2. Materials and Methods
2.3. Data Preprocessing
In this study, we developed a preprocessing pipeline specifically tailored to the structure of ICU data and the clinical context of vancomycin use. All predictor variables were extracted based on the most recent available measurement prior to the first vancomycin dose, ensuring strict temporal validity for prospective risk modeling.
We applied different imputation strategies to continuous and categorical variables. For continuous variables, including laboratory and physiological measures such as
Phosphate and
Anion Gap, missing values were imputed using the median from the training set [
19]. This method is robust to outliers and skewed distributions, which are common in critical care laboratory data. For categorical variables such as
Richmond-RAS Scale,
Braden Mobility, and presence of an
Arterial Line, mode imputation was used to preserve the most frequent clinical state and reduce noise from documentation inconsistencies. To ensure data quality and avoid unreliable imputations, variables with more than 20% missingness were removed during preprocessing.
Because our study aimed to predict renal outcomes based on patient status at the time of drug initiation, we did not compute temporal summary statistics (e.g., max, min, or mean over 24 h). Instead, each variable was represented by a single value—the latest recorded measurement prior to vancomycin administration. This approach mirrors clinical practice and enhances the interpretability of predictions in real-time applications.
To ensure scale comparability across variables, min–max normalization was applied to all continuous variables. This transformation mapped raw values to the interval and prevented features with large numerical ranges, such as lactate or platelet count, from dominating learning algorithms. Binary variables, including indicator flags for devices, procedures, and comorbidities, were retained in their native 0/1 form to preserve clinical meaning and compatibility with tree-based and linear models.
The dataset exhibited a moderate class imbalance, with substantially fewer patients meeting the criteria for vancomycin-associated renal injury. To address this, we applied the Synthetic Minority Over-sampling Technique to the training folds during cross-validation [
20]. SMOTE generated synthetic minority-class samples by interpolating between existing observations, helping the model learn boundary regions more effectively while avoiding overfitting. Specifically, for a minority-class instance
and one of its
k-nearest neighbors
, a synthetic sample
is created using:
where
is a random scalar drawn from a uniform distribution. This oversampling step was restricted to the training data and excluded from test folds to ensure unbiased evaluation. We tested alternative imbalance-handling strategies, including class weighting and focal loss; however, these approaches produced less stable performance across validation folds, likely due to the small number of positive nephrotoxicity cases and resultant probability compression effects. SMOTE was therefore selected as the primary strategy because it generated the most consistent improvement in both AUROC while preserving physiologically plausible feature relationships by operating directly within the minority-class feature space.
All preprocessing steps—including imputation, normalization, encoding, and oversampling—were conducted independently on training data and subsequently applied to the corresponding validation sets. This design avoided information leakage and ensured that model evaluation reflected performance on unseen data. The resulting feature matrix provided a temporally aligned, numerically stable, and clinically interpretable basis for predictive modeling.
2.4. Feature Selection
We initially compiled a comprehensive set of candidate predictors from MIMIC-IV’s three primary clinical data tables, each representing distinct aspects of ICU patient monitoring and care. This systematic approach ensured comprehensive coverage of physiological systems associated with vancomycin-induced nephrotoxicity while maintaining clinical interpretability.
The chartevents table contains real-time physiological measurements and clinical assessments documented at the bedside, representing continuous patient monitoring data. This table captures vital signs, neurological assessments, and point-of-care laboratory values that reflect immediate patient status. From this domain, we extracted features including Richmond-RAS Scale, Total Bilirubin, Arterial Base Excess, AST, Braden Mobility, and Mean Airway Pressure, along with additional real-time observations such as Heart Rate, Non-Invasive Blood Pressure, and SpO2 that were initially considered. These variables represent the dynamic physiological state and acute illness severity that may predispose patients to vancomycin nephrotoxicity.
The
labevents table encompasses formal laboratory test results processed in hospital laboratories, providing biochemical markers of organ function and metabolic status. This systematic laboratory evaluation offers objective measures of renal function, electrolyte balance, coagulation status, and metabolic derangements. Key features from this domain included
Phosphate, Anion Gap, Magnesium, Lactate, PTT, Platelet Count, White Blood Cells, and
Glucose, as well as additional candidates like
BUN, INR, and
Calcium that were reviewed during initial screening [
21]. These laboratory parameters are critical for assessing baseline renal vulnerability and identifying patients at higher risk for drug-induced nephrotoxicity.
We initially compiled a comprehensive set of candidate predictors from MIMIC-IV’s three primary clinical data tables, with each table representing distinct aspects of ICU patient monitoring and care. This systematic approach ensured comprehensive coverage of physiological systems associated with creatinine elevation following vancomycin administration while maintaining clinical interpretability.
Real-time physiological measurements and clinical assessments documented at the bedside are contained within the chartevents table, which represents continuous patient monitoring data. Vital signs, neurological assessments, and point-of-care laboratory values that reflect immediate patient status are captured by this table. From this domain, we extracted features including Richmond-RAS Scale, Total Bilirubin, Arterial Base Excess, AST, Braden Mobility, and Mean Airway Pressure, along with additional real-time observations such as Heart Rate, Non-Invasive Blood Pressure, and SpO2 that were initially considered. The dynamic physiological state and acute illness severity that may be associated with creatinine elevation following vancomycin treatment are represented by these variables.
Formal laboratory test results processed in hospital laboratories are encompassed by the
labevents table, which provides biochemical markers of organ function and metabolic status. Objective measures of renal function, electrolyte balance, coagulation status, and metabolic derangements are offered through this systematic laboratory evaluation. Key features from this domain included
Phosphate, Anion Gap, Magnesium, Lactate, PTT, Platelet Count, White Blood Cells, and
Glucose, as well as additional candidates like
BUN, INR, and
Calcium that were reviewed during initial screening [
21]. Assessing baseline renal vulnerability and identifying patients at higher risk for creatinine elevation after vancomycin administration are critical functions of these laboratory parameters.
The
procedureevents table documents invasive procedures and interventions performed during ICU care, reflecting both illness severity and therapeutic intensity. Procedural data indicates the level of medical intervention required and serves as a proxy for critical illness severity. From this domain, we included features such as the presence of an
Arterial Line, central line insertion and
mechanical ventilation initiation [
22]. These interventional markers provide insights into hemodynamic monitoring needs, vascular access requirements, and respiratory support intensity, all of which correlate with nephrotoxicity risk in critically ill patients receiving vancomycin.
This clinically grounded three-domain approach reflects the fundamental pillars of ICU care—continuous physiologic monitoring, systematic biochemical evaluation, and therapeutic intervention intensity—enabling structured feature selection while preserving alignment with clinical decision-making processes for renal risk assessment.
Our feature selection employed a two-stage approach combining statistical filtering with machine learning-based importance ranking to identify the most predictive variables for vancomycin-associated nephrotoxicity while minimizing overfitting and maintaining clinical interpretability.
Stage 1—Statistical filtering (SelectKBest): The initial stage utilized univariate statistical testing to reduce dimensionality and eliminate non-informative features. We applied the F-statistic from ANOVA F-test via SelectKBest with f_classif scoring function to evaluate each feature’s individual association with nephrotoxicity outcome [
23]:
where
k represents the number of groups (nephrotoxicity vs. no nephrotoxicity),
is the sample size of group
i, and
N is the total sample size. This approach selected the top 30 features with the highest F-statistics, effectively filtering variables that demonstrated significant group differences in vancomycin-treated ICU patients.
Stage 2—Machine learning-based ranking (Random Forest importance): From the 30 statistically significant features, we applied Random Forest feature importance to identify the most predictive subset for nephrotoxicity risk [
24]. Random Forest importance quantifies each feature’s contribution to prediction accuracy by measuring the mean decrease in node impurity across all trees [
25]:
where
T is the number of trees,
represents nodes in tree
t that split on feature
,
is the proportion of samples reaching node
v, and
is the Gini impurity decrease from splitting on feature
at node
v. This method has proven effective in medical prediction tasks, particularly for identifying key risk factors in critical care settings [
26].
This two-stage approach selected the final 15 most important features, balancing statistical significance with predictive utility while reducing overfitting risk. The combination of univariate filtering and ensemble-based ranking ensures both statistical validity and clinical relevance for vancomycin nephrotoxicity prediction, following established practices in medical machine learning applications [
27].
The ordered importance of the selected variables is shown in the
Table 1.
In addition to the three clinical event categories, we incorporated four key admission-level characteristics that establish baseline patient risk profiles.
Age serves as a fundamental predictor of vancomycin nephrotoxicity due to reduced renal reserve and altered drug clearance in older patients [
28].
Emergency Department (ED) duration reflects clinical acuity and complexity prior to ICU admission, potentially indicating hemodynamic instability that may predispose to renal injury.
Charlson Comorbidity Index quantifies pre-existing chronic disease burden using admission diagnoses, providing standardized baseline health status assessment that influences nephrotoxicity susceptibility [
29].
Acute Physiology Score III (APSIII) measures acute illness severity during the first 24 h of ICU admission, capturing physiological derangement that correlates with organ dysfunction risk [
30]. These admission-level features complement dynamic clinical measurements by establishing foundational risk context, enabling the model to account for both baseline vulnerability and acute physiological changes.
Beyond the three clinical event categories, we incorporated four key admission-level characteristics that establish baseline patient risk profiles.
Age constitutes a fundamental predictor of creatinine elevation following vancomycin administration, given that reduced renal reserve and altered drug clearance occur in older patients [
28].
Emergency Department (ED) duration indicates clinical acuity and complexity prior to ICU admission, which may suggest hemodynamic instability that could be associated with renal injury.
Charlson Comorbidity Index quantifies pre-existing chronic disease burden based on admission diagnoses, thereby providing standardized baseline health status assessment that influences susceptibility to creatinine elevation [
29].
Acute Physiology Score III (APSIII) evaluates acute illness severity within the first 24 h of ICU admission, thus capturing physiological derangement that correlates with organ dysfunction risk [
30]. These admission-level features complement dynamic clinical measurements by establishing foundational risk context, which allows the model to account for both baseline vulnerability and acute physiological changes.
The final selected features are summarized in
Table 2.
2.5. Modeling
To predict vancomycin-associated renal injury in ICU patients, we developed a supervised machine learning framework incorporating six representative classification algorithms, each selected for its theoretical strengths, suitability for clinical data, and complementary modeling capabilities. The dataset was randomly split into stratified training (70%) and test (30%) sets to preserve outcome distribution. All model development—including hyperparameter tuning and performance validation—was conducted using five-fold stratified cross-validation within the training set to avoid information leakage and ensure generalizability.
We employed random stratified splitting instead of temporal splitting based on admission dates for medical and data science considerations. From a medical perspective, vancomycin nephrotoxicity mechanisms involve fundamental physiological processes—oxidative stress, tubular injury, and mitochondrial dysfunction—that remain biologically consistent over time, making temporal evolution less relevant than comprehensive patient representation. From a data science perspective, random splitting maximizes model performance by incorporating the most recent clinical practices and contemporary protocols in training, while temporal splitting would force the model to learn from outdated 2008–2014 data and exclude valuable 2015–2019 insights. Our approach ensures optimal clinical applicability for current deployment while maintaining statistical rigor through robust cross-validation techniques.
We prioritized CatBoost, LightGBM, and XGBoost as core modeling algorithms due to their proven effectiveness in handling structured clinical datasets. These tree-based ensemble methods leverage gradient boosting to sequentially improve prediction accuracy while capturing non-linear feature interactions and hierarchical patterns. CatBoost was particularly advantageous for its ordered boosting strategy and native support for categorical variables, reducing overfitting risks without extensive preprocessing. LightGBM offered efficiency through histogram-based binning and leaf-wise growth, which accelerated training on high-dimensional data. XGBoost provided granular control over regularization, tree complexity, and sampling strategies, allowing for flexible bias–variance trade-offs.
For interpretability, we included logistic regression with both L1 (lasso) and L2 (ridge) penalties to serve as a transparent baseline model. Its linear structure allowed for direct inspection of coefficient weights, enabling clinicians to interpret variable effects in a familiar framework. Regularization was applied to control multicollinearity and overfitting, with hyperparameters optimized via grid search.
A Gaussian Naïve Bayes classifier was also evaluated as a probabilistic baseline. This lightweight model assumes conditional independence among predictors and models continuous variables using parametric likelihoods. While simplistic, its efficiency and interpretability make it a useful benchmark for gauging the added value of more expressive algorithms.
Lastly, we implemented a shallow feedforward neural network to explore non-linear, high-capacity representations. The architecture included a single hidden layer with ReLU activation and a sigmoid output node. Training was performed using the Adam optimizer and binary cross-entropy loss, with hyperparameters such as learning rate, dropout ratio, and batch size selected via nested tuning. Although more data-intensive and less interpretable, neural networks offer flexible function approximation that may be valuable in larger-scale or multimodal extensions of this work.
Model performance was primarily evaluated using the area under the receiver operating characteristic curve (AUROC), which provides a threshold-independent measure of discrimination. Given the class imbalance in renal injury outcomes, AUROC served as a robust metric for comparing overall model performance. To quantify variability and assess stability, 95% confidence intervals for AUROC were calculated using 2000 bootstrap replicates.
In addition to AUROC, we reported a set of complementary metrics to ensure clinical applicability. Accuracy provided a general overview of classification performance, while the F1-score balanced precision and recall—important for minimizing both false positives and false negatives. Sensitivity and specificity were included to evaluate the model’s ability to detect high-risk cases and avoid overtreatment, respectively. Positive predictive value (PPV) and negative predictive value (NPV) were also calculated to aid in clinical interpretation of the model’s predictions. This comprehensive evaluation strategy ensured a balanced assessment of both statistical performance and real-world utility.
Together, these six models span a diverse spectrum of complexity, interpretability, and inductive bias, allowing for a comprehensive assessment of machine learning paradigms in predicting early renal complications related to vancomycin administration.
2.6. Statistical Analyses
To ensure the clinical relevance, interpretability, and methodological robustness of our vancomycin-associated renal outcome prediction framework, we implemented a suite of statistical analyses tailored to five key objectives: (1) to validate the comparability of training and test cohorts; (2) to quantify the marginal contribution of each feature through ablation; (3) to uncover non-linear or threshold effects using Accumulated Local Effects; (4) to interpret individual-level predictions via SHAP; and (5) to estimate predictive uncertainty through posterior sampling. These analyses were designed to support both the statistical credibility and bedside applicability of our model for predicting early renal injury and creatinine elevation after vancomycin exposure.
We first evaluated the statistical equivalence of the training and test sets using two-sided independent-sample
t-tests across core clinical variables [
31] including laboratory results and physiologic scores. Welch’s correction was applied when unequal variances were detected. The t-statistic was computed using:
This ensured that the stratified sampling strategy did not introduce distributional bias, thereby supporting valid model generalization and downstream inference. Clinically, it confirmed that both sets of patients were comparable at baseline so that observed differences in predicted renal outcomes could be attributed to model signals rather than sampling artifacts.
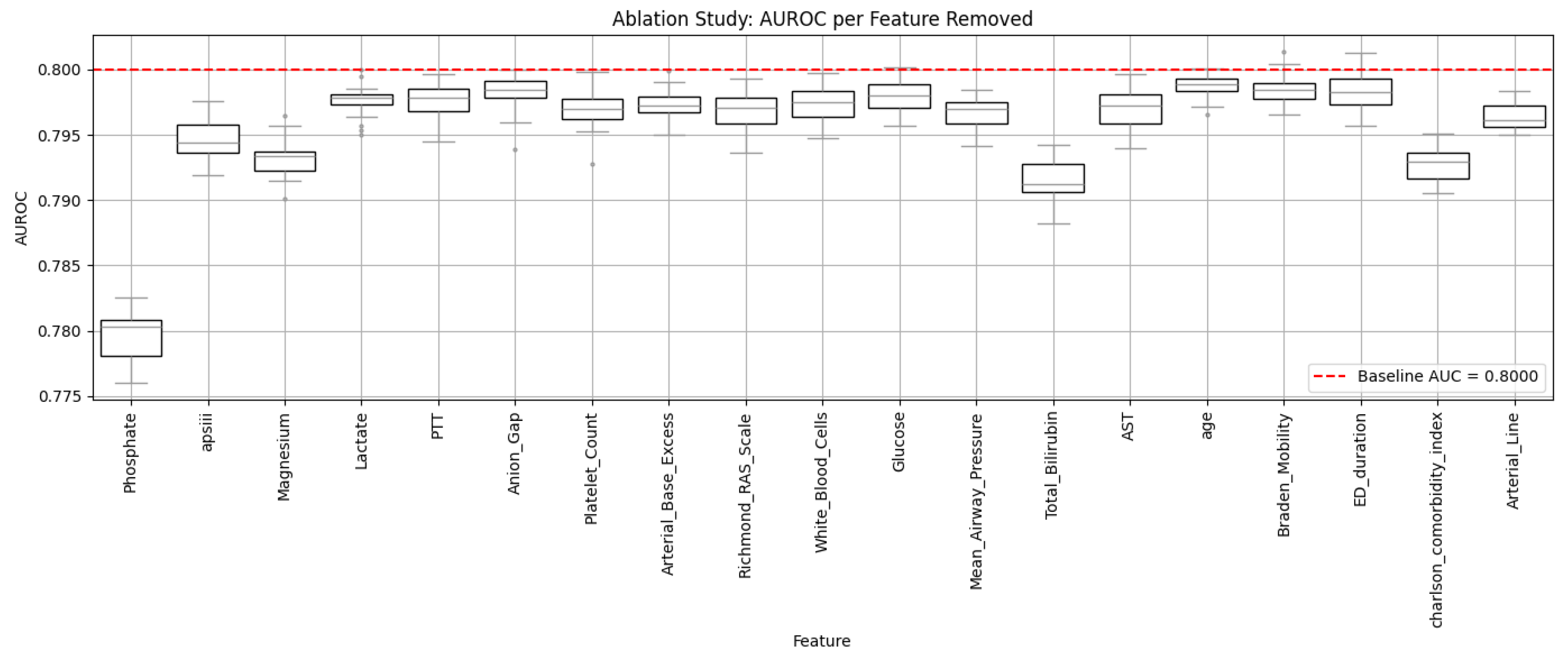
To assess the individual predictive utility of each feature, an ablation analysis was conducted by iteratively removing one variable at a time from the final model and retraining a logistic regression classifier [
32]. The impact of removal was measured by changes in AUROC, offering direct insight into each variable’s marginal contribution. This analysis helped highlight which physiologic and laboratory factors were most strongly associated with the risk of vancomycin-related renal impairment, operationalized as significant post-administration creatinine elevation.
Formally, the ablation effect
for a given feature
was computed using:
where
denotes the AUROC of the complete model including all features, and
denotes the AUROC after removing feature
. A larger
indicates greater marginal importance of the feature in predicting renal risk.
To characterize non-linear associations and clinically relevant thresholds, we applied Accumulated Local Effects for top-ranked continuous variables. ALE curves provide unbiased estimates of a feature’s local impact on model predictions while addressing multicollinearity. Formally, the ALE for a given feature
is defined as:
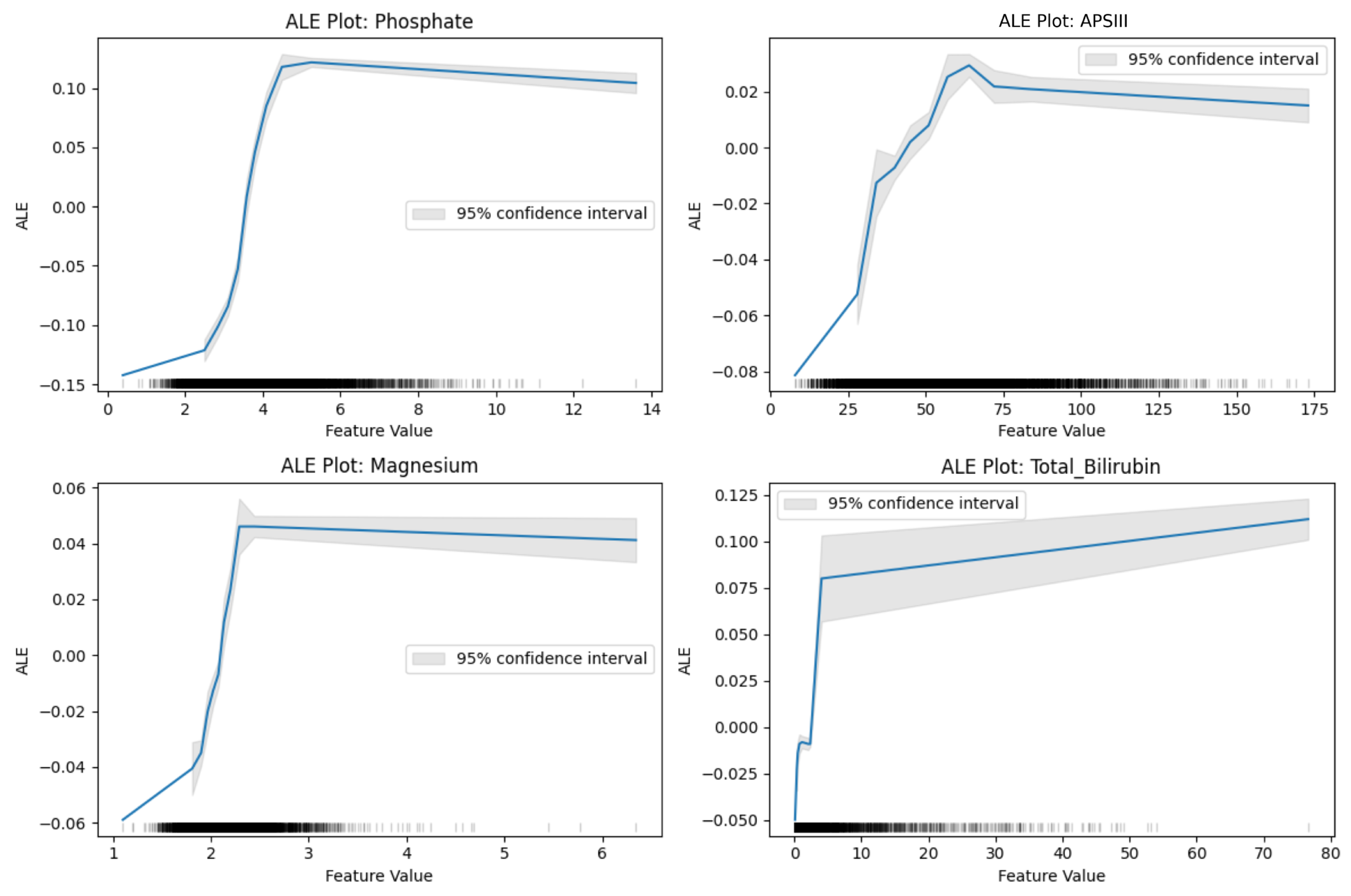
These visualizations revealed interpretable patterns—such as saturation effects in phosphate and U-shaped trends in magnesium—that aligned with known renal physiology, particularly in the context of drug-induced tubular stress and hemodynamic injury. These insights help clinicians define physiologic thresholds where risk escalates sharply, enabling more timely monitoring or adjustment of nephrotoxic therapies.
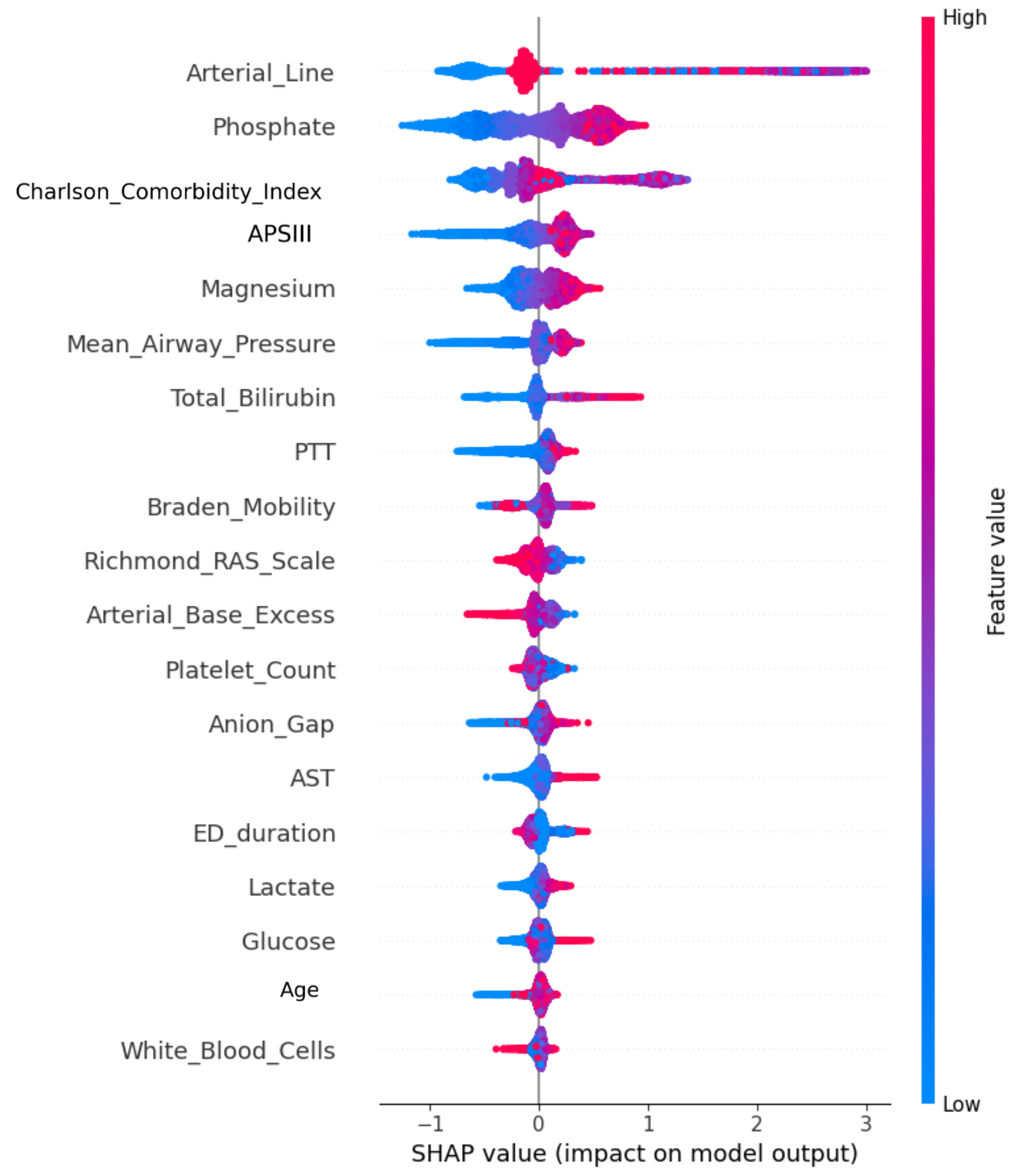
To enhance patient-level interpretability and clinical auditability, we employed SHAP to decompose predictions into additive feature contributions [
33]. Each SHAP value
represents the marginal contribution of a feature relative to all possible feature coalitions:
This allowed us to generate both global importance rankings and patient-specific explanation plots, improving model transparency and facilitating clinical trust. From a medical standpoint, SHAP enables clinicians to trace a patient’s predicted renal risk back to underlying contributing features—e.g., elevated lactate or low platelet count—offering rationale for interventions and supporting explainable AI in nephrotoxic drug management.
Finally, to quantify uncertainty in individual-level predictions, we implemented Bayesian posterior sampling using the DREAM algorithm. Unlike point estimates, this method generates a full posterior distribution for the predicted probability of vancomycin-associated creatinine elevation, allowing explicit quantification of prediction uncertainty. DREAM was incorporated as a distributional wrapper around CatBoost to capture the variability arising from model stochasticity and input perturbations, providing clinically meaningful credible intervals that help distinguish high-risk patients from those with uncertain or unstable predictions.
The posterior predictive distribution was estimated as:
where
N is the number of iterations per chain and
C is the number of parallel chains. Each
represents sampled model parameters from chain
i at iteration
j. This multi-chain, multi-iteration approach improves parameter space exploration and sampling robustness.
DREAM is particularly suited for clinical prediction due to its adaptive proposal mechanism and efficient sampling in complex, high-dimensional ICU data. Unlike basic MCMC, DREAM dynamically adjusts its sampling strategy, ensuring more reliable posterior estimation without model retraining.
In ICU practice, this uncertainty-aware prediction supports patient-specific decisions. For example, a high predicted risk with a narrow credible interval may prompt early intervention, while wide intervals may suggest close monitoring instead of immediate treatment changes. This enables more personalized and balanced renal risk management, especially for vancomycin-treated patients.
In summary, Bayesian posterior sampling with DREAM provides an efficient, interpretable way to communicate model uncertainty, enhancing the practical value of creatinine elevation risk prediction in real-time ICU settings.
Together, these statistical analyses strengthened the model’s validity, interpretability, and real-world relevance. They ensured that our framework is not only predictive but also transparent and clinically actionable for forecasting early creatinine elevation and renal risk in ICU patients receiving vancomycin.
3. Results
3.3. Model Performance on Creatinine
Elevation Risk Prediction
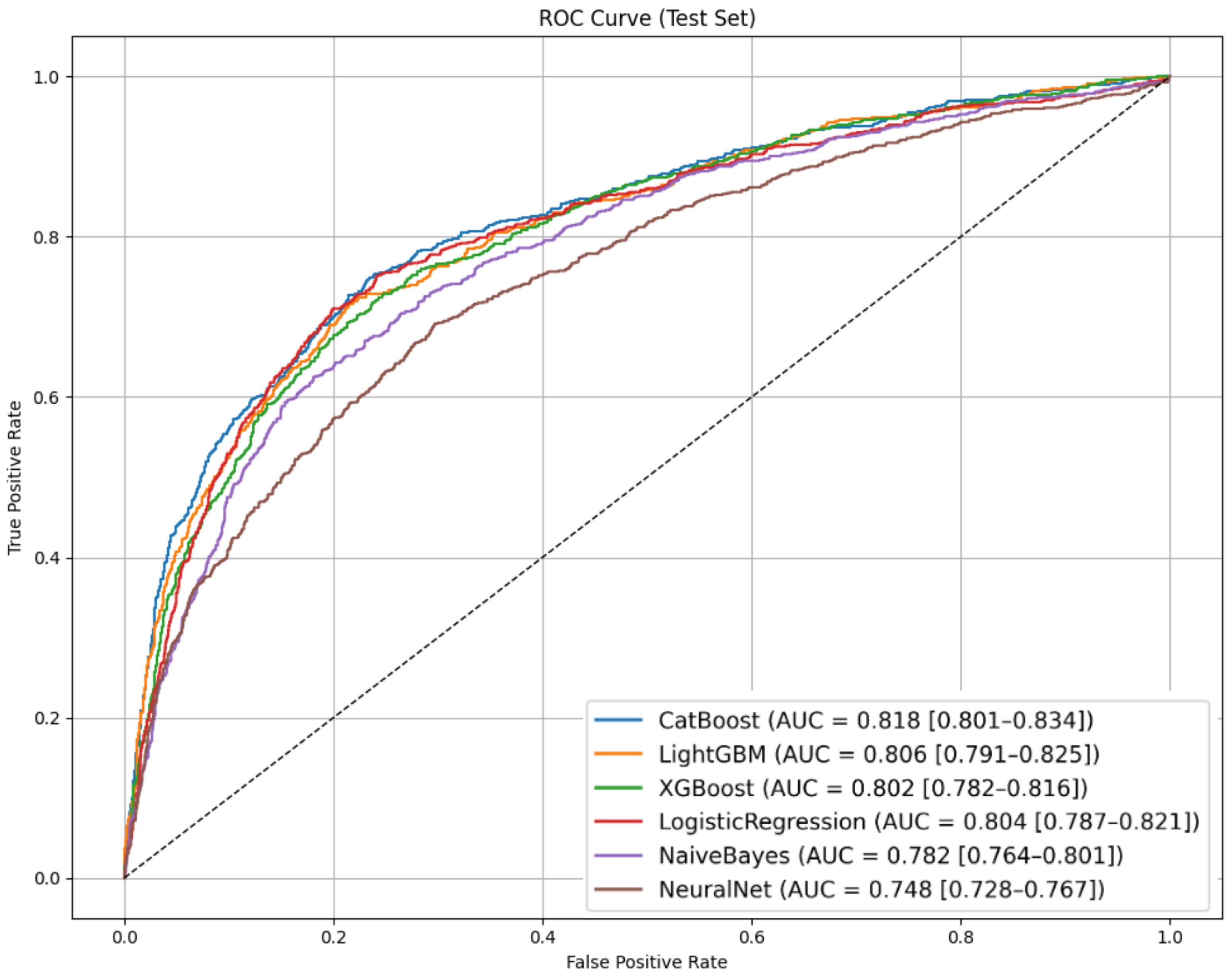
To evaluate the capacity of different algorithms to predict vancomycin-associated creatinine elevation among ICU patients, we tested six widely used machine learning models. Performance metrics on the test set—including AUROC, sensitivity, specificity, F1-score, and predictive values—are summarized in
Table 5. ROC curves for the test set are illustrated in
Figure 3.
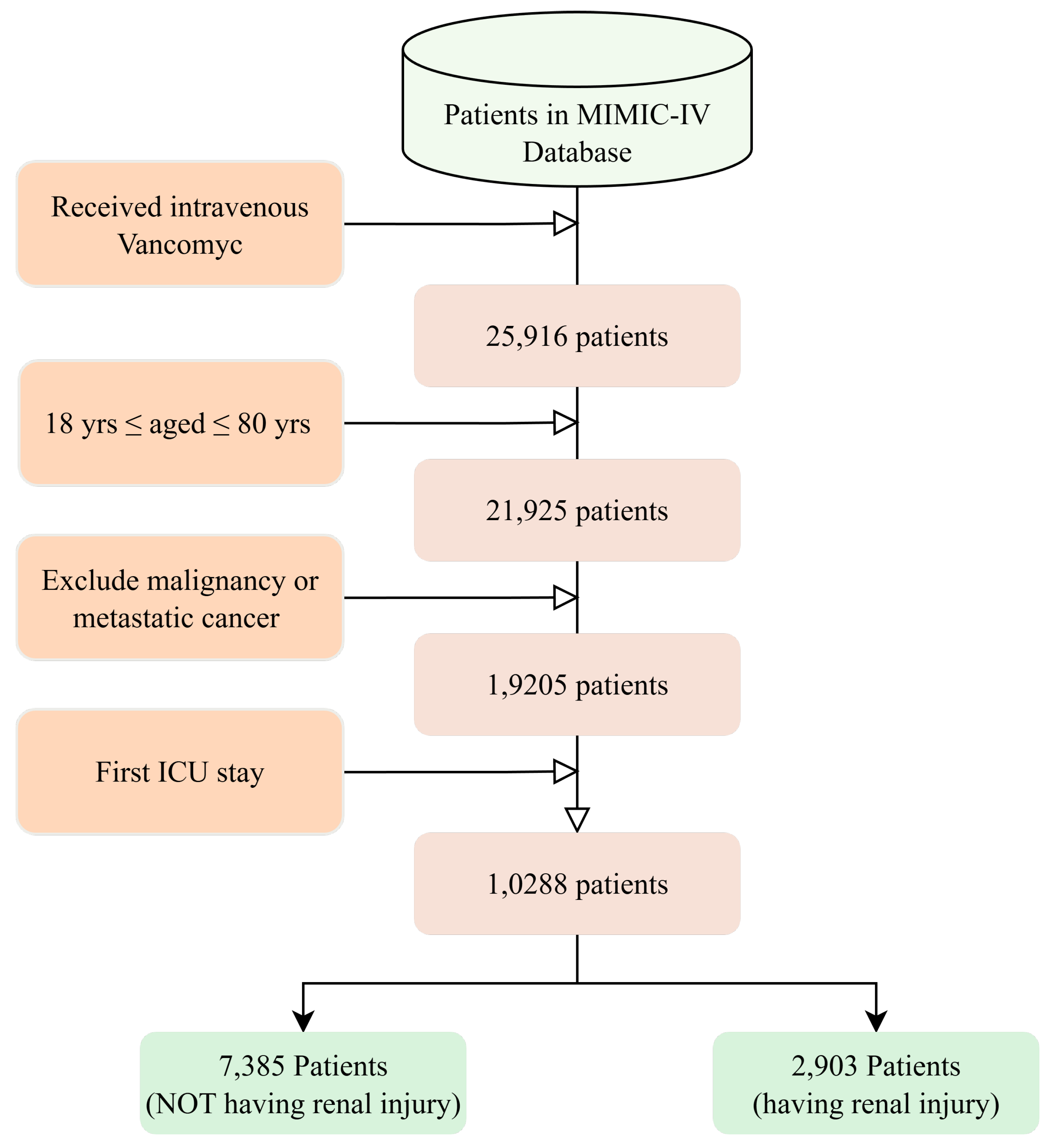
Our dataset exhibited moderate class imbalance with 28.2% nephrotoxicity cases (2903 of 10,288 patients). To ensure fair comparison across algorithms, sensitivity was manually fixed at 0.800 for all models, allowing direct evaluation of specificity and precision trade-offs at this clinically relevant threshold.
Among the models evaluated, CatBoost achieved the highest AUROC of 0.818 (95% CI: 0.801–0.834), indicating strong discriminatory ability. It also delivered the best overall accuracy (0.714), highest F1-score (0.605), and maintained a solid specificity of 0.681 at the fixed sensitivity threshold, ensuring the model captures most high-risk patients without overwhelming clinicians with false alarms. Critically, CatBoost’s performance demonstrates genuine predictive skill beyond baseline rates: its PPV of 0.486 represents a 72% improvement over the 0.282 baseline prevalence, while the NPV of 0.900 substantially exceeds the 0.718 rate a naive “always safe” classifier would achieve. This translates to correctly identifying an additional 182 low-risk patients per 1000 predictions beyond chance alone, confirming clinically meaningful risk stratification that substantially exceeds baseline performance expectations.
From a clinical perspective, this level of performance is particularly valuable in real-world ICU settings where vancomycin is commonly used to treat severe infections but carries well-documented nephrotoxic potential. Even a modest rise in creatinine can signal the early stages of renal injury in ICU patients, where rapid clinical deterioration is possible. In practice, this model can support timely risk stratification by identifying patients who may benefit from intensified renal monitoring, dose adjustment, or consideration of alternative therapies. For example, correctly flagging 80% of future creatinine elevation cases while still safely ruling out nearly 70% of low-risk patients provides actionable guidance that can directly inform bedside decisions.
The high NPV (0.900) is particularly valuable for clinical decision-making, enabling two complementary treatment strategies: confidently continuing vancomycin therapy in patients predicted as low-risk while implementing enhanced monitoring protocols for high-risk patients. This dual approach helps avoid both unnecessary treatment interruptions in safe patients and delayed intervention in vulnerable patients, optimizing antimicrobial stewardship without compromising patient safety.
CatBoost was ultimately selected as the final model not only for its superior test performance but also for its robustness to missing data and its ability to handle heterogeneous ICU feature sets. Its interpretability is strengthened by its reliance on physiologically meaningful predictors, such as phosphate, bilirubin, and comorbidity burden, which are strongly associated with renal stress mechanisms linked to vancomycin use. This ensures that the model’s outputs are transparent and clinically intuitive, even for readers without a background in machine learning.
In summary, by prioritizing high sensitivity and carefully balancing specificity, this framework provides a practical, explainable tool to support early detection of vancomycin-associated creatinine elevation and renal injury risk in ICU patients. It offers meaningful, real-time clinical support without compromising interpretability or safety.
3.6. Posterior Distribution and Prediction of Vancomycin-Related Creatinine Elevation
To incorporate uncertainty into the prediction of vancomycin-associated creatinine elevation among ICU patients, we applied the DREAM algorithm to the trained CatBoost model. Unlike traditional deterministic predictions, this approach generates a posterior distribution over the predicted probability, allowing clinicians to assess not only the most likely risk estimate but also the associated confidence range for individual patients. This is particularly valuable in ICU settings, where misjudging renal risk can lead to either delayed intervention or unnecessary treatment changes.
In this study, we set the number of iterations to 2000 and used 38 parallel chains, following the recommended practice of employing at least twice the number of model parameters (19 variables) to ensure sufficient posterior exploration. This configuration allows the DREAM algorithm to efficiently traverse the parameter space, reduce the risk of local convergence, and produce stable, reliable posterior distributions. Adequate iterations and chains are essential to capture uncertainty accurately, particularly in high-stakes ICU risk prediction tasks.
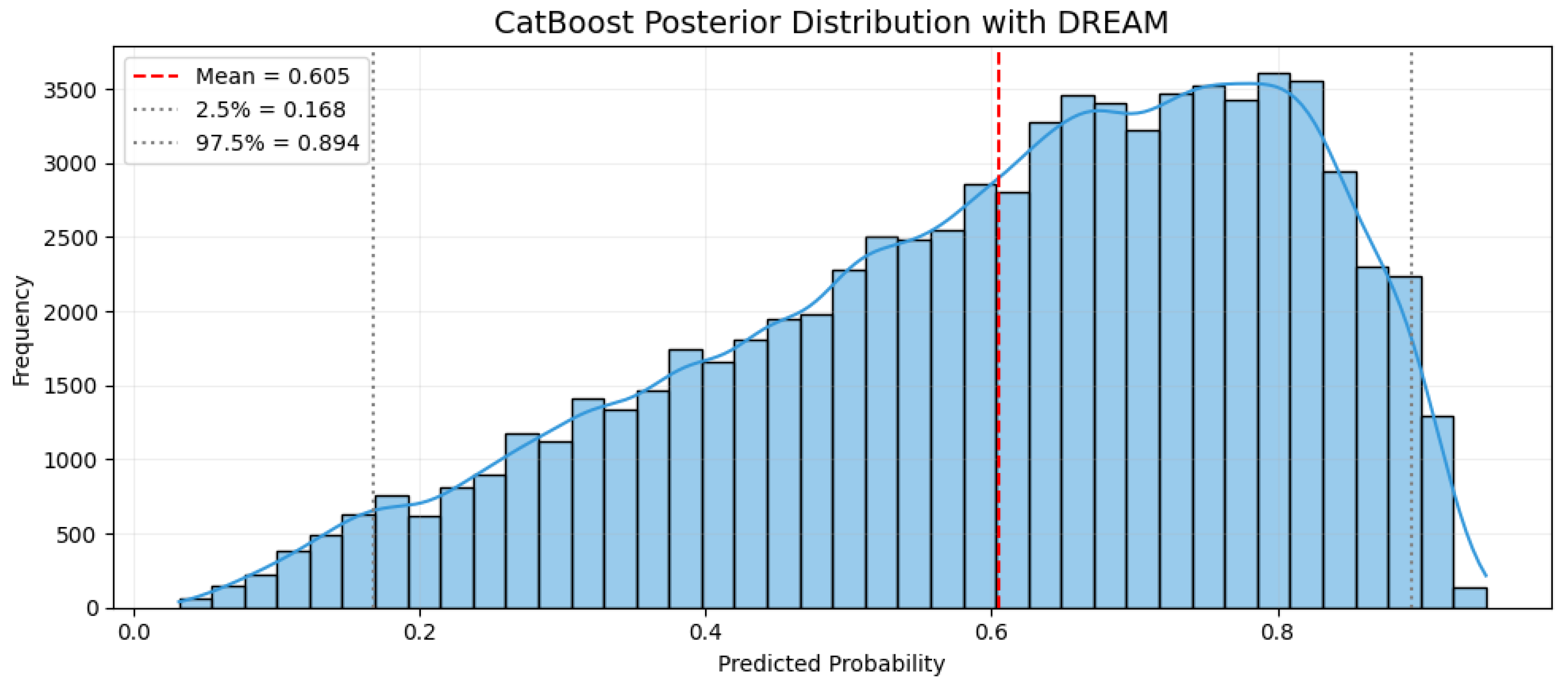
As shown in
Figure 6, the posterior distribution for a representative high-risk patient is skewed toward higher predicted probabilities, with a mean risk of 60.5% and a 95% credible interval ranging from 16.8% to 89.4%. In comparison to the overall cohort creatinine elevation rate of 28.22%, this patient demonstrates substantially increased risk. The wide credible interval highlights clinical uncertainty, indicating that while the model identifies this patient as high-risk, variability in clinical features could significantly influence the actual outcome.
The high-risk patient profile used in the posterior simulation was constructed based on the creatinine elevation subgroup described in
Table 4, which includes elevated phosphate, bilirubin, magnesium, and higher Charlson comorbidity index scores. This ensures that the sampling process is grounded in real-world ICU scenarios and reflects clinically plausible risk patterns for vancomycin-associated renal injury, rather than relying on theoretical or averaged inputs.
This probabilistic framework provides meaningful clinical nuance. Two patients may have similar mean predicted risks but differ in the width of their credible intervals—one presenting with high certainty and another with considerable uncertainty. For patients with wide intervals, clinicians may choose to prioritize enhanced monitoring over immediate medication adjustments, recognizing the potential variability in risk trajectories.
Importantly, DREAM operates without retraining the CatBoost model. It conditions posterior sampling on prior distributions derived from observed creatinine elevation cases, enabling computationally efficient uncertainty quantification that can be applied in real-time. This makes the approach practical for bedside decision support.
In summary, integrating uncertainty-aware prediction with CatBoost and DREAM offers a clinically interpretable and statistically robust framework to assess the risk of creatinine elevation during vancomycin treatment in ICU patients. This provides a clinical decision-support tool rather than establishing causal conclusions, enhancing physicians’ ability to incorporate uncertainty estimates into their clinical judgment when managing nephrotoxic therapy and optimizing renal risk surveillance.
4. Discussion
4.3. Limitations and Future Works
This study has several important limitations. First, our retrospective design using MIMIC-IV data from a single academic medical center may limit generalizability to other ICU populations with different patient demographics, clinical protocols, or resource constraints. The temporal span (2008–2019) may not reflect current clinical practices, potentially affecting model performance in contemporary settings.
Second, our nephrotoxicity definition relies solely on serum creatinine elevation, which is a delayed marker that may miss subclinical renal injury or cases where elevation is masked by clinical factors. The absence of more sensitive biomarkers (NGAL, cystatin C) or functional assessments limits our ability to detect early nephrotoxicity.
Third, methodological constraints include potential imputation bias from missing data patterns, unmeasured confounding factors (genetic predisposition, subclinical kidney disease, concomitant nephrotoxic medications such as piperacillin-tazobactam), and feature selection limitations that may have excluded clinically relevant variables such as detailed vancomycin pharmacokinetics or hemodynamic parameters.
Fourth, while our CatBoost model achieved strong performance, its ensemble nature creates interpretability challenges for clinical adoption. The evaluation focused primarily on discrimination metrics without extensive exploration of calibration or clinical utility measures, and performance thresholds were chosen arbitrarily rather than through clinical consensus.
Fifth, comprehensive model calibration assessment using metrics such as Brier score, reliability plots, and Expected Calibration Error was not performed, which would provide additional insights into the model’s predictive reliability across different probability ranges.
Moreover, the present study extracted only the most recent measurement prior to the first vancomycin administration and did not incorporate longitudinal laboratory or hemodynamic trajectories (e.g., creatinine slope, electrolyte variability, or hypotension duration). These temporal dynamics may contain clinically meaningful signals for early kidney stress, but incorporating them would require a fundamentally different modeling framework—such as sequence-based or time-series architectures—beyond the scope of the current single-time-point design. Future work will integrate longitudinal feature engineering to capture dynamic trends that may further enhance predictive performance.
Finally, clinical implementation faces significant barriers including workflow integration challenges, regulatory validation requirements, and potential algorithmic bias across demographic groups. Our model requires real-time access to multiple data streams that may not be consistently available in all clinical settings.
Future work should prioritize multi-center external validation across diverse healthcare systems to establish broader generalizability. Prospective randomized controlled trials comparing model-guided versus standard monitoring strategies are essential to demonstrate clinical utility and cost-effectiveness using meaningful endpoints such as time to nephrotoxicity detection and clinical outcomes.
Methodological enhancements should include integration of advanced biomarkers when available, development of longitudinal time-series models to capture dynamic risk evolution, and incorporation of detailed vancomycin pharmacokinetic data for more mechanistically informed predictions. Advanced analytical approaches should employ causal inference techniques, systematic fairness evaluation across demographic subgroups, and federated learning approaches for collaborative model development while preserving data privacy.
Implementation research should investigate human-AI collaboration patterns, conduct comprehensive health economic evaluations, and examine organizational barriers to clinical adoption. The development of bias mitigation strategies and systematic evaluation of model performance across diverse patient populations will be crucial for ensuring equitable clinical application.
Future success depends on addressing these limitations through rigorous validation studies, enhanced methodological approaches, and careful attention to clinical implementation challenges. Collaborative efforts among data scientists, clinicians, regulatory bodies, and healthcare systems will be essential to ensure these tools meaningfully improve patient outcomes while maintaining safety and equity in clinical care.