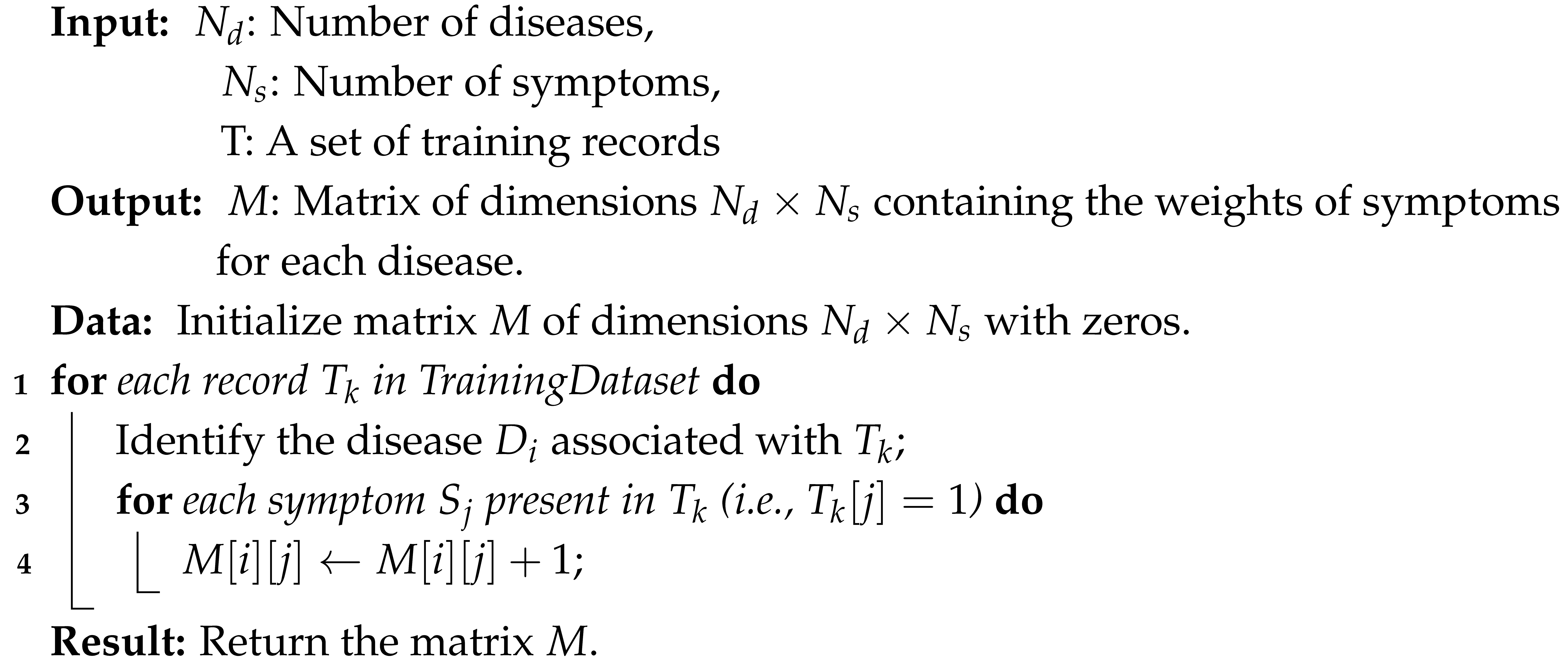1. Introduction
Medical diagnostics has undergone a remarkable transformation over the past decade. Clinicians today face an overwhelming amount of patient data, complex disease patterns, and constant pressure to make accurate decisions quickly [
1]. While traditional diagnostic approaches served medicine well for decades, they now struggle to keep pace with the sheer volume and complexity of modern healthcare data [
2,
3]. Consider a typical scenario: when evaluating a patient with multiple symptoms, a physician must consider numerous possible disease combinations. With just
n symptoms, the evaluation potentially faces
different combinations, a number that becomes unmanageable surprisingly quickly.
This computational challenge has sparked considerable interest in developing more sophisticated diagnostic tools. Researchers have explored numerous paths, each offering unique insights into improving medical diagnosis. For instance, Başçiftçi and colleagues took an interesting approach in 2018 when they applied Boolean function minimization to reduce the complexity of rule-based systems for cancer diagnosis [
4]. Their work showed that exhaustive rule evaluation is not always necessary; clever optimization can dramatically speed up the diagnostic process without sacrificing accuracy.
Meanwhile, other researchers have focused on making diagnostic tools more accessible. Sridhara’s team recently developed a mobile application that brings machine learning-powered diagnosis to remote areas where medical expertise is scarce [
5]. It serves as a reminder that sophisticated diagnostic systems mean little if they cannot reach the patients who need them most. Similarly, Singh and colleagues demonstrated in 2024 how combining machine learning association rules with rough set theory could handle the messy, incomplete data that is common in real-world medical settings [
6]. Their work on neurodevelopmental diseases showed particularly promising results, suggesting that hybrid approaches might be key to handling complex diagnostic challenges.
The question of how to manage and process vast amounts of medical data efficiently has also received significant attention. Tashkandi’s group tackled this by developing methods to perform patient similarity analysis directly within database systems, rather than extracting and processing data externally [
7]. This seemingly simple change led to substantial performance improvements. At a more fundamental level, Zhou and colleagues constructed comprehensive disease networks from biomedical literature, revealing surprising connections between symptoms, genetics, and protein interactions that were not apparent when looking at diseases in isolation [
8]. Their network-based perspective has opened new avenues for understanding disease relationships and developing more nuanced diagnostic approaches.
Perhaps one of the most promising developments has been the emergence of hybrid reasoning systems. Traditional case-based reasoning (CBR), while powerful, exhibits several limitations when used alone [
9,
10]. In medical domains, conventional CBR often struggles with large and heterogeneous datasets due to the high computational cost of retrieving similar cases, limited scalability when new cases are added, and reduced accuracy when symptoms overlap or co-occur in correlated patterns. These issues make it difficult for traditional CBR to operate efficiently in real-time or complex diagnostic environments.
Recognizing these limitations, researchers have explored hybrid approaches. Sharaf-El-Deen’s team demonstrated that combining case-based reasoning with rule-based approaches could overcome many of these constraints, particularly in breast cancer and thyroid disease diagnosis [
11]. Their system used rules to pre-filter candidate cases, reducing the computational burden of similarity calculations. Kumar and colleagues extended this concept into intensive care units, where the ability to adapt to rapidly changing patient conditions is crucial [
12]. Their hybrid system demonstrated that flexibility and adaptability are just as important as accuracy in real-world clinical settings.
Against this backdrop of ongoing innovation, this paper presents MARS (Matrix-Accelerated Reasoning System), a diagnostic methodology that takes a different approach to managing complexity. Rather than trying to brute-force through all possible disease-symptom combinations or relying solely on black-box machine learning models, MARS combines the intuitive appeal of case-based reasoning with the computational efficiency of matrix operations. The core insight is relatively straightforward: by representing disease-symptom relationships as matrices and using intelligent filtering techniques, the search space can be dramatically reduced without losing important diagnostic information.
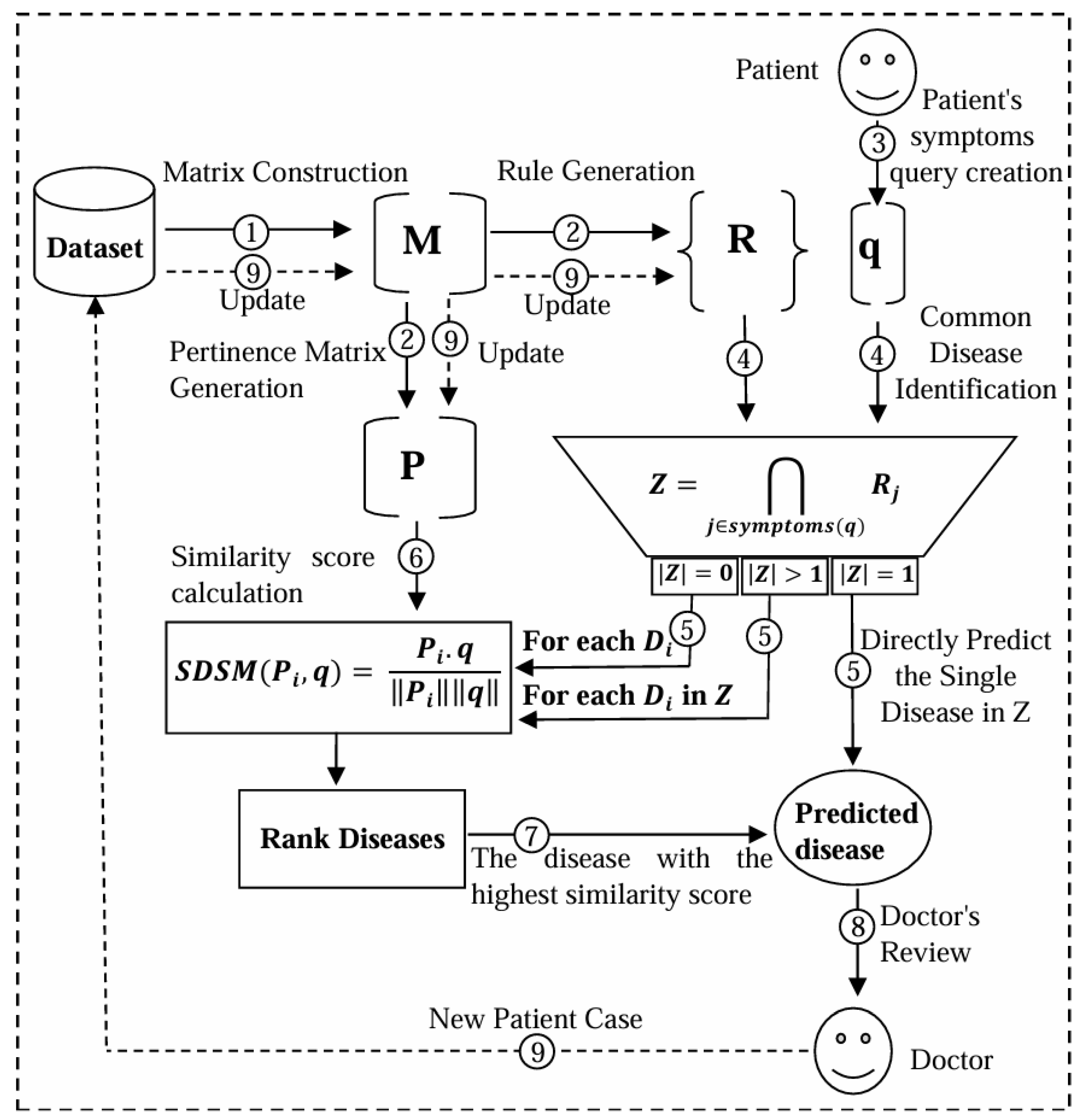
What makes this approach particularly interesting is how it handles the dynamic nature of medical knowledge. New diseases emerge, understanding of existing conditions evolves, and patient populations change over time. Traditional diagnostic systems often struggle with these changes, requiring extensive retraining or manual updates. MARS, by contrast, incorporates automatic rule generation and dynamic updating mechanisms that allow seamless adaptation as new data becomes available. The Pertinence Matrix, essentially a sophisticated weighting system that captures how relevant each symptom is for different diseases, updates automatically based on encountered cases.
Careful attention has also been paid to practical deployment aspects. The system uses set intersections to quickly narrow down the list of potential diseases for any given patient query. This might sound simple, but the effect on computational efficiency is dramatic. Where a traditional approach might need to evaluate hundreds or thousands of possibilities, this method typically considers only a handful of the most relevant cases. This efficiency does not come at the cost of accuracy; in fact, by focusing computational resources on the most promising candidates, better results are often achieved than systems that spread their analysis too thin.
To validate these claims, extensive comparative studies were conducted. The approach was evaluated against both traditional sequential methods and modern machine learning algorithms including Decision Trees, Random Forests, K-Nearest Neighbors (KNN), Support Vector Classifiers (SVCs), Bayesian classifiers (Bernoulli Naive Bayes), and neural networks (Multi-Layer Perceptron with ReLU activation). The results were encouraging: MARS consistently delivered competitive or superior accuracy while requiring significantly less computational time. Perhaps more importantly, the system maintained its performance even when scaled up to larger datasets with more symptoms and diseases.
Recent developments in explainable AI have also influenced the design philosophy [
13]. Unlike many modern diagnostic systems that operate as black boxes, the matrix-based approach provides clear insights into why particular diagnoses are suggested. Each step in the diagnostic process can be traced and understood, making it easier for clinicians to trust and validate the system’s recommendations. This transparency is crucial in medical settings where understanding the reasoning behind a diagnosis can be as important as the diagnosis itself [
14].
The implications of this work extend beyond just improving diagnostic accuracy or speed. By providing a framework that is both efficient and adaptable, this research contributes to the broader goal of making sophisticated diagnostic support available wherever it is needed. Whether in a well-equipped urban hospital or a resource-constrained rural clinic, the same underlying methodology can be applied, scaled appropriately to the available computational resources.
2. Materials and Methods
The proposed methodology integrates Case-Based Reasoning (CBR) with a matrix-based representation and rule-based filtering to predict diseases from symptoms. This combination offers a robust solution to the complexities of modern diagnostic systems by leveraging innovative matrix representations and measurement strategies. A Pertinence Matrix encodes the relationships between symptoms and diseases, while rule-based filtering refines the process by focusing on relevant cases. This approach ensures efficient handling of large datasets and high diagnostic accuracy, with dynamic updates to both the rules and the matrix for adaptability.
Figure 1 provides an overview of the steps in the proposed approach, each designed to enhance the accuracy and efficiency of disease diagnosis, particularly in complex datasets. The following subsections explain these steps with examples.
3. Results
To evaluate the effectiveness of the proposed diagnostic method, several key performance metrics were analyzed across multiple datasets. These metrics include accuracy, efficiency, and the ability to handle updates without full retraining.
3.2. Comparative Analysis with State-of-the-Art Methods
MARS was evaluated against a comprehensive range of classification approaches: traditional machine learning algorithms (Decision Tree, Random Forest, KNN, SVC), probabilistic Bayesian classifiers (Bernoulli Naive Bayes), and modern neural networks (a Multi-Layer Perceptron with two hidden layers containing 100 and 50 neurons, respectively, ReLU activation, and trained for 500 epochs). This comparison evaluates MARS against both optimization-based approaches (neural networks) and probability-based methods (Bayesian classifiers).
The primary performance metric is accuracy:
Table 4 presents the comprehensive results. All experiments were conducted on an Intel Core i9-10885H CPU.
The results highlight that MARS performs exceptionally well across all datasets. It achieves perfect accuracy (100%) on DS2 and DS3, and maintains very high performance on DS1 (99.22%) and DS4 (87.34%). These results demonstrate both robustness and adaptability across varying dataset sizes. Importantly, MARS also exhibits fast execution time for the largest dataset (DS4), confirming its scalability and practicality for large-scale diagnostic applications.
In comparison, SVC achieves comparable accuracy on DS4 (86.39%) but at a much higher computational cost. Specifically, SVC requires 4043 s (over an hour) for testing, nearly 2000× slower than MARS. This dramatic increase in processing time illustrates the scalability challenge faced by optimization-based models like SVC in large datasets, which may limit their suitability for real-time clinical use.
Among all compared approaches, neural networks are the most competitive with MARS in terms of both accuracy and efficiency. Nevertheless, MARS surpasses the neural model (87.34% vs. 85.55%) due to three key distinctions:
- i
Direct Calculation vs. Iterative Optimization: MARS constructs the Pertinence Matrix directly from symptom frequency data in a single pass (5 s for DS4), while neural networks require iterative training through multiple epochs (185 s). Moreover, MARS can incrementally update its matrix as new cases are introduced, unlike neural networks that typically require full retraining.
- ii
Explicit Rules vs. Distributed Representations: MARS produces interpretable diagnostic rules (e.g., ) that clinicians can easily validate, ensuring transparency in decision-making. Neural networks, by contrast, distribute learned information across numerous weight matrices, making their reasoning process largely opaque.
- iii
Set-Based Filtering vs. Layer-wise Transformation: MARS applies rule-based filtering, such as , to narrow down the set of possible diseases—reducing the search space in DS4 from 721 diseases to a very small subset—before computing cosine similarity (SDSM). In contrast, neural networks cannot perform such pre-filtering: the entire input is propagated through all hidden layers, and the network produces output scores for all 721 classes in the final classification layer.
Although Bayesian classifiers theoretically handle diagnostic uncertainty well, Bernoulli Naive Bayes reached only 85.49% accuracy, about 1.85 points below MARS. This is largely due to the independence assumption in Bayesian models, which rarely holds in medical contexts where symptoms are interdependent and often co-occur.
MARS’s geometric approach, leveraging cosine similarity, naturally captures these correlations via the Pertinence Matrix. When symptoms frequently appear together for a given disease, they form distinctive patterns within the disease vector, enhancing diagnostic precision.
While Decision Tree, Random Forest, and KNN achieve reasonable accuracy and fast execution times, they fall short of MARS on DS4. This suggests that although these algorithms are computationally efficient, they are less effective for large, complex diagnostic datasets.
These results validate MARS’s design: combining interpretable rule-based filtering with efficient matrix operations achieves accuracy competitive with black-box methods while maintaining transparency and computational efficiency.
3.3. Efficiency
The effectiveness of the proposed approach lies not only in its accuracy but also in the significant reduction of computational operations. This advantage is achieved through two distinct phases:
To better highlight this advantage, the following subsections provide a comprehensive breakdown of each phase.
4. Discussion
The results presented in this study illustrate the effectiveness and robustness of MARS (Matrix-Accelerated Reasoning System) in comparison to traditional algorithms across various datasets. MARS consistently outperforms or matches other approaches in terms of accuracy, particularly in large-scale and complex datasets. This is due to its ability to maintain high accuracy while significantly reducing the search space through rule-based filtering and matrix operations.
One of the most significant strengths of MARS is its ability to efficiently manage large datasets. The method excels in reducing an initial expansive search space to a more focused set, while ensuring high accuracy. This capability is crucial in medical diagnostics, where the rapid and accurate processing of extensive patient data can directly influence clinical decision-making and outcomes.
The four datasets vary widely in size, from 41 to 721 diseases and 132 to 400 symptoms, which naturally affects diagnostic accuracy. MARS achieves perfect accuracy (100%) on the smaller datasets (DS2 and DS3) and maintains high accuracy (87.34%) on the largest dataset (DS4). This slight decrease reflects the increased diagnostic complexity when more diseases and overlapping symptoms are present. Importantly, MARS’s 98.33% top-5 accuracy on DS4 demonstrates that the correct diagnosis consistently appears within a clinically manageable differential diagnosis list.
The comparison across different algorithms further underscores MARS’s superiority. In datasets where traditional methods such as Decision Trees and K-Nearest Neighbors (KNN) faced challenges, particularly in terms of accuracy, MARS consistently delivered high accuracy. When compared to modern approaches such as neural networks (85.55%), MARS achieved the highest accuracy (87.34%) on the most challenging dataset. This demonstrates MARS’s robustness in handling the inherent complexities of medical data, which often includes significant variability and noise.
Moreover, the results from dataset DS4 highlight MARS’s capacity to manage extreme cases involving large datasets. While traditional methods like the Support Vector Classifier (SVC) faced efficiency problems such as prolonged execution time (4043 s), MARS maintained high accuracy with substantially reduced processing time (2 s), further demonstrating its robustness and suitability for large-scale applications.
By maintaining high accuracy even in these challenging scenarios, MARS proves to be not only effective across a wide range of cases but also particularly suited for complex real-world medical datasets. This reliability ensures more accurate diagnoses, reducing the likelihood of errors and enhancing patient outcomes compared to traditional approaches.
Another key advantage of MARS is its flexibility in updating the dataset without requiring complete retraining. This is particularly valuable in dynamic medical environments, where new information and research findings are continually emerging. The ability to incorporate new data seamlessly through incremental Pertinence Matrix updates ensures that the diagnostic model remains up-to-date and accurate over time, which is a significant advantage over traditional models that require extensive retraining to integrate new data.
Finally, interpretability remains one of MARS’s defining strengths. The explicit rule generation (e.g., ) and transparent SDSM calculations allow clinicians to trace and validate diagnostic reasoning, a critical requirement for clinical trust and adoption. In contrast to neural networks with opaque weight distributions, MARS achieves both high accuracy and full transparency, demonstrating that interpretability and performance are not mutually exclusive in intelligent diagnostic systems.
5. Conclusions
This paper introduces MARS (Matrix-Accelerated Reasoning System), a novel diagnostic approach that seamlessly integrates matrix-based representations with rule-based methodology and advanced similarity measures, demonstrating its efficacy across diverse medical datasets. MARS excels in both accuracy and computational efficiency, consistently identifying the correct diagnosis within a significantly reduced search space across all tested datasets. The results highlight MARS’s scalability, effectively managing large-scale data while ensuring complete inclusion of the true prediction within the reduced set. This capability is particularly crucial in real-world medical applications, where both accuracy and efficiency are paramount.
The evaluation across four datasets with varying scales (41 to 721 diseases, 132 to 400 symptoms) demonstrates MARS’s robustness across different diagnostic complexities. While accuracy naturally decreases with problem scale (100% for 41-disease datasets vs. 87.34% for 721 diseases), MARS consistently outperforms all baseline methods at each scale. The 98.33% top-5 accuracy on the most complex dataset validates MARS’s capability to provide clinically useful differential diagnoses even in challenging scenarios.
One of the key strengths of this approach is its flexibility; MARS not only delivers precise results but also allows for dynamic updates to the system as new data becomes available, without necessitating complete retraining. This adaptability ensures that the system remains current and effective in rapidly evolving medical environments, addressing one of the major limitations of traditional diagnostic algorithms.
In comparative analyses, MARS consistently outperformed established algorithms such as Decision Trees, Random Forest, and K-Nearest Neighbors in terms of accuracy, particularly in complex, large-scale datasets. Furthermore, MARS demonstrated superior performance compared to modern approaches such as neural networks (85.55%), achieving 87.34% accuracy on the most challenging dataset. Its superior performance, even when faced with efficiency challenges like the prolonged execution times encountered by other algorithms such as SVC, underscores its robustness and practicality for real-world deployment.
In conclusion, MARS represents a significant advancement in the field of medical diagnostics, combining high accuracy with operational efficiency and adaptability. Its ability to handle extensive and complex datasets with remarkable performance makes it a powerful tool for enhancing diagnostic processes in healthcare, from well-equipped urban hospitals to resource-constrained rural clinics.
Future work should focus on identifying optimal similarity calculation methods and validating unknown disease detection through datasets with out-of-distribution cases to determine appropriate SDSM thresholds. These refinements will enhance MARS’s diagnostic accuracy and expand its applicability to broader medical conditions.