- Article
Lightweight Depression Detection Using 3D Facial Landmark Pseudo-Images and CNN-LSTM on DAIC-WOZ and E-DAIC
- Achraf Jallaglag,
- My Abdelouahed Sabri and
- Abdellah Aarab
- + 1 author
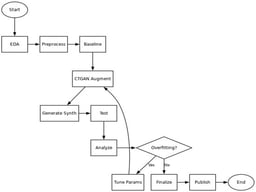
Background: Depression is a common mental disorder, and early and objective diagnosis of depression is challenging. New advances in deep learning show promise for processing audio and video content when screening for depression. Nevertheless, the majority of current methods rely on raw video processing or multimodal pipelines, which are computationally costly and challenging to understand and create privacy issues, restricting their use in actual clinical settings. Methods: Based solely on spatiotemporal 3D face landmark representations, we describe a unique, totally visual, and lightweight deep learning approach to overcome these constraints. In this paper we introduce, for the first time, a pure visual deep learning framework, based on spatiotemporal 3D facial landmarks extracted from clinical interview videos contained in the DAIC-WOZ and Extended DAIC-WOZ (E-DAIC) datasets. Our method does not use raw video or any type of semi-automated multimodal fusion. Whereas raw video streaming can be computationally expensive and is not well suited to investigating specific variables, we first take a temporal series of 3D landmarks, convert them to pseudo-images (224 × 224 × 3), and then use them within a CNN-LSTM framework. Importantly, CNN-LSTM provides the ability to analyze the spatial configuration and temporal dimensions of facial behavior. Results: The experimental results indicate macro-average F1 scores of 0.74 on DAIC-WOZ and 0.762 on E-DAIC, demonstrating robust performance under heavy class imbalances, with a variability of ±0.03 across folds. Conclusion: These results indicate that landmark-based spatiotemporal modeling represents the future of lightweight, interpretable, and scalable automatic depression detection. Second, our results suggest exciting opportunities for completely embedding ADI systems within the framework of real-world MHA.
4 February 2026