- Data Descriptor
Psoriatic Arthritis (PsA) Clinical Lipidomics Dataset with Hidden Laboratory Workflow Artifacts: A Benchmark Dataset for Data Processing Quality Control in Lipidomics
- Jörn Lötsch,
- Robert Gurke and
- Gerd Geisslinger
- + 2 authors
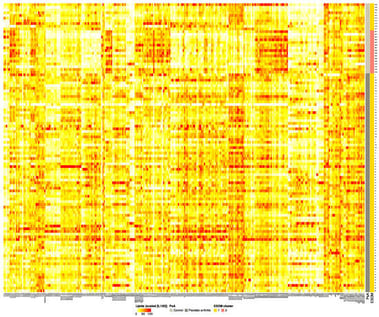
This dataset presents a real-world lipidomics resource for developing and benchmarking quality control methods, batch effect detection algorithms, and data validation workflows. The data originates from a cross-sectional clinical study of psoriatic arthritis (PsA) patients (n = 81) and healthy controls (n = 26), matched for age, sex, and body mass index, which was collected at a tertiary university rheumatology center. Subtle laboratory irregularities were detected only through advanced unsupervised analysis, after passing conventional quality control and standard analytical methods. Blood samples were processed using standardized protocols and analyzed using high-resolution and tandem mass spectrometry platforms. Both targeted and untargeted lipid assays captured lipids of several classes (including carnitines, ceramides, glycerophospholipids, sphingolipids, glycerolipids, fatty acids, sterols and esters, endocannabinoids). The dataset is organized into four comma-separated value (CSV) files: (1) Box–Cox-transformed and imputed lipidomics values; (2) outlier-cleaned and imputed values on the original scale; (3) metadata including clinical classifications, biological sex, and batch information for all assay types and control sample processing dates; and (4) a variable-level description file (readme.csv). The 292 lipid variables are named according to LIPID MAPS classification and standardized nomenclature. Complete batch documentation and FAIR-compliant data structure make this dataset valuable for testing the robustness of analytical pipelines and quality control in lipidomics and related omics fields. This unique dataset does not compete with larger lipidomics quality control datasets for comparisons of results but provides a unique, real-life lipidomics dataset displaying traces of the laboratory sample processing schedule, which can be used to challenge quality control frameworks.
3 February 2026