- Review
Artificial Intelligence in Corneal Drug Delivery Systems
- Amirhosein Panjipour,
- Soheil Sojdeh and
- Ali R. Djalilian
- + 1 author
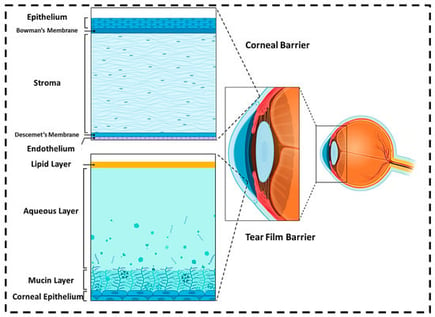
Conventional topical therapy for corneal and anterior segment diseases is limited by rapid tear clearance and multilayer corneal barriers, resulting in low bioavailability and the need for frequent dosing. Artificial intelligence (AI) is emerging as a complementary approach that learns quantitative relationships between molecular structure, formulation variables, and ocular performance. In corneal drug delivery, machine learning models have been used to optimize multicomponent formulations and processing conditions; predict key quality attributes such as particle size, zeta potential, encapsulation efficiency and release kinetics; and estimate corneal permeability, retention and ocular irritation risk, thereby reducing experimental burden and guiding safer design. AI can also be coupled with mechanistic ocular pharmacokinetic/pharmacodynamic models to translate formulation attributes into predicted tissue exposure. Finally, inverse design approaches enable the discovery of new carriers and devices, illustrated by machine learning-guided peptide carriers and smart contact lens platforms that combine sensing with on-demand drug release. Despite these advances, current datasets remain small and heterogeneous, external validation and benchmarking against conventional workflows are limited, and uncertainty quantification and interpretability must be addressed to enable clinical translation. This review summarizes corneal barriers and delivery platforms, critically evaluates where AI provides measurable value across design, characterization and performance and highlights data and validation priorities needed for trustworthy AI-enabled corneal therapeutics.
27 February 2026