H.; Ross-Hellauer, T.; Kopeinik, S.; Theiler, D.; Haberl, A.; Thalmann, S.; Kowald, D. Reproducibility in Machine-learning-based Research: Overview, Barriers, and Drivers. AI Mag. 2025, 46, e70002. [Google Scholar] [CrossRef]
- Semmelrock, H.; Kopeinik, S.; Theiler, D.; Ross-Hellauer, T.; Kowald, D. Reproducibility in Machine Learning-Driven Research. AI Mag. 2023, 46, e70002. [Google Scholar]
- Kapoor, S.; Narayanan, A. Leakage and the Reproducibility Crisis in Machine-Learning-Based Science. Patterns 2023, 4, 100804. [Google Scholar] [CrossRef]
- Devi, N.B.; Sen, S.; Pakshirajan, K. Artificial Intelligence in Synthetic Biology. In Artificial Intelligence and Biological Sciences; CRC Press: Boca Raton, FL, USA, 2025; pp. 278–300. [Google Scholar]
- Adewumi, O.O.; Oladele, E.O.; Gbenle, O.A.; Taiwo, I.A. Artificial Intelligence in Bioinformatics: Cutting-Edge Techniques and Future Prospects. Bull. Nat. Appl. Sci. 2025, 1, 79–91. [Google Scholar] [CrossRef]
- Schmitt, F.-J.; Golüke, M.; Budisa, N. Bridging the Gap: Enhancing Science Communication in Synthetic Biology with Specific Teaching Modules, School Laboratories, Performance and Theater. Front. Synth. Biol. 2024, 2, 1337860. [Google Scholar] [CrossRef]
- Brooks, S.M.; Alper, H.S. Applications, Challenges, and Needs for Employing Synthetic Biology beyond the Lab. Nat. Commun. 2021, 12, 1390. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.A. Real-World Synthetic Biology: Is It Founded on an Engineering Approach, and Should It Be? Life 2019, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Kelley, N.J.; Whelan, D.J.; Kerr, E.; Apel, A.; Beliveau, R.; Scanlon, R. Engineering Biology to Address Global Problems: Synthetic Biology Markets, Needs, and Applications. Ind. Biotechnol. 2014, 10, 140–149. [Google Scholar] [CrossRef]
- Garner, K.L. Principles of Synthetic Biology. Essays Biochem. 2021, 65, 791–811. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and Obstacles for Deep Learning in Biology and Medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Beardall, W.A.V.; Stan, G.-B.; Dunlop, M.J. Deep Learning Concepts and Applications for Synthetic Biology. GEN Biotechnol. 2022, 1, 360–371. [Google Scholar] [CrossRef]
- Kuiken, T. Artificial Intelligence in The Biological Sciences: Uses, Safety, Security, and Oversight; R47849; Congressional Research Service (CRS) Reports & Issue Briefs: Washington, DC, USA, 2023. [Google Scholar]
- Mirchandani, I.; Khandhediya, Y.; Chauhan, K. Review on Advancement of AI in Synthetic Biology. Methods Mol. Biol. 2025, 2952, 483–490. [Google Scholar] [CrossRef]
- Hynek, N. Synthetic Biology/AI Convergence (SynBioAI): Security Threats in Frontier Science and Regulatory Challenges. AI Soc. 2025, 1–18. [Google Scholar] [CrossRef]
- Iram, A.; Dong, Y.; Ignea, C. Synthetic Biology Advances towards a Bio-Based Society in the Era of Artificial Intelligence. Curr. Opin. Biotechnol. 2024, 87, 103143. [Google Scholar] [CrossRef] [PubMed]
- Ameyaw, S.A.; Boateng, J.; Afari, D.A. Artificial Intelligence Algorithms for Designing Synthetic Biological Systems and Predicting Their Behavior. OSF Prepr. 2025. [Google Scholar] [CrossRef]
- Gaeta, A.; Zulkower, V.; Stracquadanio, G. Design and Assembly of DNA Molecules Using Multi-Objective Optimization. Synth. Biol. 2021, 6, ysab026. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Si, L.; Zhang, X.; Cheng, R.; He, C.; Tan, K.C.; Jin, Y. Evolutionary Large-Scale Multi-Objective Optimization: A Survey. ACM Comput. Surv. 2022, 54, 1–34. [Google Scholar] [CrossRef]
- Collins, T.K.; Zakirov, A.; Brown, J.A.; Houghten, S. Single-Objective and Multi-Objective Genetic Algorithms for Compression of Biological Networks. In Proceedings of the 2017 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology, CIBCB, Manchester, UK, 23–25 August 2017. [Google Scholar] [CrossRef]
- Patane, A.; Santoro, A.; Costanza, J.; Carapezza, G.; Nicosia, G. Pareto Optimal Design for Synthetic Biology. IEEE Trans. Biomed. Circuits Syst. 2015, 9, 555–571. [Google Scholar] [CrossRef]
- Boada, Y.; Reynoso-Meza, G.; Picó, J.; Vignoni, A. Multi-Objective Optimization Framework to Obtain Model-Based Guidelines for Tuning Biological Synthetic Devices: An Adaptive Network Case. BMC Syst. Biol. 2016, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Fernau, S.; Dabrock, P. (Re-)Designing Nature? An Overview and Outlook on the Ethical and Societal Challenges in Synthetic Biology. Adv. Biosyst. 2019, 3, 1800326. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, A.; Lastra, L.; Nguyen, B.; Chen, Y.-J.; Nivala, J.; Ceze, L.; Strauss, K. Physical Laboratory Automation in Synthetic Biology. ACS Synth. Biol. 2023, 12, 3156–3169. [Google Scholar] [CrossRef] [PubMed]
- Anh Tuan, D.; Uyen, P.V.N.; Masak, J. Hybrid Quorum Sensing and Machine Learning Systems for Adaptive Synthetic Biology: Toward Autonomous Gene Regulation and Precision Therapies. Preprints 2024, 2024101551. [Google Scholar] [CrossRef]
| DBTL Stage | Traditional Approach/Challenge | AI/ML/DL Contribution | Impact/Benefit | Representative Studies |
|---|---|---|---|---|
| Design | Ad hoc, intuition-driven methods: Designers often rely on trial-and-error due to the inherent complexity and non-linear interactions within biological systems. This approach is time-consuming and lacks predictability in output behavior. Difficulty arises in predicting the precise behavior of novel genetic circuits or pathways. | Predictive modeling & generative AI: Predictive modeling of genetic circuits, metabolic pathways, and cellular responses allows in silico validation before experimental work. De novo sequence generation for proteins, RNA, and DNA elements (e.g., promoters, RBS using generative models (e.g., GANs, VAEs) that adhere to biological constraints. Optimization of DNA sequences for expression, stability, or specific functionalities. AI-driven recommendations for optimal experimental designs and component selections, exploring vast design spaces far beyond human capacity. | Accelerated design & enhanced success rates: Significantly reduced design iterations, minimizing costly and time-consuming experimental cycles. Higher probability of successful outcomes due to in silico validation and optimized component selection. Enhanced precision and predictability in desired biological functions. Exploration and discovery of non-intuitive or counter-intuitive designs that would be inaccessible through traditional, heuristic approaches. Expedited time-to-market for novel biological products and therapies. | DL for gene/promoter design: DL models (transformers, CNNs) accurately predict gene expression and optimize sequences in silico. De novo rational design; transformer models for synthetic promoter design [14]. CNNs to design N-terminal coding sequences, resulting in increase in protein expression [15]. RL for protein design [16] |
| Build | Manual and labor-intensive assembly: The physical construction of genetic constructs is often performed manually, making it labor-intensive, prone to human error, and limited in throughput. This constraint severely restricts the diversity of genetic libraries that can be synthesized and screened. | Automated synthesis & robotic assembly: Automated DNA synthesis and cloning platforms leveraging high-throughput robotics for constructing gene circuits and pathways. Robotic assembly of genetic constructs (e.g., Golden Gate, Gibson Assembly) minimizes manual intervention and improves consistency. AI-driven scheduling and optimization of robotic workflows to maximize efficiency and minimize material waste. Integration with liquid handling systems for precise reagent dispensing and reaction setup. | Increased throughput & reduced costs: Substantially faster assembly times for complex genetic constructs. Significant reduction in labor costs and human-induced errors, leading to higher reliability. Vastly increased throughput and library diversity, enabling the exploration of a much broader design space. Improved consistency and reproducibility of built components, leading to more reliable experimental outcomes. | ML-guided design–build automation: Bayesian optimization and RL integrate with robotic platforms to automatically determine and execute the most efficient DNA construction protocols. Minimizes human error and labor; accelerates construction time for large genetic libraries by intelligently optimizing assembly protocols [17]. |
| Test | Laborious and low-throughput screening: Experimental testing involves laborious screening processes and often manual analysis of microscopy or flow cytometry data. This creates a limited capacity for handling large datasets and extracting meaningful insights efficiently. | High-throughput data analysis & automated phenotyping: High-throughput data analysis (e.g., genomics, transcriptomics, proteomics, metabolomics data) using advanced ML/DL algorithms for pattern recognition and feature extraction. Automated image and microscopy data analysis for rapid quantification of cellular phenotypes, morphology, and protein localization. Rapid identification of subtle patterns and anomalies in large, complex datasets that are imperceptible to human analysis. Real-time monitoring and anomaly detection in bioreactors and cell cultures. | Rapid insights & efficient evaluation: Accelerated insights from experimental results, enabling faster decision-making and iteration. Massively increased experimental throughput, allowing for parallel testing of numerous designs. More efficient and accurate evaluation of phenotypes and functional outputs. Early identification of successful or problematic designs, minimizing resources spent on unproductive pathways. Discovery of unexpected relationships within experimental data. | DL-enabled high-throughput analysis: CNN and GNN models analyze massive datasets (microscopy, multi-omics) to rapidly quantify phenotypes, classify cell states, and extract performance metrics [18]. Accelerates data-to-insight; enables rapid and objective evaluation of vast numbers of constructs; uncovers subtle or novel phenotypes otherwise missed by manual inspection. |
| Learn | Limited model-based refinement: Iterative refinement is often based on limited empirical models, leading to unpredictable performance and a high number of design-build-test cycles. The lack of a robust feedback loop slows down the overall discovery process. | AL and RL for iterative improvement: AL strategies guide subsequent experiments by selecting the most informative data points to optimize model training and reduce experimental burden. RL frameworks enable autonomous optimization of experimental parameters and processes based on observed outcomes. AI-driven feedback loops that automatically analyze test data, update design models, and recommend parameters for the next iteration. Identification of underlying trends, optimal parameters, and governing rules from vast experimental data, facilitating true biological understanding. | Accelerated discovery & optimized cycles: Significantly accelerated discovery cycles by intelligently guiding subsequent experiments. Smarter, data-driven feedback loops replace manual guesswork. Optimized experimental conditions and resource allocation. Minimized trial-and-error, leading to more efficient resource utilization and faster convergence to desired designs. Continuous improvement of synthetic biology platforms and processes through automated knowledge extraction. | AI-driven closed-loop optimization: RL and AL systematically retrain models on new results to propose the most informative subsequent experiment, iteratively refining designs [19]. AL accelerates metabolic pathway optimization. Minimizes trial-and-error; guides the search space intelligently, leading to a faster convergence on optimal designs; accelerates scientific discovery by automating the hypothesis generation step. |
| DBTL integration | Not applicable as a traditional stage/challenge, but the lack of true autonomy is the underlying issue. | End-to-end AI frameworks coupling all DBTL stages: Comprehensive closed-loop AI pipelines orchestrate and connect the automated design, build, and test steps, creating self-optimizing bioengineering systems | Achieves autonomous science: Realizes the full potential of high-throughput bioengineering; Dramatically reduces cycle time and cost for industrial-scale pathway and product development. | AI integrated DBTL platforms for autonomous genetic circuit optimization [20]. This dramatically reduces cycle time and cost for industrial-scale pathway and product development. |
| Challenge Area | Description | Implications | Current Gaps/Needs |
|---|---|---|---|
| Interpretable AI models | AI models must provide mechanistic, explainable predictions that align with biological reasoning. | Enhances scientific understanding and regulatory transparency. | Lack of models with biologically meaningful interpretability; limited tools for hypothesis generation. |
| Data & protocol standardization | Harmonization of datasets, experimental protocols, and performance metrics across the field. | Enables reproducibility, benchmarking, and fair evaluation of AI models. | Fragmented datasets, inconsistent formats, lack of agreed-upon evaluation metrics. |
| Multi-objective optimization | Integration of frameworks that balance biological performance with manufacturability, safety, and cost-effectiveness. | Supports practical translation of AI designs into real-world applications. | Limited optimization tools that handle diverse biological and engineering constraints simultaneously. |
| Uncertainty quantification | Development of AI tools that assess the confidence or reliability of predictions within their operational domain. | Increases user trust and identifies when models are making extrapolative or risky suggestions. | Absence of robust, domain-specific uncertainty quantification methods in synthetic biology contexts. |
| Regulatory frameworks | Establish policies and standards that can assess and approve AI-designed biological products without compromising safety. | Critical for commercialization, public trust, and long-term viability of AI-enabled products. | Regulatory ambiguity; lack of precedent and guidelines for AI-derived biological constructs. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manan, A.; Qayyum, N.; Ramachandran, R.; Qayyum, N.; Ilyas, S. Digital to Biological Translation: How the Algorithmic Data-Driven Design Reshapes Synthetic Biology. SynBio 2025, 3, 17. https://doi.org/10.3390/synbio3040017
Manan A, Qayyum N, Ramachandran R, Qayyum N, Ilyas S. Digital to Biological Translation: How the Algorithmic Data-Driven Design Reshapes Synthetic Biology. SynBio. 2025; 3(4):17. https://doi.org/10.3390/synbio3040017
Chicago/Turabian StyleManan, Abdul, Nabila Qayyum, Rajath Ramachandran, Naila Qayyum, and Sidra Ilyas. 2025. "Digital to Biological Translation: How the Algorithmic Data-Driven Design Reshapes Synthetic Biology" SynBio 3, no. 4: 17. https://doi.org/10.3390/synbio3040017
APA StyleManan, A., Qayyum, N., Ramachandran, R., Qayyum, N., & Ilyas, S. (2025). Digital to Biological Translation: How the Algorithmic Data-Driven Design Reshapes Synthetic Biology. SynBio, 3(4), 17. https://doi.org/10.3390/synbio3040017







