- Article
Isolation, Identification, and Molecular Characterization of Mycoplasma bovis from Beef Cattle in Kunming, and Development of a SYBR Green qPCR Assay
- Guojun Wang,
- Yuqing Li and
- Wengui Li
- + 6 authors
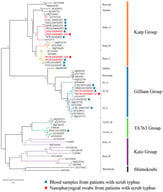
Mycoplasma bovis (M. bovis) is a major pathogen responsible for bovine respiratory disease, mastitis, and arthritis, causing significant economic losses to the cattle industry worldwide. To elucidate the genetic and biological characteristics of M. bovis circulating in Yunnan Province, China, twenty PCR-positive bovine respiratory samples were collected from cattle farms in Kunming; three isolates—M.bo-YNXD-1, A1, and A8—were successfully cultured and identified through colony morphology, biochemical assays, and molecular characterization. Antimicrobial susceptibility testing showed that M.bo-YNXD-1 exhibited multidrug resistance to six antibiotics, including ciprofloxacin and lincomycin, while A1 and A8 were resistant to one or two agents, respectively. Multilocus sequence typing (MLST) analysis revealed that isolates M.bo-YNXD-1 and M.bo-YNXD-A8 belonged to sequence type ST52, whereas isolate M.bo-YNXD-A1 was assigned to ST90, indicating the coexistence of distinct genetic lineages in this region. Virulence gene screening showed that isolate M.bo-YNXD-A8 was positive for VspX and p81, whereas all three isolates were positive for p48 and Vpam. A SYBR Green I-based quantitative PCR (qPCR) assay targeting the oppD/F gene was established, exhibiting high specificity, a detection limit of 10 copies/μL, and intra-/inter-assay variation below 3%. Validation using clinical samples demonstrated superior sensitivity compared with conventional PCR. Taken together, these findings indicate the presence of distinct MLST genotypes and virulence-associated genetic heterogeneity among regional Mycoplasma bovis isolates, and introduce a rapid, sensitive, and reliable qPCR assay for early detection and epidemiological surveillance. This study provides critical insights for rational antimicrobial use and targeted control strategies against M. bovis infections.
2 February 2026



![Schematic of the Flaviviridae virus particle. The left particle is a cross-section with the viral components labeled. Members of the old genera Orthoflavivirus, Hepacivirus, and Pegivirus have two E proteins (E dimers), whereas the genus Pestivirus has three (E trimers). The right particle shows the surface proteins arranged in an icosahedral-like symmetry. (Reproduced from [57]. Source: SwissBioPics. The images are licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) License https://creativecommons.org/licenses/by/4.0/).](https://mdpi-res.com/cdn-cgi/image/w=281,h=192/https://mdpi-res.com/pathogens/pathogens-15-00160/article_deploy/html/images/pathogens-15-00160-ag-550.jpg)