- Article
Development and Validation of a CatBoost-Based Model for Predicting Significant Creatinine Elevation in ICU Patients Receiving Vancomycin Therapy
- Junyi Fan,
- Li Sun and
- Shuheng Chen
- + 3 authors
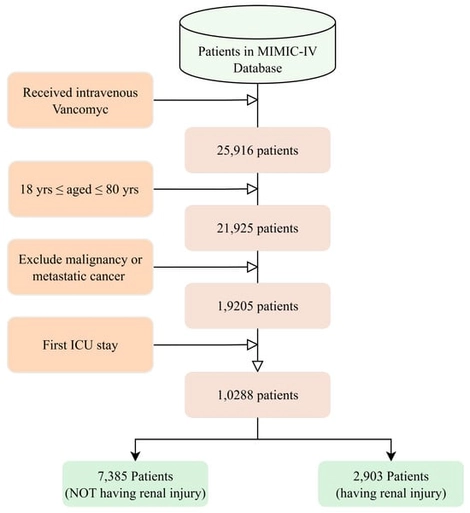
Vancomycin remains a cornerstone for severe Gram-positive infections in the ICU, yet creatinine elevation—a sensitive marker of early renal stress—occurs frequently and complicates therapy. We developed a machine learning model to predict vancomycin-associated creatinine elevation using routinely available clinical data, enabling preemptive risk stratification. In this retrospective MIMIC-IV cohort study (
10 December 2025