Abstract
Recent findings have sparked great interest in the putative magnetic receptor protein MagR. However, in vivo experiments have revealed no magnetic moment of MagR at room temperature. Nevertheless, the interaction of MagR and MagR fusion proteins with silica-coated magnetite beads have proven useful for protein purification. In this study, we recombinantly produced two different MagR proteins in Escherichia coli BL21(DE3) to (1) expand earlier protein purification studies, (2) test if MagR can magnetize whole E. coli cells once it is expressed to a high cytosolic, soluble titer, and (3) investigate the MagR-expressing E. coli cells’ magnetic properties at low temperatures. Our results show that MagR induces no measurable, permanent magnetic moment in cells at low temperatures, indicating no usability for cell magnetization. Furthermore, we show the limited usability for magnetic bead-based protein purification, thus closing the current knowledge gap between theoretical considerations and empirical data on the MagR protein.
1. Introduction
Iron–sulfur (Fe–S) cluster proteins are important for numerous physiological processes and are present in most known prokaryotic and eukaryotic cells [1,2,3]. The iron atoms in [2Fe–2S] clusters have been reported to interact through antiferromagnetic coupling [4]. Only recently, the Fe–S cluster protein MagR (magnetic receptor) came into spotlight [5]. The authors proposed a possible answer to the question on navigation of migratory animals. They reported that MagR, a small (~14 kDa) [2Fe–2S] protein from pigeons with homologs in numerous species, forms a ferrimagnetic, multimeric complex that responds to magnetic fields in vitro. Qin et al. also showed that the MagR protein and a MagR/Cryptochrome complex can be isolated and enriched from a complex matrix by silica-coated magnetite (SiO2–Fe3O4) beads [5]. Later, MagR fusion proteins were successfully captured from a complex matrix [6,7].
Since its discovery, the physical capabilities of MagR have been intensively questioned. When MagR constructs were subjected to magnetic stimuli in mammalian cells, they were not able to induce significant membrane channel activity in a magnetic field [8], in contrast to previous results [9]. The biologist Markus Meister considered the plausibility of magnetic sensing of MagR by calculations based on simple physical principles [10]. He found the number of iron atoms in the postulated assembly of MagR proteins [5] to be too low to even sense magnetic fields sufficiently [10]. Then, Winklhofer and Mouritsen argued that the weak exchange interactions among [2Fe–2S] clusters of adjacent proteins may only lead to spontaneous magnetization only below a few Kelvin, but not around room temperature [11]. Interestingly, one recent theory states that radical pairs might enable sensing of magnetic fields through induction of magnetic fluctuation in the MagR structure rather than permanent magnetism [12].
Until now, the magnetic behavior of MagR has not been tested at low temperatures, which could give clearer indications on a potential magnetic behavior. Additionally, the stated usability of MagR fusion proteins for protein capture with magnetic beads [6,7] requires further characterization and comparison to state-of-the-art affinity downstream processing methods to reveal potential drawbacks or benefits. In this study, we deepened the investigation on MagR in two different aspects. First, we analyzed magnetic bead capture using recombinant MagR from the pigeon Columbia livia (clMagR) and MagR from Drosophila melanogaster (dMagR) [5,6,7]. Secondly, we tested if highly expressed MagR (>15% total intracellular soluble protein) would yield a magnetic moment in Escherichia coli cells at different temperatures to investigate if MagR expression would be sufficient to magnetize cells in vivo for diverse applications [13]. Our results close the existing knowledge gap between theoretical considerations [10,11,12] and empirical data [6,7,8,9] on the magnetic characteristics and the usability of MagR.
2. Results
2.1. Evaluation of MagR Capture from a Complex Matrix
Overexpression of hexa-histidine-tagged (his-tag) dMagR and clMagR in E. coli was clearly visible with bands around 14 kDa in SDS-PAGE analysis (Figure 1a). Despite codon optimization, clMagR-his was mainly produced as insoluble inclusion bodies and could not be further investigated (Figure 1a). Binding studies with dMagR-his on SiO2-Fe3O4 beads showed that the protein was enriched from E. coli lysates. However, many host-cell proteins also adsorbed nonspecifically to the beads (Figure 1a). When we compared the efficiency of the magnetic bead capture with a state-of-the-art IMAC capture, we found that the IMAC capture was much more specific, and SDS-PAGE indicated a product with higher purity (Figure 1b). High absorption of dMagR-his at 320 nm clearly indicated the presence of Fe–S clusters in the protein. Binding studies with dMagR without his-tag underlined that protein binding occurred also without his-tag on beads, but again with many host-cell protein impurities (Supplementary Figure S1). To shed more light on the binding conditions of MagR on beads, we performed binding studies with IMAC-purified dMagR-his in different buffers. We found that dMagR-his bound to magnetic beads between pH 5–11 in the presence of up to 2 M NaCl or 1 M (NH4)2SO4 (Supplementary Figure S2). Binding was only hindered at pH 12. Based on these results, we hypothesize very strong ionic interactions to be the reason for MagR binding, rather than specific magnetic interactions.
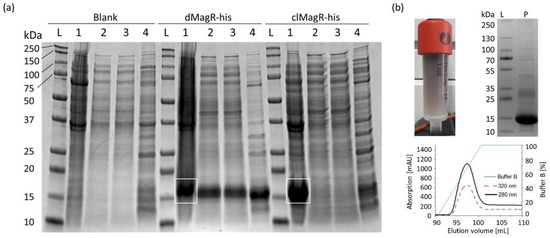
Figure 1.
Evaluation of MagR purification from a complex matrix. (a) SDS-PAGE analysis of magnetic bead purification of Blank, dMagR-his and clMagR-his from cell disruption supernatant. White rectangles show respective target proteins in the applied cell pellet. Equivalent volumes of sample were applied for each respective lane (1–4). The following samples per lane are seen for each target sample: lane L: protein ladder; lane 1 (3 µL): solubilized cell pellet; lane 2 (10 µL): cell-free supernatant after cell disruption; lane 3 (10 µL): supernatant after magnetite bead precipitation; lane 4 (6 µL): bead-precipitated proteins after washing of beads. (b) Purification of dMagR-his and clMagR-his by IMAC. Coloration of IMAC column of dMagR-his is shown together with IMAC elution profile and the respective SDS-PAGE analysis of the elution pool. SDS-PAGE shows the standard protein ladder in lane L and 10 µL of the respective IMAC elution pool in lane P. Elution profiles of IMAC show absorption at 280 nm (black line), absorption at 320 nm for iron–sulfur cluster proteins (red dashed line) and relative concentration of elution buffer B (blue line).
2.2. Potential of MagR to Magnetize Bacterial Cells
For magnetization studies, we overexpressed the Fe–S protein dMagR without his-tag to approximately 17% of total soluble protein in E. coli (Figure 2a and Figure S3). This high intracellular content was also visible as a black–brown coloration of BL21-dMagR cell biomass and its supernatant after cell disruption (Figure 2b). Quantification by SDS-PAGE densitometry (non-MagR impurities at around 14 kDa were excluded based on a respective negative control) yielded an approximate intracellular, soluble dMagR concentration of 54 mg g−1 dry cell weight (DCW) or 5.12 pg cell−1 (1 cell~9.5·× 10−13 g DCW [14]) equivalent to 2.20·× 106 dMagR molecules cell−1. However, placing a strong neodymium magnet (50 × 50 × 12.5 mm) near the BL21-dMagR biomass suspension at room temperature resulted in no observable movement of cells towards the magnet.
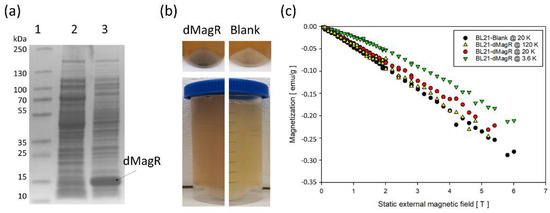
Figure 2.
Potential of dMagR to magnetize bacterial cells. (a) SDS-PAGE of cell-free supernatants after cell disruption, with a clear band around 14 kDa for dMagR in Lane 3. Lane 1: Ladder; Lane 2: Supernatant of BL21-Blank; Lane 3: Supernatant of BL21-dMagR. (b) Upper part compares biomass coloration of BL21-dMagR and BL21-Blank cells. Lower part compares coloration of cell free supernatant after cell disruption. BL21-dMagR biomass and cell free supernatant show clear brownish coloration due to dMagR. (c) Isothermal, magnetic field-dependent SQUID magnetometry measurements of lyophilized BL21-Blank and BL21-dMagR at 3.6, 20 and 120 K. Conversion of emu g−1 = Am2 kg−1.
We further analyzed magnetization behavior with lyophilized cells by superconducting quantum interference device (SQUID) magnetometry. Based on the vague knowledge about MagR and its applicability in cells to interact with magnetic fields at ambient conditions [8,9], we hypothesized that measurements at low temperatures of only 3.6, 20 and 120 K would give a clearer indication on a potential applicability in cells. That is due to the known temperature-dependent magnetic susceptibility of magnetic materials. The field-dependent isothermal magnetization measurements revealed a dominant diamagnetic response of BL21-Blank and BL21-dMagR cells in a static external magnetic field (emu/g = electromagnetic unit per gram DCW; emu = 10−3 Am2; emu g−1 = Am2 kg−1) (Figure 2c). The comparison of 20 K isothermal magnetization data of BL21-dMagR with corresponding BL21-Blank data revealed a rather small additional paramagnetic contribution of the former, which likely results from dMagR-bound iron in BL21-dMagR cells. As expected, this paramagnetic contribution increases with decreasing temperature, as found for BL21-dMagR cells at 3.6 K (Figure 2c). However, our results clearly show that overexpression of intracellular dMagR does not exhibit a sufficiently strong magnetic contribution to overcome the diamagnetic character of the E. coli cell, even at only 3.6 K.
3. Discussion
In 2016, the proposal of a magnetic biocompass in animals for geomagnetic sensing sparked great interest in MagR [5]. However, this study was heavily criticized [10,11] and in vivo experiments attributed no clear picture on a potential permanent magnetic moment to MagR at ambient conditions [8,9]. Interestingly, some authors published successful capture of MagR and MagR fusion proteins with magnetic SiO2–Fe3O4 beads [5,6,7].
In this study, we recombinantly produced two different MagR proteins, namely clMagR and dMagR, in E. coli BL21(DE3) to further investigate capture with magnetic beads and to measure the magnetization of dMagR-overexpressing E. coli cells at very low temperatures. As we used a similar recombinant production strategy as in the recent studies by Wang et al. and Jiang et al. [6,7], we hypothesize that the recombinant MagR protein in our study was also correctly folded and that the Fe–S clusters were properly assembled in the protein.
Although stated in the initial study on MagR [5], we could not verify that his-tagged MagR from the pigeon C. livia (clMagR-his) can be sufficiently expressed recombinantly in its soluble form in E.coli, despite codon optimization. This is likely a result of its theoretical isoelectric point of pH 6.82 (Table 1), which is near the cytosolic pH of E. coli [15]. However, recombinant expression of soluble his-tagged MagR from D. melanogaster (dMagR-his) was successful. Our binding studies on magnetic beads and the comparison to a state-of-the-art IMAC capture step revealed limitations of using dMagR for magnetic protein separation. Binding of dMagR to magnetic beads was not quantitative, and many E. coli host-cell proteins adsorbed nonspecifically. Due to binding experiments in different buffers and results from SQUID magnetometry, we hypothesize that dMagR binds to SiO2–Fe3O4 beads rather through strong ionic interactions than magnetism. Concerning this nonmagnetic interaction of dMagR and beads, it is known that some proteins can naturally adsorb to silica surfaces through charge interactions [16]. The strong binding of dMagR under different conditions revealed yet another limitation of this protein for capture with beads, namely its elution. Successful elution of a binding partner from a dMagR fusion is dependent on an enzymatic cleavage step, such as thrombin, as shown before [6]. Therefore, we do not see a benefit in using dMagR compared to already existing cleavable fusion tags for affinity chromatography. Additionally, our SQUID magnetometry results provide sufficient data to show that overexpression of soluble dMagR to ~17% of total soluble protein in E. coli yields no spontaneous magnetization of dMagR, and therefore no usable magnetization of cells even at only 3.6 K. This underlines that dMagR and likely also its variants from different species are not suitable for studies that require substantial sensing of static external magnetic fields, which is in agreement with previous conclusions drawn from theoretical calculations [10].

Table 1.
Overview of protein specific data.
4. Materials and Methods
4.1. Chemicals
Chemicals were purchased from Carl Roth (Vienna, Austria), if not stated otherwise. MiniPrep was performed with the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Vilnius, Lithuania).
4.2. Putative Magnetic Proteins
Two magnetic receptor proteins (MagR) were used in this study, comparable to the study of Jiang et al. [6]. MagR from the pigeon (Columbia livia, clMagR) and from the fly (Drosophila melanogaster, dMagR) were examined. Both proteins are [2Fe–2S] iron–sulfur cluster proteins and structural homologs of the mammalian Fe–S Cluster Assembly 1 (ISCA1) [18] and bacterial IscA [19]. The protein-specific characteristics are compared in Table 1. Amino acid and nucleotide sequences of the target proteins are accessible in the Supplementary Information.
4.3. Bacterial Strains, Plasmids and Cloning
Both MagR genes, dMagR and clMagR, were ordered codon-optimized with and without C-terminal his-tag in a pET-21a(+) plasmid with an ampicillin resistance gene (AmpR) from GenScript (Nanjing, China) (Supplementary Information). A pET-21d(+) plasmid with the gene for pyranose 2-oxidase (P2Ox) was used as negative control [20]. The respective plasmids were heat-shock transformed into chemically competent E. coli BL21(DE3) cells from New England BioLabs. Clones were picked from Luria-Bertani (LB) medium plates with carbenicillin (50 µg mL−1). Constructs were verified by sequencing (Microsynth GmbH, Vienna, Austria). Cryogenic cultures were prepared for each strain (Table 2) from LB-carbenicillin shake-flask cultivations at 37 °C and stored in 25% (v/v) glycerol at −80 °C.

Table 2.
Overview of recombinant E. coli strains used in this study.
4.4. Expression of Target Proteins
BL21-dMagR-his, BL21-clMagR-his, BL21-dMagR and BL21-Blank strains were cultivated in 2.5 L ultra-yield shake flasks (Thomson Instrument Company, Oceanside, Cleveland, OH, USA) in 0.5 L of LB-carbenicillin medium with 1 g L−1 ferric citrate H2O at 37 °C at 230 rpm to an optical density at 600 nm (OD600) around 0.8. Then, the temperature was set to 20 °C and isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 20 µM. Cells were cultivated for 24 h and then harvested by centrifugation. The resulting biomass was stored at −20 °C.
4.5. Capture of Target Proteins by Magnetite Beads
Frozen biomass of BL21-dMagR-his, BL21-clMagR-his, BL21-dMagR and BL21-Blank was resuspended with a T10 basic ULTRA-TURRAX® (IKA, Staufen, Germany) in TBS buffer (20 mM TRIS; 150 mM NaCl, pH 7.5) to around 10 g L−1 dry cell weight (DCW). Cell disruption was performed with an ultrasonication lance (SONOPULS HD 2070.2 with MS 73 probe from Bandelin, Berlin, Germany). The lance was placed into the respective ice-cooled cell suspension and cell disruption was performed at 75% power output with an alternating sequence (20 s pulse; 40 s pause) for 15 min. The resulting suspension was centrifuged (15,650× g, 4 °C, 20 min) and the supernatant was used for binding experiments. Plain SiO2–Fe3O4 (sicastar®-M) nanoparticles with a mean diameter of 350 nm were used (micromod Partikeltechnologie GmbH; Rostock; Germany). A total of 50 µL of a 25 mg mL−1 iron oxide bead suspension was mixed with 1.4 mL of cell-free supernatant in a polypropylene tube. The suspension was incubated at room temperature under light agitation for 30 min. Then, the beads were centrifuged at 500× g for 1 min to remove nonadsorptive proteins before they were washed six times with 0.5 mL TBS buffer. Afterwards, the precipitated beads were resuspended with 30 µL TBS buffer. The collected supernatant and the final bead suspension were mixed with 4× denaturation buffer (1 L contained 250 mL 1 M TRIS; pH 6.8, 100 g SDS, 500 mL pure glycerol, 2.5 g bromophenol blue and 5 mL 2-mercaptoethanol) for analysis by SDS-PAGE.
Additional testing of different binding conditions was performed with 150 µL IMAC purified 0.75 g L−1 dMagR-his in various buffers: (a) 20 mM TRIS, 150 mM NaCl, pH 7.5; (b) 20 mM citric acid, 150 mM NaCl, pH 5.0; (c) 20 mM TRIS, 150 mM NaCl, pH 9.0; (d) 20 mM KH2PO4, 150 mM NaCl, pH 11.0; (e) 20 mM KH2PO4, 150 mM NaCl, pH 12.0 150 mM; (f) 20 mM TRIS, pH 7.5; (g) 20 mM TRIS, 1 M NaCl, pH 7.5; (h) 20 mM TRIS, 2 M NaCl, pH 7.5; (i) 20 mM TRIS, 1 M (NH4)2SO4, pH 7.5. The pH was set with 1 M HCl or 1 M NaOH, respectively.
4.6. Purification of Target Proteins by Immobilized Metal Affinity Chromatography
Frozen biomass of BL21-dMagR-his was resuspended with a T10 basic ULTRA-TURRAX® (IKA, Staufen, Germany) in IMAC buffer A (100 mM TRIS; 500 mM NaCl; 20 mM Imidazole; pH 7.4) to 20 g L−1 DCW. Cell disruption was performed with an ultrasonication lance as described above. The resulting suspension was centrifuged (15,650× g, 4 °C, 20 min) and the supernatant was used for immobilized metal affinity chromatography (IMAC). Therefore, an ÄKTA pure (GE Healthcare, Tiefenbach, Austria) equipped with a 1 mL HisTrapFF IMAC column (GE Healthcare, Austria) was used. The column was equilibrated with 10 column volumes (CV) IMAC buffer A. Afterwards, the column was loaded with cell-free supernatant. Elution was performed with a linear gradient to 100% IMAC buffer B (100 mM TRIS; 500 mM NaCl; 500 mM Imidazole; pH 7.4) over 10 CV with a flow rate of 156 cm h−1. Absorption was measured at 280 nm and 320 nm. The respective elution peak was analyzed by SDS-PAGE.
4.7. Expression of dMagR and Preparation for Cell Magnetization Measurement
BL21-Blank and BL21-dMagR cells were cultivated in 2.5 L ultra-yield shake flasks (Thomson Instrument Company, Oceanside, Cleveland, OH, USA), as described above. Then, the broth was transferred to a glass beaker and a strong neodymium magnet (N35 50 × 50 × 12.5 mm; Webcraft GmbH, Gottmadingen, Germany) was used to test for magnetic attraction of suspended cells at room temperature. Afterwards, cells were centrifuged and washed twice with deionized water to remove salts and were prepared for lyophilization by washing them twice with 15% sucrose in deionized water. The final cell pellet was mixed with 5 mL deionized water, transferred to a 50 mL polypropylene tube and sucrose was added as lyoprotectant to yield an approximate 15% (w/w) sucrose to biomass ratio. Only clean plastic or glass materials were used to avoid metallic contaminations. The suspension was then poured into a prechilled glass beaker and frozen in liquid N2. Lyophilization of cells was conducted with a Labconco™ Freezone™ 2.5 L table freeze-dryer (FisherScientific GmbH, Vienna, Austria) for 24 h and cells were stored at −80 °C until further use. Intracellular protein concentration was measured after cell disruption by high-pressure homogenization (HPH) of a lyophilized BL21-dMagR biomass aliquot resuspended to 12 g L−1 DCW in TBS buffer. HPH was performed with a PandaPLUS 2000 (GEA Mechanical Equipment, Parma, Italy). The biomass was processed at 1400 bar for 3 passages under cooling. After centrifugation (15,650× g, 4 °C, 20 min), the supernatant was used for total, intracellular soluble protein analysis by SDS-PAGE (Supplementary Figure S3). The dMagR content per cell was calculated through densitometry by the following steps with the Image Lab 6.0.1 Software (Bio-Rad Laboratories, Vienna, Austria): (1) An equine cytochrome c protein standard row was applied to SDS-PAGE. (2) The band volumes of the respective bands around 14 kDa were extracted from BL21-Blank and BL21-dMagR. (3) The volumes of the bands were correlated to the standard row to yield the approximate concentration of proteins with a size around 14 kDa. (4) Results of BL21-Blank were subtracted from BL21-dMagR to yield the dMagR concentration; (5) Based on the disrupted DCW mass and applied sample volume, a ratio of dMagR and biomass was calculated. (6) Further, the dMagR mass per cell was evaluated (1 g DCW = 1.05·× 1012 E. coli cells (1 cell = 9.5·× 10−13 g from BNID 103904 [13])).
4.8. Magnetization Measurements of Whole Cells
Magnetization measurements were performed with a superconducting quantum interference device (SQUID) magnetometer that measures the total dipolar magnetic moment of a sample, including all atomic and molecular magnetic contributions. Measurements were performed on a commercial 6 T SQUID magnetometer (CRYOGENIC Limited, London, UK). All samples were prepared in the same manner. The respective lyophilized biomass–sucrose sample was weighed with a plastic spatula in a polycarbonate capsule (Quantum Design, Darmstadt, Germany) and placed in a straw sample holder. When we subtracted the sucrose content, a dry cell mass of 12 mg was used for each measurement, which represented 1.26 × 1010 cells. Sample powder was held in place by filling the remaining space of the capsule with cotton. Therefore, the total sample in the SQUID magnetometer consisted of a lyophilized biomass–sucrose powder, cotton, a capsule, a straw and a small amount of vacuum grease to hold the capsule in place inside the straw. Field dependent measurements were conducted under isothermal conditions at 3.6, 20 and 120 K up to 6 T.
4.9. Gel Electrophoresis
All gels were run with 1× SDS-PAGE running buffer (3 g L−1 TRIS, 14 g L−1 glycine, 1 g L−1 SDS). All samples were prepared with 4× denaturation buffer and heated at 95 °C for 10 min before use. We either used 4–15% or 8–16% gradient gels (Mini-PROTEAN TGX Stain-Free Gels; Bio-Rad Laboratories, Vienna, Austria), which were run at 160 V for 40 min. The Precision Plus Protein Dual Color protein ladder standard (Bio-Rad Laboratories, Vienna, Austria), the PageRuler Plus Prestained protein ladder (Bio-Rad Laboratories, Vienna, Austria) and Precision Plus Protein All Blue Prestained Protein ladder (Bio-Rad Laboratories, Vienna, Austria) were used. Gels were visualized with Molecular Imager®Gel Doc™ XR System (Bio-Rad Laboratories, Vienna, Austria) and analyzed with the Image Lab 6.0.1 Software (Bio-Rad Laboratories, Vienna, Austria).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/magnetochemistry7110147/s1, The data that support the findings of this study are available in the supplementary material and from the corresponding author upon reasonable request. Figure S1: SDS-PAGE analysis to test dMagR capture with beads., Figure S2: SDS-PAGE analysis of dMagR-his capture with magnetic beads under different conditions., Figure S3: SDS-PAGE analysis for quantification of dMagR.
Author Contributions
A.P. wrote the manuscript, planned and conducted the experiments. H.M. supervised and helped in conducting the SQUID experiments including raw data transformation and interpretation. O.S. supervised the study and corrected the manuscript draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Konhauser, K.O.; Kappler, A.; Roden, E.E. IRON IN MICROBIAL METABOLISMS. Elements 2011, 7, 89–93. [Google Scholar] [CrossRef]
- Lill, R.; Hoffmann, B.; Molik, S.; Pierik, A.J.; Rietzschel, N.; Stehling, O.; Uzarska, M.A.; Webert, H.; Wilbrecht, C.; Mühlenhoff, U. The role of mitochondria in cellular iron–sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1823, 1491–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, V.D.; Lill, R. Biogenesis of cytosolic and nuclear iron–sulfur proteins and their role in genome stability. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1853, 1528–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandelia, M.-E.; Lanz, N.; Booker, S.J.; Krebs, C. Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1853, 1395–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, S.; Yin, H.; Yang, C.; Dou, Y.; Liu, Z.; Zhang, P.; Yu, H.; Huang, Y.; Feng, J.; Hao, J.; et al. A magnetic protein biocompass. Nat. Mater. 2016, 15, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, L.; Wang, F.; Zhang, J.; Liu, G.; Gao, B.; Wei, D. Novel Application of Magnetic Protein: Convenient One-Step Purification and Immobilization of Proteins. Sci. Rep. 2017, 7, 13329. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xu, H.; Liu, Z.; Sun, T.; Yuan, C.; Yang, Y.; Guo, J.; Xie, H. Magnetic immobilization of a quorum sensing signal hydrolase, AiiA. Microbiologyopen 2019, 8, e00797. [Google Scholar] [CrossRef] [Green Version]
- Pang, K.; You, H.; Chen, Y.; Chu, P.; Hu, M.; Shen, J.; Guo, W.; Xie, C.; Lu, B. MagR Alone Is Insufficient to Confer Cellular Calcium Responses to Magnetic Stimulation. Front. Neural Circuits 2017, 11, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, X.; Ye, J.; Zhao, D.; Zhang, S.-J. Magnetogenetics: Remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Sci. Bull. 2015, 60, 2107–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, M. Physical limits to magnetogenetics. eLife 2016, 5, e17210. [Google Scholar] [CrossRef] [PubMed]
- Winklhofer, M.; Mouritsen, H. A room-temperature ferrimagnet made of metallo-proteins? bioRxiv 2016, 8, e094607. [Google Scholar]
- Xiao, D.-W.; Hu, W.-H.; Cai, Y.; Zhao, N. Magnetic Noise Enabled Biocompass. Phys. Rev. Lett. 2020, 124, 128101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pekarsky, A.; Spadiut, O. Intrinsically Magnetic Cells: A Review on Their Natural Occurrence and Synthetic Generation. Front. Bioeng. Biotechnol. 2020, 8, 573183. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010, 38, D750–D753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilks, J.C.; Slonczewski, J.L. pH of the Cytoplasm and Periplasm of Escherichia coli: Rapid Measurement by Green Fluorescent Protein Fluorimetry. J. Bacteriol. 2007, 189, 5601–5607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moerz, S.T.; Huber, P. pH-Dependent Selective Protein Adsorption into Mesoporous Silica. J. Phys. Chem. C 2015, 119, 27072–27079. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Beilschmidt, L.K.; De Choudens, S.O.; Fournier, M.; Sanakis, I.; Hograindleur, M.-A.; Clémancey, M.; Blondin, G.; Schmucker, S.; Eisenmann, A.; Weiss, A.; et al. ISCA1 is essential for mitochondrial Fe4S4 biogenesis In Vivo. Nat. Commun. 2017, 8, 15124. [Google Scholar] [CrossRef] [PubMed]
- Ollagnier-de-Choudens, S.; Mattioli, T.; Takahashi, Y.; Fontecave, M. Iron-sulfur cluster assembly: Characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem. 2001, 276, 22604–22607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spadiut, O.; Leitner, C.; Salaheddin, C.; Varga, B.; Vertessy, B.G.; Tan, T.-C.; Divne, C.; Haltrich, D. Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design. FEBS J. 2009, 276, 776–792. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).