- Article
SPINET-KSP: A Multi-Modal LLM-Graph Foundation Model for Contextual Prediction of Kinase-Substrate-Phosphatase Triads
- Michael Olaolu Arowolo,
- Marian Emmanuel Okon and
- Sulaiman Olaniyi Abdulsalam
- + 2 authors
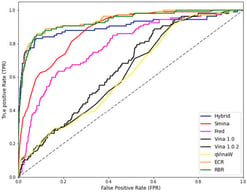
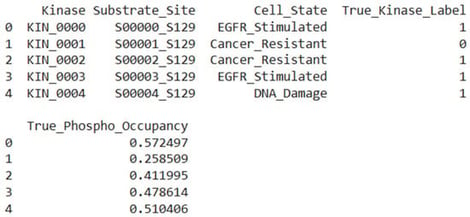
Reversible protein phosphorylation is an important regulatory mechanism in cellular signalling and disease, regulated by the opposing actions of kinases and phosphatases. Modern computer methods predict kinase–substrate or phosphatase–substrate interactions in isolation and lack specificity for biological conditions, neglecting triadic regulation. We present SPINET-KSP, a multi-modal LLM–Graph foundation model engineered for the prediction of kinase–substrate–phosphatase (KSP) triads with contextual awareness. SPINET-KSP integrates high-confidence interactomes (SIGNOR, BioGRID, STRING), structural contacts obtained from AlphaFold3, ESM-3 sequence embeddings, and a 512-dimensional cell-state manifold with 1612 quantitative phosphoproteomic conditions. A heterogeneous KSP graph is examined utilising a cross-attention Graphormer with Reversible Triad Attention to mimic kinase–phosphatase antagonism. SPINET-KSP, pre-trained on 3.41 million validated phospho-sites utilising masked phosphorylation modelling and contrastive cell-state learning, achieves an AUROC of 0.852 for kinase-family classification (sensitivity 0.821, specificity 0.834, MCC 0.655) and a Pearson correlation coefficient of 0.712 for phospho-occupancy prediction. In distinct 2025 mass spectrometry datasets, it identifies 72% of acknowledged cancer-resistance triads within the top 10 rankings and uncovers 247 supplementary triads validated using orthogonal proteomics. SPINET-KSP is the first foundational model for simulating context-dependent reversible phosphorylation, enabling the targeting of dysregulated kinase-phosphatase pathways in diseases.
22 January 2026



![Ligand-binding to primary and secondary sites of CK2α’ and CK2α. (a) Surface representation of a CK2α’Cys336Ser/3,4-dichlorophenethylamine (DPA) complex structure [19] (gray: N-terminal domain; yellow: C-terminal domain) with superimposed complex structures of CK2α’Cys336Ser/4p (PDB_ID 7AT9), CK2α’Cys336Ser/ARC-780 (PDB_ID 8Q77), CK2α’Cys336Ser/compound 12 (PDB_ID 9FBI), and CK2α1-335/heparin (PDB_IDs 7B8H and 7B8I); for the superimposed complex structures, only the ligands (in the case of ARC-780, only the middle part, which was defined by electron density) were drawn in order to indicate the various binding sites, as indicated in the figure. (b) Differences between CK2α’ (yellow C-atoms and bonds) and CK2α (magenta-colored C-atoms and bonds) in the hydrophobic shell of the αD pocket: the side chain difference Leu140/Ile141 correlates with a 180-degree rotation of DPA, the αD pocket anchor group of the bivalent inhibitor KN2 [19]. The figure was prepared with PyMOL, version 1.7.0.3 [20].](https://mdpi-res.com/cdn-cgi/image/w=281,h=192/https://mdpi-res.com/kinasesphosphatases/kinasesphosphatases-04-00001/article_deploy/html/images/kinasesphosphatases-04-00001-ag-550.jpg)