Abstract
The natural fermentation of antibiotics, along with semi-synthetic and synthetic approaches, is one of the most important methods for their production. The majority of the antibiotic market comes from the fermentation of high-yielding (HY) fungal strains. These strains have been obtained since the 1950s from wild-type (WT) isolates as a result of classical strain improvement (CSI) programs primarily involving multi-round random mutagenesis and screening. However, the molecular basis leading to high-yield production was unknown. In recent years, due to the application of multiomic approaches, key changes that occur in CSI programs, with WT strains that become HY industrial producers of a particular antibiotic, have begun to be understood. It becomes obvious that, during CSI, certain universal events are selected, which lead both to a direct increase in the production of the target metabolite and affect other vital processes of the cell (side mutations). These key events include: the upregulation of the target biosynthetic gene cluster (BGC), changes in the system of global regulation, disruption of alternative BGCs, the rearrangement of energy fluxes in favor of the target SM (secondary metabolite), changes in the regulation of the response to stress, and the redirection of primary metabolic pathways to obtain more precursors for target production. This knowledge opens up the possibility of both introducing targeted changes using genetic engineering methods when creating new producers and increasing the production of CSI strains as a result of fermentation with low-molecular compounds, targeted to compensate for the effects of side mutations.
1. Introduction
The discovery of antibiotics and their widespread introduction into medical practice has radically changed the life of mankind, since they made it possible to save human lives in situations that were previously hopeless for patients [1]. Since the 1900s, numerous attempts have been made to create drugs that satisfy Paul Ehrlich’s “magic bullet” (Zauberkugel, ger.) concept, where a compound selectively kills disease-causing microorganisms without harming the human body [2,3]. Ehrlich carried over the term “magic bullet” from Carl Maria von Weber’s popular 1821 opera Der Freischütz (The Freeshooter), which is based on the German legend of magic bullets that fly not according to the laws of ballistics, but according to the will of the shooter himself. The shooter buys seven magic bullets from the devil in exchange for a soul. For the first six bullets, he chooses the target himself, and the last one is controlled by the devil. Ehrlich obtained the first magic bullets, the anti-syphilis arsenical-based drugs salvarsan (arsphenamine, compound 606) and its less toxic analogue neosalvarsan (compound 914), against the spirochete Treponema pallidum by chemical synthesis [4,5] (Figure 1). Although these first organic antimicrobials (the first antibiotics), salvarsan and neosalvarsan, were commercialized in the early 1910s (by Hoechst AG), their structural formulas were only established a hundred years later, and the mechanism of action is still not clear [1,6]. The concept of the “magic bullet” formulated by Ehrlich and the introduction of the first chemically synthesized antimicrobial drugs into medical practice not only ushered in the era of chemotherapy, but also became a key event that marked the beginning of the evolution of antibiotics [4]. For 30 years, many laboratories around the world actively searched for similar magic bullets, until in the 1940s arsenical-based drugs were replaced as treatments for syphilis by less toxic penicillins, which were obtained as a result of natural fermentation of the fungus Penicillium chrysogenum (later reclassified as Penicillium rubens) [7]. Since then, the fermentation of natural fungal and bacterial strains has become, along with chemical synthesis, the most important industrial method for obtaining antibiotics [8].
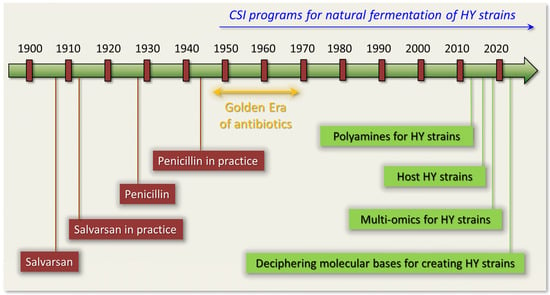
Figure 1.
Antibiotic Era timeline focused on classical strain improvement (CSI) programs for high-yielding (HY) antibiotic producers.
The revolution in the field of antimicrobials occurred after the realization that, among the numerous synthesized low molecular weight metabolites, some molds (filamentous fungi) and bacteria can produce compounds that effectively destroy other microorganisms [9,10,11]. These so-called natural products, or secondary metabolites (SMs), play an important ecological role, enabling a particular microorganism to occupy a certain ecological niche by inhibiting the growth of other microorganisms [12,13,14]. Since such compounds are directed against microorganisms that differ significantly at the molecular level from humans, some of them are able to selectively kill pathogenic microbes [15,16]. In other words, nature itself creates magic bullets, casting them in the furnaces of the evolution of microbiological communities [17,18,19,20,21]. The main task in the so-called Golden Age of antibiotics (the period from the beginning of the 1940s to the end of the 1960s) was treasure hunting, the search for magic bullets created by nature: natural antibiotics [1,22] (Figure 1). The first and most important discovery of such a bullet was the discovery by Fleming in 1928 of penicillin, which began to be widely introduced into pharmaceutical practice from 1942 [7].
It turned out that, for the production of antibiotics, special pathways of secondary metabolism are involved, using non-ribosomal peptide synthesis, polyketide synthases, terpene cyclases, cytochromes P450s, and many other biosynthetic enzymes [23,24,25,26,27,28,29]. The final highly active product is obtained as a result of several tens (and even hundreds) of stereospecific reactions [30,31,32]. After deciphering the structures of such biologically active compounds, synthetic chemists developed numerous schemes for their chemical (or, so called, total) synthesis [33,34]. The total synthesis of antibiotics and other bioactive natural products, according to Kuniaki Tatsuta, one of the pioneers in this field, “is a display of beauty, achieving a status of ‘art’ in organic chemistry” [35]. Therefore, the main source for obtaining the discovered pharmaceutically significant SMs was the fermentation of microbial strains, since total synthesis proved too complicated or too expensive [36,37]. Along with this, for the production of some antibiotics, schemes of multi-stage chemical synthesis (or total synthesis) were developed, leading to a yield comparable in economic costs [38]. For example, the broad-spectrum antibiotic chloramphenicol, isolated from Streptomyces venezuelae in 1947, is produced in the industry by total synthesis [39,40].
In natural isolates, the yield of the required antibiotics was not sufficient for industrial production and did not exceed several tens of milligrams per liter [7,41,42,43]. Therefore, on the basis of wild-type (WT) strains, improved high-yielding (HY) producers were obtained, in which the yield of the target SM was increased by 100–1000 or more times [44,45,46,47,48,49]. As a result of the widespread introduction of antibiotics discovered during the Golden Era, many pathogenic strains began to develop resistance, which was expressed in a significant or complete loss of sensitivity to antibiotics of many classes [50,51]. This ultimately led to the end of the Golden Age of antibiotics in the late 1960s–early 1970s [52,53].
One of the main methods of combating microbial resistance, since the end of the Golden Era of antibiotics, is the modification of compounds that have become inactive, which makes it possible to overcome resistance [36]. For example, in the UK in 1961, the so-called methicillin-resistant Staphylococcus aureus (MRSA), resistant both directly to methicillin and to other beta-lactam antibiotics, including penicillins, cephalosporins, and carbapenems, was first recorded. The mechanism of resistance is due to the presence of the mecA gene, encoding penicillin-binding protein 2a (PBP2a), which, unlike other penicillin-binding proteins, does not bind to beta-lactams. Introduced in the early 2010s, ceftaroline (Teflaro®, Teva Pharmaceuticals, Tel Aviv, Israel is sold in USA; and Zinforo®, AstraZeneca, Cambridge, UK is sold in Europe), the 5th generation cephalosporin, has become active against MRSA strains because it binds and inhibits the PBP2a enzyme [54]. To create modified natural products, both total chemical synthesis and in vitro modifications of compounds obtained after natural fermentation (semisynthetic synthesis) are used [55,56,57,58]. As a result, antibiotics of several generations appear within the same class. For example, five generations of cephalosporins, four generations of aminoglycosides, four generations of fluoroquinolones and so on are currently on the market [59,60,61,62]. Such a struggle between humanity, which creates new generations of antibiotics that overcome resistance, and microorganisms, which become resistant to newer and newer generations of antibiotics, is reminiscent of the arms race of the Cold War period of the second half of the 20th century [63,64].
In the following decades, after the end of the Golden Era, until the early 2000s, the number of new classes of antibiotics that were introduced into medical practice dropped sharply [1]. This period is characterized as an innovative gap in the development of antibiotics [65]. Apparently, this was due to the fact that all the magic bullets lying on the surface were found during the Golden Era using the methods available at that time, and new technologies were required to create novel antibiotics in the 21st century [66,67,68]. At the turn of the 21st century, breakthroughs in the field of genetic engineering made it possible to understand the molecular basis of antibiotic production, associated with so-called biosynthetic gene clusters (BGCs) [24]. These discoveries, combined with new technologies in recent decades, made it possible to close the innovation gap in the development of antibiotics, and new classes of antibiotics began to enter the market [61,69,70,71,72]. Along with this, there is now a situation in which the discovery and development of antibiotics is no longer cost-effective when using traditional cost recovery models [73]. This leads to a reduction in the number of companies and laboratories dedicated to delivering new antibiotics, which, against the backdrop of a constant increase in pathogen resistance to the antibiotics used, could lead to catastrophic consequences in the future [73]. However, the antibiotics currently available play an enormous role in protecting human health, and the amount of antibiotics consumed is steadily increasing [74].
In this review, we will focus on the natural fermentation of improved microbial strains for obtaining antibiotics and the potential use of magic bullets, not for selective action to kill a pathogen, but for a selective increase in the yield of the synthesized antibiotic. Such a type of magic bullet can be introduced, because, when working with natural antibiotics, not only are the mechanisms of their action on pathogenic microorganisms studied, but also the changes that occur inside the producer organism, which can significantly increase the target yield. At present, all industrial antibiotic-producing strains were obtained as a result of the so-called classical strains improvement (CSI) methods, which involve multi-round random mutagenesis and screening [75,76,77,78,79]. During these procedures, the fungal genome is shot, not with single aimed bullets, but with shrapnel; that is, with numerous random bullets [41]. Then, several hundreds or thousands of surviving clones are screened for a desired phenotype (an increase in antibiotic production) determined by a set of random mutations. Some of these mutations are critical for increasing the yield of antibiotics, while others are byproducts of mutagenesis. A selected improved strain is used for the next round of random mutagenesis, after which positive (and side) mutations are again selected for antibiotic production. As a result, high-yielding (HY) strains are obtained, which have an improved antibiotic production by several hundred or thousand times, compared to the original wild-type (WT) strains. However, for these HY phenotypes, the molecular basis leading to improvement is unknown [80]. Currently, comparison of multi-omics data between initial WT and improved HY strains makes it possible to reveal key changes at the genome, transcriptome, proteome, and metabolome levels, driving high-yielding antibiotic production [81,82,83,84,85,86,87] (Figure 1). It turned out that the key magic bullets for the improving the fungal genome lead to: (i) the upregulation of genes in the target BGC (which may, but not necessarily, be accompanied by BGC duplications); (ii) knock outs of the genes responsible for the production of alternative secondary metabolites; (iii) the rearrangement of energy fluxes in favor of target secondary metabolism; (iv) the redirection of metabolic pathways to obtain more precursors; (v) changes in the regulation of the stress response; and (vi) an effect on the global system of regulation of secondary metabolism [41,43,79,88,89]. However, recent work has shown that, for strains that have reached their technological limit of improvement by classical methods, there is an additional possibility of increasing production: the seventh and diabolical (in terms of the Freeshooter mythologeme) magic bullet [90,91,92]. It turned out that, during the fermentation of HY strains, it is possible to increase the yield of the target SM by 15–50%, by adding specific low-molecular compounds (such as polyamines) aimed at compensating for the effects of side mutations that have arisen during mutagenesis or stimulated other processes [24,93] (Figure 1).
Knowing the laws that lead to the improvement in strains at the molecular level, it is possible to shoot with genetic engineering methods [44,94]. Genetic engineering itself is a gun for magic bullets that introduce targeted mutations.
2. Types of Antibiotics by Production Method
According to the industrial production method, there are three main types of antibiotics currently used: (i) natural products (obtained by the fermentation of improved strains of bacteria and fungi) [37,95]; (ii) semisynthetic antibiotics (based on in vitro modifications of natural products) [96,97]; synthetic antibiotics (obtained by chemical synthesis) [34,98] (Figure 2).
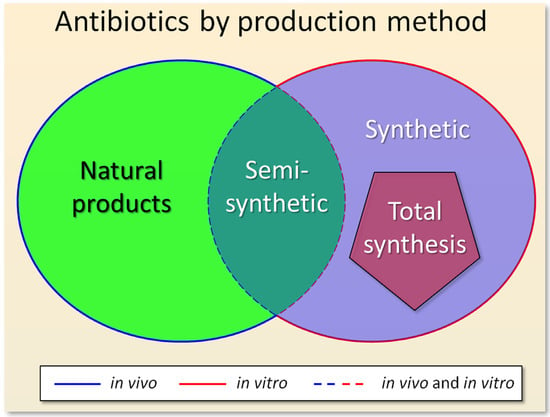
Figure 2.
Types of antibiotics according to the method of their industrial production: (i) natural products (obtained by the fermentation of improved microbial strains); (ii) semisynthetic antibiotics (obtained from natural products after fermentation and subsequent modification in vitro); (iii) synthetic antibiotics (obtained by chemical synthesis). Total synthesis is highlighted in the pentagon, as a special case of chemical synthesis, which serves to create structural analogues of natural products. Stages of antibiotic synthesis occurring in vivo, or in vitro, or both in vivo and in vitro, are indicated by a solid blue line, or a solid red line, or dotted blue and red lines, respectively.
Chemical synthesis makes it possible to obtain both new highly active compounds that are not found in nature, and structural analogues of natural antibiotics (as a result of total synthesis). Along with innovative changes in chemical and biocatalytic instrumentation, the fermentation of improved industrial strains remains one of the most important approaches for antibiotic production [19,99]. Using this method, it is economically more profitable to obtain raw materials for subsequent modifications of semisynthetic antibiotics [56]. For example, as a result of the fermentation of Acremonium chrysogenum (in 2023 reclassified as Hapsidospora chrysogena [100]), the beta-lactam antibiotic cephalosporin C is obtained, which is the initial substrate for the synthesis of several dozen cephalosporin antibiotics of the first–fifth generations [92,101]. Moreover, the vast majority of antibiotics on the market since the 2000s are microbial products and are still produced by fermentation [37]. For example, to obtain the glycopeptide teicoplanin, a high-yielding strain of Actinoplanes teichomyceticus is used [34], and all modern schemes for the total synthesis of this compound were not introduced into industry due to the high cost [37,102]. Among the new synthetic antibiotics, only oxazolidinones entered the market [103].
Currently, the vast majority of commercial antibiotics, with the exception of sulfa drugs, quinolones, and oxazolidinones, are natural or semisynthetic [104].
3. Biosynthesis of Secondary Metabolites (SMs) in Filamentous Fungi
Filamentous fungi (or mold) are a morphologically heterogeneous group of Ascomycota and Zygomycota with a multicellular mycelial structure called hyphae and the ability to produce airborne spores or conidia [105]. Currently, about 80 thousand filamentous fungi are known; it is estimated that there are several million of them on Earth [78,106]. Some of their representatives have unusual or unique metabolic pathways—in particular, secondary metabolic pathways—and thus serve as an important source for drug discovery [24,68]. The most valuable fungal SMs used in medicine are antibiotics, which inhibit the growth of competing microorganisms in nature and are capable of selectively killing pathogenic microorganisms in patients [107]. At the global level, the introduction of antibiotics into medical practice has become one of the main factors in a significant increase in the average life expectancy of a person. Thus, between 1950 and 2017, life expectancy increased from 48.1 to 70.5 years for men and from 52.9 to 75.6 years for women [108]. Individual representatives of fungi are capable of producing about 100 or more SMs, which are necessary both for development (transition from one morphological form to another) and for interaction with the environment [109,110]. For example, 215 SMs are currently described for Stachybotrys chartarum, and 180 SMs are described for Fusarium oxysporum [111,112]. However, under normal physiological conditions, most of these compounds are not synthesized [113]. The synthesis of one or another SM is triggered by the corresponding external or internal signal [24]. This process functions through the coordinated regulation of several dozen so-called biosynthetic gene clusters (BGCs) of secondary metabolism [23]. BGCs assemble genes responsible for both the biosynthesis of the corresponding secondary metabolite and its transport between cell compartments, pathway-specific regulation, and resistance against it (in cases where the synthesized product is toxic to the host organism) [24,114]. However, some of these genes may be localized outside of their BGCs [115]. Currently, the corresponding BGCs for known natural antibiotics have been determined. Along with this, bioinformatic analysis makes it possible to predict loci containing BGCs for unknown and potentially medically useful SMs [116]. To build the core structure of most fungal SMs, so-called central (or backbone) enzymes are used, such as non-ribosomal peptide synthetase (NRPS), polyketide synthase (PKS), and terpene cyclase (TPC) [117,118,119]. They have characteristic conserved motifs, and the corresponding genes are relatively easy to identify in silico [120]. At a close distance from the genes of the backbone enzymes, the genes of tailoring enzymes for modifying the core structure are usually localized, as well as genes encoding transporters of intermediate and/or final compounds, pathway-specific regulators (transcription factors that regulate the expression of the BGC in which their genes are localized), and genes for resistance to the final metabolite (if the synthesized substance is toxic to the cell itself) [24]. Such organization of BGCs allows for, after finding genes encoding backbone enzymes, the screening of nearby areas for assembled genes and the rough delineation of the boundaries of the cluster [121]. Currently, after genome sequencing, various tools are used, such as antiSMASH and MIBiG, to find potential BGCs that are mosaically distributed in the genome [116,122,123]. As a result, it becomes possible to assess the biosynthetic potential of a particular organism, in terms of the production of SMs, and compare its BGCs discovered in silico with previously characterized ones, including clusters for which the final products of biosynthesis are known [121]. This makes it possible to assess the feasibility of developing a platform for the subsequent awakening of silent BGCs and the establishment of their latent products in a specific organism [124,125,126].
In particular, antibiotics begin to be actively synthesized when signals are detected in the environment associated with the appearance of microorganisms competing for the substrate [127]. In this case, the corresponding external signals lead to the activation of previously silent corresponding BGCs [128]. This process is accompanied by chromatin remodeling (the conversion of required chromosomal loci containing BGCs from heterochromatin to euchromatin) under the influence of the system of global regulation of fungal secondary metabolism [129]. During chromatin remodeling, individual components of protein complexes act as readers, writers (methyltransferases and acetyltransferases), or erasers (demethylases and deacetylases) [129]. As a result, the transcription of biosynthetic and other genes necessary for the functioning of the antibiotic is activated, which leads to its accumulation in the environment to eliminate competing microorganisms [130]. At the present stage of development of science, when creating preparations for the targeted destruction of pathogenic microorganisms, both natural and human developments are used (Figure 3).
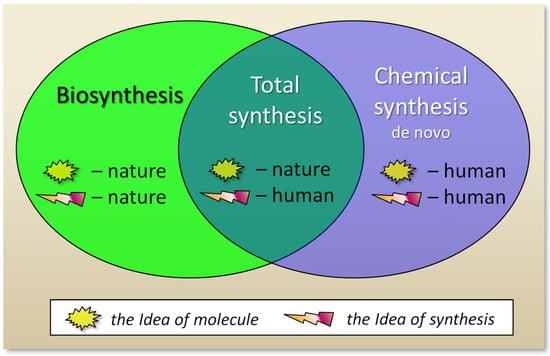
Figure 3.
The impact of humans and nature according to different ways of creating antibiotics. During biosynthesis, nature itself synthesizes the molecule it has invented. In the course of total synthesis, a human invents a path of chemical reactions to obtain a molecule invented by nature. In de novo chemical synthesis, a human invents both a new molecule that did not previously exist in nature, and a method for its synthesis.
If antibiotics are classified according to the method of their production, then semisynthetic compounds are at the junction of natural products and synthetic antibiotics, since both in vivo and in vitro processes are involved in their production (Figure 2). At the same time, antibiotics obtained as a result of total synthesis are not linked with natural products, despite the fact that the idea of their structure was taken from nature. In this regard, one can also classify antibiotics based on the ideas about their structure and synthesis (Figure 3). The structure of natural products came from nature, which receives these compounds in the process of biosynthesis. In the case of compounds obtained as a result of total synthesis, nature created the idea (invented the structure) of such compounds, and man, as a result of elegant chemical transformations, at the level of art, came up with alternative schemes for their production [131]. And for de novo synthesized compounds, man came up with new structures, for compounds that did not previously exist in nature, and also came up with a way of chemical reactions for their synthesis (Figure 3).
4. Classical Strain Improvement (CSI) for Industrial Production of Antibiotics in Fungi
In order to force a fungal strain to synthesize much more of a natural product than is intended by its physiology (100–1000 or more times), there are a number of biotechnological tools, such as: (i) random mutagenesis with physical and/or chemical mutagens and screening [78,109,132]; (ii) sexual crossing [133,134,135,136]; (iii) somatic crossing (including parasexual recombination) [137,138,139]; and (iv) genetic engineering [76,140,141]. The first three methods belong to the so-called classical strain improvement (CSI) methods. The core of such improvements is based on random mutagenesis and the subsequent screening of surviving clones for the desired phenotype, which may result in increased production of the target enzyme, primary metabolite, or secondary metabolite, including antibiotics [142] (Figure 4). This is due to the fact that sexual breeding is not available for most industrially valuable fungi (except for members of the genera Claviceps, Emericellopsis, and Aspergillus), and the parasexual cycle in biotechnology has proved less useful than it was, at one time, predicted to be [137,143]. The powerful genetic engineering tool has great potential in terms of targeting and the diversity of its capabilities [124]. However, none of them led to the creation of an industrial antibiotic-producing fungal strain (as distinct from the production of primary metabolites or enzymes) [142,144,145,146].
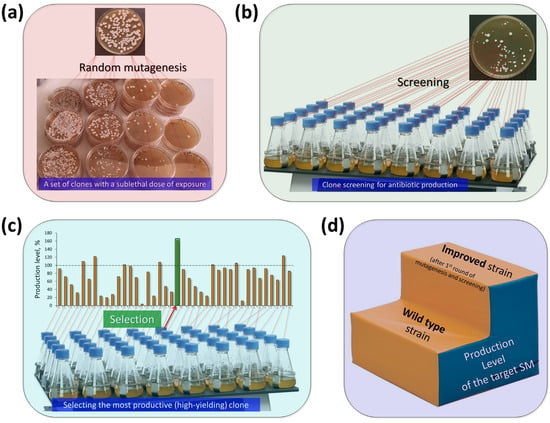
Figure 4.
Classical strain improvement for antibiotic production in fungi. (a) The wild type (WT) strain is subjected to random mutagenesis at a sub-lethal level. Surviving clones carry random mutations, some of which may result in increased production of the targeted antibiotic. (b) The obtained clones are then screened (as a result of fermentation, plate tests, or other methods) for the level of antibiotic production. (c) The level of antibiotic production in most of the clones obtained as a result of mutagenesis is less than or equal to that of the initial WT strain. However, clones are found in which antibiotic production is increased; they are selected. (d) The result is an improved strain that, after the first round of mutagenesis, has a higher yield than the WT strain. This strain is then used for a new round of random mutagenesis and selection.
Natural isolates, i.e., WT strains, as a rule produce the required SM in an amount of several tens of mg per liter or less [88], which is not enough for industrial fermentation. HY producers derived from such WT strains can produce several tens of grams per liter [48,147]. Such a tremendous leap in increasing production is currently achieved as a result of the sequential selection of the desired phenotype occurring at successive stages of mutagenesis. In this selection, mutational changes, as a rule, do not lead to an increase in production more than several times per stage [47].
In the first stage of improvement, a natural isolate that produces the required SM is subjected to sublethal mutagenic action, which leads to mutations in the fungal genome, but does not kill all cells (Figure 4a). Single cells that survive such an impact multiply and give rise to clones; genetically homogeneous colonies. All cells of a particular clone contain specific sets of mutations from the surviving parent cell. Then, the clones are screened on nutrient media for the level of production of the required antibiotic (Figure 4b). Generally, most clones exhibit lower or similar levels of production compared to the parental strain. However, as numerous studies with strains of microorganisms since the 1950s have shown, after mutagenesis, clones with increased production of the target secondary metabolite also arise (Figure 4c). The most active clone is selected and can be used as a strain for further manipulations; this ends the first round of mutagenesis (Figure 4d).
The resulting increase in production determines the effectiveness of mutagenesis at one or another stage of strain improvement. Typically, after the first round, the CSI program does not end. The improved strain obtained at the first stage is subjected to a new mutagenesis, as a result of which new clones arise, after the screening of which it is again possible to obtain an even more active strain. The process continues until the next mutagenesis makes it possible to select clones with an increased yield of the required SM. This stage of mutagenesis corresponds to the technological limit of the program for improving a particular fungal producer. In many programs, it was possible to implement 10–50 or more rounds of mutagenesis until the technological limit of the method is reached [47,48,84,147].
It turns out that mold strains are extremely convenient objects for CSI, for the following reasons: (i) they have amazing resistance to mutagenesis, maintaining cellular fitness and high-yield production even after dramatic chromosomal rearrangements; (ii) they produce haploid conidia, which are an excellent material for mutagenesis, since it is enough to knock out only one allele to immediately screen for the phenotype; and (iii) fungal mutants generally have good stability [89,143,148,149]. These properties made it possible to carry out numerous CSI programs with them, during which, at various rounds of mutagenesis, “correctly surviving” fungal strains were selected; that is, those carrying mutations necessary for improvement, as well as side random mutations that do not cover the positive effect.
5. Industrial Production of Antibiotics in Fungi
Improved fungal strains produce key beta-lactam antibiotics (penicillins and cephalosporins), which account for nearly half of the antibiotic market [150,151,152] (Figure 5).
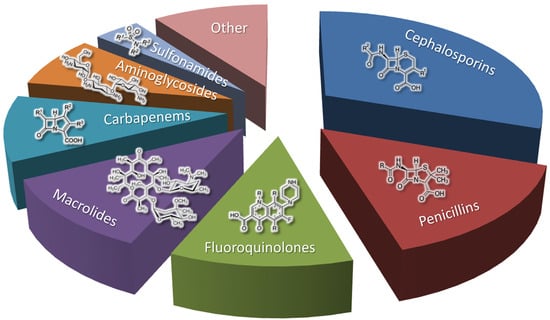
Figure 5.
Antibiotic market share by drug class, according to the open access data from [150].
Fungal strains are also used to obtain fusidanes, echinocandins, griseofulvins, and other classes of antibiotics [153]. All of them are significantly inferior to beta-lactams in terms of market share and occupy part of the “Other” sector in Figure 4. However, these minor (in terms of market share) antibiotics are extremely in demand in a number of cases [153]. In addition, the fermentation of improved fungal strains is used to produce not only antibiotics, but also a number of other pharmaceutically significant SMs, such as statins (cholesterol-lowering drugs), immunosuppressants, and antitumor drugs [8].
5.1. Fermentation of Beta-Lactam Antibiotics in Fungi
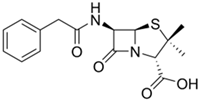
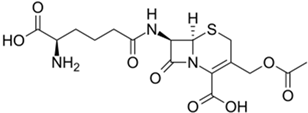
The fermented penicillin G (PenG) and cephalosporin C (CepC) are modified in vitro to create drugs used in medicine (PenG is also used unmodified) [152]. Obtaining PenG and CepC in improved fungal strains is more economical than in vitro production [154]. In fungi, the biosynthesis of these beta-lactams is under the control of a complex hierarchical regulatory system, modified in the process of strain improvement, and is accompanied by a series of biosynthetic reactions, with a significantly increased intensity in HY producers [94,155]. In these reactions at the first stage, LLD-ACV tripeptide δ-(L-α-Aminoadipoyl)-L-cysteinyl-D-valine is obtained as a result of non-ribosomal peptide synthesis by the enzyme PcbAB (EC: 6.3.2.26). This tripeptide is then cyclized to isopenicillin N (IPN) through a dioxygenase reaction catalyzed by PcbC (isopenicillin N-synthase (EC: 1.21.3.1)). The IPN core structure is then modified by tailoring the enzyme PenDE (IAT, isopenicillin-N N-acyltransferase [EC:2.3.1.164]) to produce penicillin G, or by sequential reactions of the tailoring enzymes to produce cephalosporin C (CefD1, isopenicillin N-CoA synthetase (EC: 5.1.1.17), CefD2 (isopenicillin N-CoA epimerase (EC: 5.1.1.17), CefEF (penicillin N expandase, EC: 1.14.20.1)/deacetoxycephalosporin C hydroxylase, EC: 1.14.11.26), and CefG (deacetylcephalosporin-C acetyltransferase (EC: 2.3. 1.175)).
5.1.1. Fermentation of P. chrysogenum for Penicillin G (PenG) Production
The main industrial producer of penicillin G (PenG, or benzylpenicillin, and “Peanut Butter Shot” in military slang) is P. chrysogenum (formerly known as Penicillium notatum), which belongs to the class Eurotiomycetes and the division Ascomycota [156,157,158]. Currently, a number of molds are known that are capable of producing PenG, for example from the genera Arthroderma, Aspergillus, Penicillium, and Trichophyton; however, only P. chrysogenum strains are used for the industrial production of this important antibiotic [159,160,161,162]. PenG is the first industrially produced natural antibiotic, which was accompanied by an innovative breakthrough in the development of deep fermentation [163]. From the early 1940s to the present day, PenG has been produced worldwide from the fermentation of improved strains of the P. chrysogenum NRRL 1951 initial isolate [79,164,165]. This strain was discovered in 1943 growing on a cantaloupe at the local market in Peoria (IL, USA) [166]. It turned out that P. chrysogenum NRRL 1951 produces more penicillin than the original Fleming isolate (which produces mostly the unstable and difficult to isolate penicillin F, named after its discoverer) and other P. chrysogenum industrial strains used at that time for commercial production of PenG (such as P. chrysogenum NRRL 1249-B21) [7,167,168]. NRRL 1951 was adapted for commercial use; CSI programs were carried out on its basis, which made it possible to increase the PenG yield by more than 1000 times [41,101,169,170] (Table 1). Seventy-seven years after its discovery, P. chrysogenum NRRL 1951 received the status of “State Microbe of Illinois” [166].

Table 1.
Production level of the most important antibiotics from fungi of wild type (WT) and high-yielding (HY) strains derived from them.
Currently, the P. chrysogenum strains used for PenG production (both the original Fleming strain and the strains used to obtain improved industrial producers) have again been reclassified as P. rubens [183]. At the same time, the name P. chrysogenum was retained for a number of strains that are not used for the industrial production of penicillins and differ from P. rubens in some characteristics, such as their production of secondary metabolites [183]. However, in the vast majority of recent publications, it is still common to use the name P. chrysogenum for PenG producers, in order to avoid confusion [7,156]. PenG can be used as a final drug [184] or can serve as a raw material for the production of semisynthetic penicillin antibiotics (Figure 6a). To achieve this, at the first stage, penicillin nucleus or 6-aminopenicillanic acid (6-APA) is obtained from PenG as a result of the deacylation reaction in vitro by amidase enzymes of bacterial origin (penicillin G amidase, EC 3.5.1.11) [56,185]. Then, various semisynthetic penicillins are obtained by grafting different side chains onto 6-APA [186,187,188] (Figure 6a).
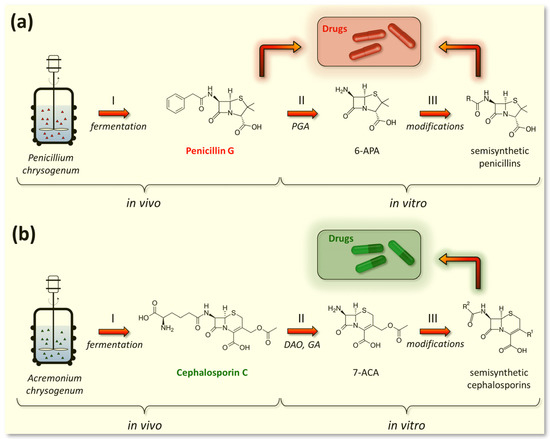
Figure 6.
Industrial production of beta-lactam antibiotics. (a) Obtaining of penicillin drugs; both penicillin G (a direct fermentation product of Penicillium chrysogenum) and its semisynthetic derivatives are used in pharmaceutical industry. (b) Obtaining of cephalosporin drugs; cephalosporin C (a fermentation product of Acremonium chrysogenum) has low activity and is not used in the pharmaceutical industry, unlike its semisynthetic derivatives. Curly brackets indicate the stages occurring in vivo (stage I—fermentation of fungal strains) and in vitro (stage II—production of core structures and stage III—production of semisynthetic antibiotics). PGA—penicillin G amidase (EC 3.5.1.11); DAO—D-amino acid oxidase (EC 1.4.3.3); GA—glutaryl-7-aminocephalosporanic acid acylase (EC 3.5.1.93); 6-APA—6-aminopenicillanic acid, 7-ACA—7-aminocephalosporanic acid.
5.1.2. Fermentation of Penicillins Other Than PenG
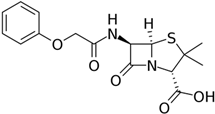
Since the 1950s, intensive research has been carried out to produce PenG analogues with altered activity profiles by fermenting P. chrysogenum with various mono-substituted acetic acids [189]. As a result, industrial strains of P. chrysogenum, in addition to benzylpenicillin (PenG), also produce another pharmacologically important compound, phenoxymethylpenicillin (penicillin V, PenV) [171,190]. PenG is produced if phenylacetic acid is added to the culture medium as a side chain precursor, and PenV is synthesized if phenoxyacetic acid is added [185]. To effectively produce PenV in industrial strains, multi-round mutagenesis and selection are also carried out to reduce the level of by-products, such as para-hydroxypenicillin V [191]. PenV can be used either as a final drug or as a raw material to obtain semisynthetic penicillins through 6-APA (Figure 6). Currently, only two penicillins produced during fermentation are of industrial importance, namely PenG and PenV [189]. Other natural penicillins, such as PenF (Fleming’s penicillin), PenK, PenN, PenO, PenU1, PenU6, or PenX, are not currently used in medical practice.
5.1.3. Fermentation of A. chrysogenum for Cephalosporin C (CefC) Production
The exclusive industrial producer of CefC is A. chrysogenum (formerly known as Cephalosporium acremonium), which belongs to the class Sordariomycetes, division Ascomycota (Table 1) [101]. In contrast to numerous PenG producers, CefC production is found only in a few fungal species. In addition to A. chrysogenum, the biosynthesis of CefC was also found in Pochonia chlamydosporia, Kallichroma tethys, and Paecilomyces persicinus, where it is not used for industrial production [159,162]. This relatively rare distribution of the biosynthetic pathway for this beta-lactam antibiotic in fungi is apparently associated with the appearance of additional genes, the origin of which is discussed in [7,192,193]. In particular, for the biosynthesis of CefC, part of the additional genes is assembled into the so-called “late” beta-lactam BGC, which is located on a different chromosome relative to the localization of the “early” beta-lactam BGC [89,193]. Currently, in 2023, the A. chrysogenum have again been reclassified as Hapsidospora chrysogena [100]. Since Acremonium chrysogenum has been used for a long time to refer the CPC producer, we use this name in current review to avoid confusion. Current industrial producers of CefC are derived from the Brotzu initial isolate A. chrysogenum ATCC 11550 as a result of CSI programs [194]. Italian pharmacologist Giuseppe Brotzu isolated A. chrysogenum ATCC 11550 in 1948 from seawater near Sardinia, Italy [195]. Industrial descendants of this strain produce several hundred times more CefC [101,172,173]. CefC itself, unlike PenG, has weak antibacterial activity and is not used as a final drug in medical practice (Figure 6b) [196]. The key role of cephalosporin C is associated with its use as a starting substance for the production of all pharmaceutically important cephalosporin antibiotics [197]. To achieve this, at the first stage, cephalosporin nucleus or 7-aminocephalosporanic acid (7-ACA) is obtained in vitro from CefC, either chemically or enzymatically (using D-amino acid oxidase (EC 1.4.3.3) and glutaryl-7-aminocephalosporanic acid acylase (EC 3.5.1.93)) [56,198,199]. The side groups of 7-ACA are then modified by various chemical or enzymatic methods to produce cephalosporin drugs [56,169,200,201].
5.2. Fermentation of Non-Beta-Lactam Antibiotics in Fungi
The fermentation of fungal strains is used to produce a number of non-beta-lactam antibiotics for medical use. Some of them, such as fusidic acid and griseofulvin, were introduced to the market during the Golden Age of antibiotics and are still used today [202,203]. Other antibiotics, such as pleuromutilins, were discovered in the early 1950s, but only entered the market as semi-synthetic derivatives in 2007 [204,205,206]. The emergence of new classes of antibiotics from fungi, which entered the market in the last 20–25 years, was one of the most important factors that made it possible to fill the innovative gap in the introduction of antibiotics that arose after the end of the Golden Era of antibiotics [65]. For example, relatively recently discovered antibiotics, from the classes echinocandins and enfumafungins, were introduced into medical practice at the beginning of the 21st century [207,208]. There are also antibiotics that were previously obtained using fermentation in fungal strains, but that have now been discontinued due to discovered toxicity. For example, fusafungine, which belongs to the enniatin class of antibiotics, was withdrawn from the market in 2016 due to toxicity [209]. Although this review focuses only on antibiotics produced by fungal fermentation, it should be noted that improved bacterial strains are also widely used for the industrial production of non-beta-lactam antibiotics [210,211,212]. For example, some major market antibiotics, such as macrolides and aminoglycosides, and some minor market antibiotics, such as tetracyclines and glycopeptides, were originally isolated from actinomycetes, especially from the genus Streptomyces (Figure 5) [210]. Bacteria are also a promising source for obtaining new antibiotics, for example, antimicrobial peptides [213]. On the other hand, there are vast classes of non-beta-lactam antibiotics, such as fluoroquinolones and sulfonamides, which are obtained by chemical synthesis [214,215].
5.2.1. Fusidanes
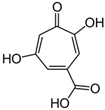
Steroid antibiotics from the fusidane class, such as fusidic acid, helvolic acid, and cephalosporin P1, are produced by various filamentous fungi [153]. Although these antibiotics have been known since the 1960s, they have drawn renewed attention, due to the fact that they have no cross-resistance to generally used antibiotics [174,216]. Fusidic acid, the key fusidane antibiotic, is obtained from the fermentation of improved strains of Fusidium coccineum [176]. The production level in the improvement process was increased by a hundred times (Table 1). Other fusidanes, helvolic acid from Aspergillus fumigatus and cephalosporin P1 from A. chrysogenum, do not currently have such extensive use in medicine (Table 2) [217,218].

Table 2.
Some examples of antibiotic production by fungal strains.
5.2.2. Griseofulvin
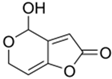
Griseofulvin, an ascomycete polyketide metabolite that interferes with the polymerization of fungal microtubules, was marketed in 1959 [230]. It was first isolated in 1939 and is still widely used in fields ranging from medicine to agriculture [230,231]. The industrial fermentation of griseofulvin is carried out on the basis of improved strains of Penicillium griseofulvum (reclassified from Penicillium patulum) [84,232]. The production level is several tens of grams per liter; however, specific numbers and methods of classical improvement are described in the relevant patents and are successfully masked [179,230].
5.2.3. Pleuromutilins
Pleuromutilins are SMs of the terpene type [24]. Despite the fact that pleuromutilin, the first representative of this class of antibiotics, was discovered back in 1951, pleuromutilins only recently entered the market as a medicine for humans [204]; retapamulin, the first antibiotic of this class, which is approved for humans, began to be used in 2007 [233,234]. To obtain the semisynthetic antibiotic retapamulin, pleuromutilin is used, which is produced as a result of fermentation of Clitophilus scyphoides and other basidiomycetes [205,235]. In 2019, lefamulin, another semisynthetic antibiotic from the pleuromutilin class, was approved for systemic use in humans [209,236,237,238]. An important characteristic of pleuromutilins is the lack of cross-resistance with other antibiotics, such as the macrolides, which act on a similar target, the 50S subunit of the prokaryotic ribosome [239].
5.2.4. Echinocandins
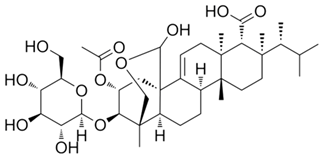
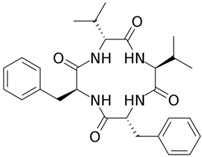
Currently, more than 20 echinocandins, lipopeptide antibiotics, have been isolated from fungi, the fermentation of three of which—echinocandin B, pneumocandin B0, and FR901379—has been commercialized [240]. They are used as raw materials in semisynthetic synthesis to obtain anidulafungin, caspofungin, and micafungin, respectively, which were introduced into medical practice in the 2000s [207,241,242,243]. These compounds are also called “antifungal penicillins” because they act on the biosynthesis of the cell wall of fungi, just as penicillins destroy the cell wall of bacteria [244]. Echinocandin B was first isolated in 1974 from A. nidulans, becoming the first antibiotic belonging to the echinocandin class [245]. Currently, it is produced by improved strains of A. nidulans and is used as a raw material for the semisynthetic antibiotic anidulafungin, used in medical practice [246,247]. Industrial strains of A. nidulans produce approximately a thousand times more echinocandin B than WT strains [181]. Caspofungin, another semisynthetic lipopeptide widely used in medicine, is derived from pneumocandin B0, which is produced by fermentation of the ascomycete Glarea lozoyensis [248]. Wild-type strains of G. lozoyensis predominantly synthesize pneumocandin A0, approximately 10 times more than pneumocandin B0 [240]. As a result of CSI programs, HY producers of pneumocandin B0 were obtained, which in its structure is less similar to echinocandin B than pneumocandin A0 [180,249,250,251]. Now, industrial strains produce hundreds of times more pneumocandin B0 compared to WT strains [180]. One of the most important antibiotics of this class, semisynthetic micafungin, is derived from the natural product FR901379 (via the enzymatic synthesis of compound FR179642) [252,253]. Lipopeptide FR901379 is obtained as a result of the natural fermentation of the improved strains of the ascomycete Coleophoma empetri [254,255]. Recently, the industrial producer C. empetri MEFC009, using a genetic engineering approach, was able to further increase the yield of FR901379 [182,256].
5.2.5. Enfumafungins
Enfumafungin is a glycosylated triterpenoid that acts on the fungal cell wall [257]. This SM, like the echinocandins, targets β-1,3-glucan synthase, but appears to bind this multiprotein complex at a different site [258]. Due to this, there is a lack of cross-resistance between these two classes of antibiotics, and pathogenic fungi that have acquired resistance to echinocandins show sensitivity to enfumafungins [259,260]. Enfumafungin is obtained from the ascomycete Hormonema carpetanum [220]. In 2021, ibrexafungerp, a semi-synthetic derivative of enfumafungin, was introduced into medical practice, representing the first non-azole oral antifungal drug to be approved in the United States for the treatment of vaginal yeast infections [208,261] (Table 2).
5.2.6. Enniatins
Enniatins are cyclohexadepsipeptides that are produced by Fusarium, Verticillium, Halosarpheia, and several other fungal genera [262]. Currently, about thirty natural compounds of this class have been characterized [219]. The first enniatins were discovered more than 70 years ago [263]. Since then, the antibiotic fusafungine has been introduced into clinical practice [264]. It is a cocktail of enniatins resulting from the fermentation of Fusarium lateritium strains, due to partially the non-specific work of non-ribosomal multifunctional enzyme enniatin synthetase (NRPS) [219,265,266,267]. The European Medicines Agency, in 2016, recommended the withdrawal of fusafungine from the market due to possible rare but severe allergic reactions [209,268].
5.3. Fungal Secondary Metabolites for the Development of Novel Drugs
Fungi produce significant amounts of SMs with antimicrobial properties [71,269]. Only a few of them have been introduced into medical practice (Table 1 and Table 2) [71,203]. However, there are a significant number of compounds that have demonstrated antibiotic or other medically important activity [23]. A case in point is a literature survey showing that, among 1500 compounds isolated from fungi in the 1990s, more than half exhibited antibacterial, antifungal, or antitumor activity [270]. Modern approaches that combine advances in the fields of genetic engineering and bioinformatics have made it possible to significantly increase the number of fungal SMs that are potentially significant for human use [271,272]. Some of these compounds and their semisynthetic derivatives are currently in clinical trials at phase I or greater [71,273]. Thus, the semisynthetic beta-lactam antibiotics benapenem, sulopenem, and its prodrug sulopenem etzadroxil, which act on the cell wall of bacteria, are in phase-III clinical trials [273,274,275,276]. Semisynthetic beta-lactams sanfetrinem and its prodrug sanfetrinem cilexetil are in phase-II clinical trials [273,277,278]. Several fungal antibiotics are currently being tested in clinical trials for the treatment of other diseases, such as cancer. For example, wortmannin, an antifungal furanosteroid antibiotic isolated in 1975 from Talaromyces wortmannii (formerly Penicillium wortmannii) [279], was then also shown to have inhibitory activity against phosphatidylinositol 3-kinase [280]. Currently, a semisynthetic compound PX-866 has been developed based on wortmannin, which has entered phase II clinical trials for recurrent glioblastoma [281]. Fungal SMs are being tested in clinical trials for the treatment of a wide range of diseases [71]. For example, muscimol, an isoxazole from Amanita pantherina, is an ionotropic GABAA-R receptor agonist and was developed for the treatment of drug-resistant epilepsy (phase I) [282]. Psilocybin from hallucinogenic fungus Psilocybe mexicana binds to the 5-HT2A serotonergic receptor and is developed for treatment-resistant depression [283,284]. Antroquinonol was isolated in 2007 from Taiwanofungus camphoratus (formerly Antrodia camphorata), a parasitic fungus indigenous to Taiwan [285]. It inhibits isoprenyl transferase, an enzyme that is involved in the post-translational processing of Ras and Rho proteins, which leads to the inhibition of Ras and Rho signaling, and that has antiviral, anti-inflammatory, anti-fibrotic, anti-cancer, and anti-hyperlipidemia properties [286]. This ubiquinone derivative was entered into phase II clinical trials for treating non-small-cell lung cancer [287]. The introduction, in recent years, of new classes of antibiotics from fungi (and their semi-synthetic derivatives), such as echinocandins, enfumafungins, and pleuromutilins, indicates significant potential for the development of novel drugs based on fungal SMs.
6. Role of Low Molecular Weight Inductors in the Production of Antibiotics in High-Yielding Strains
The biosynthesis of SMs in fungi can be stimulated either by external inducers (e.g., abiotic factors, plant-derived external elicitors, inducers from co-culture, and synthetic epigenetic modifiers) [288] or by internal inducers, which can act similarly to bacterial autoinducers [289,290]. Among these multiple influences for the production of SMs, various low molecular weight compounds (LMWCs) play an important role. Their effect, on the one hand, is observed in nature during interspecies communication, and on the other hand, can be used to increase antibiotic production during fermentation [24]. Some of these compounds may act as inducers and stimulate the production of specific SMs [90,291], while others inhibit the biosynthesis of SMs [292,293]. Moreover, it was shown that, depending on the cultivation conditions, the same LMWC can act as both an inducer and an inhibitor of the biosynthesis of a particular secondary metabolite in the same microorganism [92,294].
6.1. Effect of Multispecies Communication on Biosynthesis of Fungal Secondary Metabolites
In nature, microorganisms can communicate through specific LMWCs [295]. Their effect can be expressed in the targeted activation or inhibition of the biosynthesis of particular fungal SMs. So, in fungal–plant interactions, on the one hand, symbiosis can stimulate the production of fungal SMs, for example, to protect the plant from insect pests [296]. Thus, the interaction between the soil fungi Trichoderma harzianum and Zea mays leads to the resistance of this plant to the scarab beetle Phyllophaga vetula [297]. On the other hand, SMs of phytopathogenic fungi can serve as virulence factors, and inhibition of their production leads to a decrease in pathogenicity against the host plant [298]. For example, LMWCs from an extract of the medicinal plant Ephedra major can inhibit the production of the polyketide aflatoxin B1 in Aspergillus parasiticus [299]. Another study identified the active substance in the inhibitory extract from Betula alba; it turned out that methyl syringate works as a specific inhibitor of aflatoxins biosynthesis in both A. parasiticus and Aspergillus flavus [300]. Another example of an LMWC for chemical microbial communication is jasmine acid and its derivatives, jasmonates [301]. These lipid-derived signaling molecules are produced by plants and fungi. The addition of exogenous jasmonates stimulates the production of a number of SMs in plant cell suspension cultures, for example, paclitaxel and baccatin in Taxus sp., or camptothecin in Ophiorrhiza mungos L. [302,303]. In this regard, jasmonates can be promising inducers of the biosynthesis of a number of SMs in fungi.
Another example of an LMWC involved in the communication of microorganisms is short-lived hydrophobic gas nitric oxide (NO) [304,305]. Several studies have shown that nitric oxide can affect the production of SMs in fungi [306]. For example, in Aspergillus nidulans, deletion of the flavohemoglobin gene fhbA (the product of which is involved in reducing NO levels) led to a decrease in the production of sterigmatocystin, a polyketide mycotoxin [307]. And supplementation with a nitric oxide-releasing compound led to the restoration of the production level of this SM [307]. It has also been shown that treatment with lipopolysaccharide (LPS) from Gram-negative bacterial cells of Penicillium sp. and Aspergillus sp. also leads to an increase in NO production, which is accompanied by an increase in the production of a number of SMs [308]. The addition of an NO donor (sodium nitroprusside) to the mold Shiraia sp., which is used in traditional Chinese medicine, led to an increase in the production of perylenequinone pigments, in particular hypocrellin A [309,310,311], a promising photosensitizer for anticancer photodynamic therapy [312]. In addition, enzymes that control cellular NO content, such as P450nor (cytochrome P450 nitric oxide reductase), may also affect the biosynthesis of SMs in fungi [313].
Studying the role of LMWCs in microbial communication can be challenging because such interactions sometimes involve such small numbers of molecules that they are beyond the sensitivity of currently available assays [314]. In addition, under certain culture conditions, the microorganism typically produces only a small portion of its LMWC. Under simulated natural conditions based on co-culture, silent fungal BGCs can be awakened to biosynthesize cryptic SMs [290]. To achieve this, the following cultures can be used: (i) fungal–fungal co-culture, (ii) fungal–bacterial co-culture (in this case the fungus is usually used as the host strain and the bacterium as the guest), and (iii) fungal–host co-culture (in this case, plants are used as the host, and an endophytic fungus is used as the guest) [290]. By the end of 2022, as a result of fungal–fungal co-cultivation, 109 new SMs were discovered, 75 SMs were obtained by liquid state fermentation (LSF) and 34 SMs were obtained by solid state fermentation (SSF); due to fungal–bacterial co-cultivation, 42 new SMs were discovered, 23 SMs were isolated from LSF co-cultures and 19 SMs were isolated from SSF co-cultures, and six new SMs were isolated from endophyte–host co-cultures [290]. Despite significant advances in the use of co-culture technology to extract novel SMs, the exact mechanism for awakening silent BGCs is not yet fully understood [290,295]. It is assumed that the activation of BGCs occurs because specific receptors on the surface of the fungal cell recognize the attachment of another organism and/or LMWC secreted during the interaction [295]. Recent work has shown that the signal, arising from fungal–fungal co-cultivation, affects the system of global regulation of secondary metabolism. Thus, the partial loss-of-function VeA1 protein, but not VeA, was associated with the widespread SM changes in both A. nidulans and A. fumigatus during co-cultivation [126]. In this case, there was both a decrease in the content of some SMs and an increase in the content of other SMs, the structures of some of which are unknown. In addition, it was found that VeA1 regulation required the transcription factor SclB and the members of the velvet complex, such as LaeA and VelB, for production of aspernidines in A. nidulans [126].
6.2. Effect of Polyamines on Secondary Metabolism in Fungi
Aliphatic polyamines, such as 1,3-diaminopropane (1,3-DAP), putrescine, spermidine, and spermine, are ubiquitous in living nature [315,316] (Figure 7). These compounds are also found in viruses as part of virions and can participate in the life cycle of viruses [317,318,319]. Cells maintain a strict homeostasis of polyamine content [320]. This is ensured by the presence of both enzymes of biosynthesis and catabolism of polyamines, and polyamines content is controlled at the levels of transcription, translation, and the lifetime of these enzymes [321,322,323].
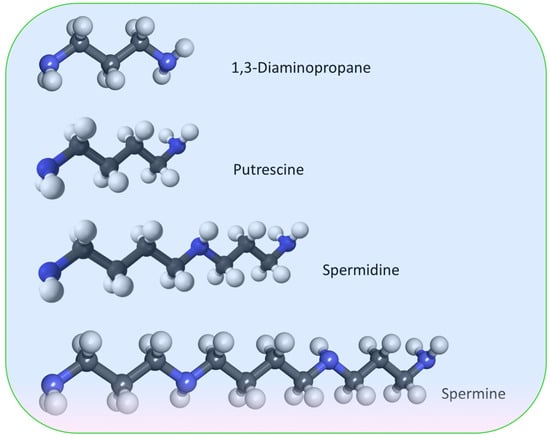
Figure 7.
Structural formulas of the most widespread aliphatic polyamines of fungi.
Work with plants demonstrated polyamine-induced NO biosynthesis, suggesting that NO is a signaling intermediate in PA action [324]. In particular, in Arabidopsis thaliana, treatment with 1 mM spermidine and spermine greatly increased NO release in the seedlings [325]. It is assumed that NO is formed as a result of the work of polyamine oxidases [324]. About 10 years ago, it was shown that the addition of 1,3-DAP and spermidine can further increase production of PenG in a reference strain of P. chrysogenum Wisconsin 54-1255, obtained by CSI [90]. Initially, as a result of a plate test with an overlay of Bacillus subtilis ATCC 6633, it was shown that the addition of conditioned culture broths from P. chrysogenum and A. chrysogenum stimulates PenG production in P. chrysogenum Wis 54-1255 [289]. At the same time, no induction effects were found with the addition of N-acyl homoserine lactones, γ-butyrolactone, jasmonic acid, or the PenG precursor LLD-ACV tripeptide. The inducer molecule from conditioned culture broths of P. chrysogenum and A. chrysogenum was characterized as 1,3-DAP [289]. In this regard, the submerged fermentation of Wis 54-1255 was carried out with the addition of 1,3-DAP in the concentration range of 1–10 mM. It turned out that the strongest effect was caused by the addition of 5 mM or 10 mM 1,3-DAP, with an increase in PenG production of approximately 100%. Fermentation was also carried out with the addition of other aliphatic polyamines (putrescine, spermidine, and spermine). Among these compounds, only spermidine, at a concentration of 5–10 mM, had a stimulating effect on PenG production; this effect was close to that of the addition of 1,3-DAP. The addition of putrescine and spermine did not change the production of the target antibiotic [289].
The results obtained were scaled for 5-L stirred tank fermenters, where an additional increase in PenG production was also obtained with the addition of 5 mM 1,3-DAP or 5 mM spermidine (but not putrescine) [90]. It was also shown that the addition of 1,3-DAP or spermidine upregulates all PenG biosynthesis genes (pcbAB, pcbC, and penDE), and the addition of putrescine upregulates pcbAB and pcbC, but does not affect the expression of penDE. The possible mechanisms of the effect of polyamines on the upregulation of beta-lactam BGCs were also studied. It turned out that the addition of 1,3-DAP and spermidine did not affect the expression of pacC, a gene of the global pH-stress regulator, thus excluding a modification of the pH control mechanism [326]. On the other hand, the addition of these polyamines led to upregulation of the laeA gene [90]. LaeA is the global regulator of fungal secondary metabolism, S-adenosyl-L-methionine-dependent histone methylase, which acts epigenetically through chromatin remodeling [327]. In order to understand the mechanism of action of exogenous polyamines leading to an increase in PenG production, we also conducted a comparative proteomic analysis [328]. It turned out that the addition of both 1,3-DAP and spermidine led to fairly similar rearrangements at the proteome level. In both cases, the following was observed: (i) the overproduction of IAT, the last enzyme of the penicillin pathway, for which, in addition to the main form, an alternative isoform was also newly detected; (ii) the overproduction of enzymes involved in the biosynthesis of valine and other precursors (e.g., coenzyme A) of PenG; and (iii) a decrease in the production of two enzymes involved in the degradation of phenylacetic acid, the precursor of PenG [328].
Subsequent work showed that the addition of 1,3-DAP or spermidine was able to further increase lovastatin production, by 25–45%, in a HY strain of Aspergillus terreus, for which the CSI program had reached the technological limit of improvement [91]. This was accompanied by upregulation of the lov-BGC and laeA genes. In addition to LaeA, the biosynthesis of lovastatin is under the control of the pathway-specific regulator LovE [42]. Moreover, there is mutual regulation between two positive regulators of the lovastatin biosynthesis pathway, LaeA and LovE, contributing to the production of lovastatin under the influence of exogenous polyamines [329]. It was also shown that, in A. terreus, the HY strain has acquired an increased resistance to α-difluoromethylornithine (DFMO) and 1-aminooxy-3-aminopropane (APA), inhibitors of a key enzyme in polyamine biosynthesis, ornithine decarboxylase (ODC; EC 4.1.1.17) [49]. Wherein the genes for polyamine biosynthesis were upregulated and the oaz gene for ornithine decarboxylase antizyme, that reduces polyamine content, was downregulated. The authors suggest that the increased resistance inhibitors of polyamine biosynthesis in this HY lovastatin producer may be associated with the increased production of polyamines themselves [49].
Another study also demonstrated an increase in CefC production, by 15–20%, in an A. chrysogenum HY strain, obtained as a result of the CSI program [92]. This was accompanied by the upregulation of genes from both “early” and “late” beta-lactam BGCs. Moreover, on a complex nutrient medium optimized for fermentation, no percentage increase in either the intermediate products of CefC biosynthesis or impure secondary metabolites was observed [92]. At the same time, using a specially selected synthetic medium, it was shown that the addition of 1,3-DAP led to an increase in CefC production, and the addition of spermidine led to its decrease [294]. This was due to the fact that, when 1,3-DAP, and not spermidine, was added to the synthetic medium, the accumulation of the CefC metabolic pathway precursor, deacetylcephalosporin C (DAC), occurred. The total amount of cephems (DAC and CefC) was the same with the addition of as 1,3-DAP, as spermidine. However, in both cases, the upregulation of all genes for CefC biosynthesis (including cefG, responsible for the last stage of CefC biosynthesis) was at the same level compared to the control. Apparently, this differential effect is associated with the intersection of the last stage of cephalosporin C biosynthesis and the catabolism of polyamines at the level of a common substrate, acetyl coenzyme A (acetyl-CoA). N1-acetylation is much more efficient during spermidine catabolism than for 1,3-diaminopropane. The addition of spermidine, but not 11,3-DAP, depleted the pool of acetyl-CoA by more than two times compared to the control, which could lead to the accumulation of DAC [294]. It turned out that an A. chrysogenum HY strain also exhibits increased resistance to ODC inhibitors, such as DFMO and APA [93]. Moreover, the content of polyamines in this strain is approximately five times higher than in the original wild-type strain that was used for CSI. The data obtained indicate that, at least in some CSI-derived HY fungal strains, the addition of polyamines can result in increased production of the target SM (Figure 8).
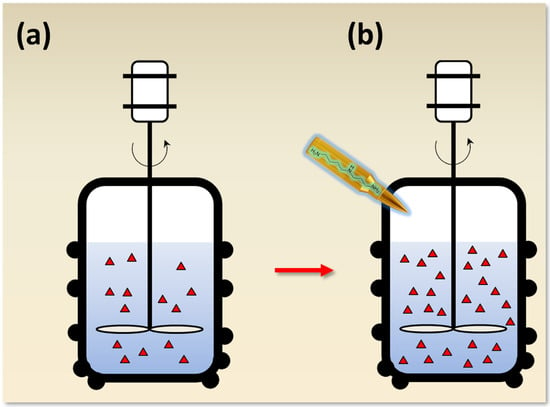
Figure 8.
Increasing antibiotic yield in improved fungal strains with the magic bullet of low molecular weight inductors. (a) For the industrial fermentation of antibiotics and other secondary metabolites (SMs), high-yielding (HY) strains obtained as a result of classical strain improvement (CSI) programs are used. CSI programs have upper thresholds for the yield of target SMs, after which it cannot be further increased. (b) Supplementation with a number of low molecule weight inducers (LMWIs) could potentially be used to further increase production. The addition of such LMWIs can compensate for incidental mutations arising during CSI programs; in this regard, they can be considered as magic bullets, shot to improve metabolism. Fermentation under the influence of a magic bullet (i.e., adding a LMWI) leads to an increase in the yield of the SM. As an example, the magic bullet contains the formula of spermidine, which has been shown to stimulate several HY fungal producers [90,91,92]. Red triangles indicate the relative amount of SM produced without additives and with the addition of an inducer.
The content of polyamines in such improved strains is increased compared to the original WT strains; this is surprising, given the strict homeostasis of polyamine content for certain cells and organisms [320]. Such a shift in polyamine content may be due to mutations affecting polyamine metabolism that were selected as accompanying in the CSI programs. It is known that polymers are able to protect cells from free-radical damage of DNA by reacting directly with the reactive oxygen species [330] and participating in homology-directed DNA repair with RAD51 recombinase [331], which can occur during mutagenesis [332]. Polyamines are also able to help the cell survive under conditions of stress that arise both during mutagenesis and highly active production of the target metabolite [333]. Therefore, an increase in the production of polyamines could help cells survive mutagenic effects at a sublethal level and overproduction of the target SM. Mutations in the metabolism of polyamines, leading to an increase in their content, were selected as co-occurring during CSI programs. However, increased polyamine synthesis utilizes additional cellular resources that could potentially be redirected to produce the target SM. In this regard, it is possible to explain the effect of increasing target production due to the addition of exogenous polyamines. The exogenous introduction of polyamines leads to the inhibition of their biosynthesis in the cell via a feedback mechanism, which is known for polyamines [323,334]. The released cell resources can be redirected to the production of the target metabolite, which is expressed in an additional increase in its production (Figure 8).
7. Key Molecular Events Leading to High-Yield Production of Secondary Metabolites in Fungi
Since classical enhancement methods, involving random mutagenesis and screening, have been used to obtain improved secondary metabolite producers in fungi from the dawn of the Golden Age of antibiotics to the present, the molecular basis leading to the production of HY strains has remained uncharacterized. The current approaches provide insight into some of the key changes that must occur in the genome of a WT strain for it to become a high-yielding producer (Figure 9).

Figure 9.
The Freeshooter magic bullets for improving the production of target secondary metabolites (SMs) in filamentous fungi. The term “magic bullet” (Zauberkugel, ger.) was coined by Paul Ehrlich from Carl Maria von Weber’s popular 1821 opera Der Freischütz (The Freeshooter) to designate compounds that specifically kill pathogenic microorganisms and do not harm the human body. The opera Freeshooter is based on the German legend of magic bullets that fly not according to the laws of ballistics, but according to the will of the shooter himself. The shooter buys seven magic bullets from the devil in exchange for a soul. For the first six bullets, he chooses the target himself, and the last one is controlled by the devil. Following Ehrlich, the term “magic bullets” is used here to highlight key mutations leading to the development of high-yielding (HY) fungal producers of SMs. The genomic target stands on a Petri dish with Czapek Dox agar medium, on which Penicillium chrysogenum STG-117 (MW556011.1) is cultivated. Magic bullets are aimed at corresponding changes in genomes that lead to: (i) target BGC upregulation—an increase in the levels of mRNA expression of genes belonging to the biosynthetic gene cluster (BGC) for the production of the SM of interest; (ii) global regulation—changes in the system of global regulation of secondary metabolism; (iii) other BGCs disruption—inactivation of the production of alternative secondary metabolites by knocking out key BGCs genes; (iv) energy fluxes—the rearrangement of energy fluxes in favor of target SM production; (v) stress response—changes in regulation in response to stress during high-yield production; and (vi) metabolic pathways—the redirection of primary metabolic pathways to obtain more precursors for target production. (vii) Also, an additional increase in production during the fermentation of HY strains obtained in classical strain improvement (CSI) programs is possible, with the addition of a number of low-molecular weight compounds that compensate for the effect of side mutations common to CSI programs. As an example, on a magic bullet located near the center of the genomic target, the structural formula of the spermidine, which has been shown to have a stimulating effect on fungal strains, is drawn.
Early work to characterize key differences centered on BGCs responsible for the production of target metabolites [335]. It turned out that, in HY strains, the target BGCs are upregulated tens to hundreds of times compared to the original natural isolates [42,43]. Moreover, BGC duplications were observed in some HY strains, whereas no increase in gene dosage occurred in others [42,156,335,336,337,338,339]. It has been shown that target production does not have a linear correlation with the BGC copy number [340], suggesting that some other modification is also important [88]. Also, changes in production levels could be accompanied by other chromosomal rearrangements, such as translocation [89,156]. Then, as a result of comparative omics analyses of the original isolates and high-yielding strains obtained from them, it was possible to understand some other patterns [341]. For example, a comparative genomic analysis of P. chrysogenum WT (NRRL195) and the HY strain derived from it (DS17690) revealed a wide spread of mutations, that statistically did not result in an over- or underrepresentation of specific gene classes [41]. However, the authors identified some key changes as mutations in genes of global regulators of secondary metabolism, such as velA and laeA, as well as mutations that led to the inactivation of some alternative BGCs. It is known that, during BGC duplication, only one of them can be expressed; the rest are silent. Thus, in a detailed study of the industrial strain P. chrysogenum P2niaD18, which has two copies of the penicillin gene cluster, it was shown that PenG production is not dependent on the copy number of BGCs [342]. DS17690 contains eight PenG-BGC copies. It is possible that mutations in the velA and laeA genes, obtained through random mutagenesis, allowed PenG-BGCs to “escape” epigenetic regulation, which allowed for increased target production.
However, when the system of global regulation of secondary metabolites is disrupted, an increase in the production of alternative secondary metabolites is also possible [343]. Therefore, their inactivation to reduce the levels of impurities and the consumption of cell resources on non-target products is important, which, apparently, can also be selected in SCI programs. Thus, in the improved PenG producer with mutations in global regulation genes, eight out of thirty-one secondary metabolite genes (twenty polyketide synthases and eleven non-ribosomal peptide synthetizes) were targeted, with a corresponding and progressive loss in the production of a range of SMs unrelated to β-lactam production [41]. A comparative transcriptomic analysis was also carried out to determine changes in two phylogenetically distant HY fungal strains, such as P. chrysogenum, belonging to the class of Eurotiomycetes, and A. chrysogenum, belonging to the class of Sordariomycetes [87]. HY strains were compared with the corresponding WT strains and their HY strain lacking the gene for velA gene of global transcriptional regulator. The analysis of RNA-seq data showed that differential gene expression occurred in distinct functional categories in both industrial strains. Moreover, velA knockouts lead to similar transcriptomic rearrangements. The attempts made to understand the general patterns of improvement in these two strains, which are quite far away in evolutionary terms, demonstrate the presence of a certain universal effect on the global regulatory system, which is required and selected during the improvements in fungal HY producers of secondary metabolites.
Another important aspect is the redistribution of energy flows in HY producers in favor of targeted secondary metabolism. For example, the HY strain of A. chrysogenum has approximately 2-fold reduced activity of the plasma membrane H+-ATPase (PMA), compared to the original WT strain [344]. An increase in PMA activity in recombinant HY strains (expressing an additional copy of the Pma1 gene from S. cerevisiae), up to the level of activity in the WT strain and higher, led to a decrease in the production of the target secondary metabolite, CefC. Moreover, a clear correlation was observed: the greater the PMA activity, the more the level of CefC production decreased [344]. These data may be explained by an overlap in the level of ATP consumption by the PMA enzyme and the biosynthetic pathway of CefC. PMA is the main membrane protein of filamentous fungi; it consumes 20–50% of cell ATP [345,346]. CefC biosynthesis in A. chrysogenum also requires ATP [347]. In the improved producer, reduced PMA activity released ATP for CefC biosynthesis. When PMA activity in the recombinant HY clones increased to the level in the original WT strain, CefC production fell approximately 15-fold [344]. Rearrangement of the internal energetic fluxes through the glycolysis and pentose phosphate pathways to the PenG biosynthetic pathway was also shown in a comparative proteomic analysis of P. chrysogenum strains WT (NRRL 1951) and HY (AS-P-78) [88,348].
In addition, a comparison of the proteomes of the two strains showed a significant change in the production of stress response proteins [348]. In the P. chrysogenum HY strain, several proteins involved in the oxidative stress response are overexpressed, such as glutathione reductase, glutathione S-transferase, quinone oxidoreductase, etc. [88]. The authors suggest that the oxidative stress response may serve as an adaptation mechanism for penicillin overproduction [348]. Important rearrangements also occurred in the metabolism of precursor compounds for the PenG biosynthase. On the one hand, a decrease in the production of enzymes involved in their catabolism was observed (cystathionine β-synthase for cysteine catabolism, methylmalonate-semialdehyde dehydrogenase for valine catabolism, and phytanoyl-CoA dioxygenase family protein for fatty acid degradation). On the other hand, there was an increase in the number of enzymes of their biosynthesis (cysteine synthase for cysteine synthesis, branched-chain amino acid aminotransferase for valine synthesis, and dihydroxy-acid dehydratase for branched amino acid synthesis) [88]. Modification also occurs in the metabolic fluxes through the homogentisate pathway for phenylacetic acid catabolism [88].
The listed changes (associated with: (i) the upregulation of the target BGC, (ii) changes in the system of global regulation of secondary metabolism, (iii) the disruption of non-target BGCs, the rearrangement of (iv) energy fluxes and (v) metabolic pathways in favor of target SM production, (vi) the overexpression of proteins involved in the oxidative stress response), by analogy with the German legend of Freeshooter, can be correlated with the result of shooting the first six magic bullets, which led to the production of HY strains (Figure 9). In this analogy, the devil shot the seventh bullet instead of us, and in doing so, the accompanying unwanted mutations were selected. However, recent knowledge associated with the use of targeted supplements of low molecular weight compounds may allow us to compensate for the results of this diabolical shooting and further increase the production of SMs. Understanding at the molecular level the key events that must occur for a fungal strain to become a high-yielding secondary metabolite producer opens up the possibility of the targeted creation of phenotypes of industrial strains [85,349].
8. Perspectives
Fungi produce a significant number of secondary metabolites with antimicrobial properties. Most of them are not commercialized but have significant latent potential for use as an antibiotic agent. The search for new natural antibiotics is an urgent task, due to their widespread consumption and the emergence of resistance. Current research methods enable us, on the one hand, to understand at the molecular level the mechanisms of action of known antibiotics, the emergence of resistance to them, and to make attempts to overcome this resistance in new-generation drugs, and, on the other hand, to intensify the search for new antibiotics.
Also, current approaches, including multiomic, open up the opportunity to understand the key changes that occurred against the background of numerous random mutations during the production of industrial producers of antibiotics. Such knowledge is important for the targeted creation of industrial strains using genetic engineering methods, which, ideally, will not have the “genetic load” that arises in highly active producers due to concomitant mutations.
Understanding the key changes in industrial fungal producers of secondary metabolites also opens up the possibility of selecting targeted additives aimed at compensating the negative effects of a number of associated mutations. One of these compounds seems to be aliphatic polyamines, the exogenous introduction of which can further increase the production of beta-lactams and lovastatin and upregulate the corresponding BGCs genes and laeA, the gene of global regulator of fungal secondary metabolism.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The author declares no conflict of interest.
References
- Aminov, R.I. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, P. Experimental Researches on Specific Therapy: On Immunity with special Reference to the Relationship between Distribution and Action of Antigens: FIRST HARBEN LECTURE. In The Collected Papers of Paul Ehrlich; Elsevier: Amsterdam, The Netherlands, 1960; pp. 106–117. [Google Scholar] [CrossRef]
- Bosch, F.; Rosich, L. The Contributions of Paul Ehrlich to Pharmacology: A Tribute on the Occasion of the Centenary of His Nobel Prize. Pharmacology 2008, 82, 171. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J. The introduction of “chemotherapy” using arsphenamine—the first magic bullet. J. R. Soc. Med. 2009, 102, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Strebhardt, K.; Ullrich, A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat. Rev. Cancer 2008, 8, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, N.C.; Morgan, H.W.; Nicholson, B.K.; Ronimus, R.S. The composition of Ehrlich’s salvarsan: Resolution of a century-old debate. Angew. Chem. Int. Ed. Engl. 2005, 44, 941–944. [Google Scholar] [CrossRef]
- Bayer, E.A.; Fierro, F.; Vaca, I.; Castillo, N.I.; Ovidio García-Rico, R.; Chávez, R. Penicillium chrysogenum, a Vintage Model with a Cutting-Edge Profile in Biotechnology. Microorganisms 2022, 10, 573. [Google Scholar] [CrossRef]
- Sanchez, S.; Demain, A.L. Bioactive Products from Fungi. In Food Bioactives; Springer: Cham, Switzerland, 2017; p. 59. [Google Scholar] [CrossRef]
- Hadacek, F.; Bachmann, G. Low-molecular-weight metabolite systems chemistry. Front. Environ. Sci. 2015, 3, 128574. [Google Scholar] [CrossRef]
- Fouillaud, M.; Dufossé, L. Microbial Secondary Metabolism and Biotechnology. Microorganisms 2022, 10, 123. [Google Scholar] [CrossRef]
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 449147. [Google Scholar] [CrossRef]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 2000, 69, 1–39. [Google Scholar] [CrossRef]
- Maresca, M.; Muchagato Mauricio, E.; Brito, L.; Conrado, R.; Colombo Gomes, T.; Sales Calaço Roque, G.; Olívia De Souza, A. Overview of Bioactive Fungal Secondary Metabolites: Cytotoxic and Antimicrobial Compounds. Antibiotics 2022, 11, 1604. [Google Scholar] [CrossRef]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Brandi, L.; Fabbretti, A.; Pon, C.L. (Eds.) Antibiotics: Targets, Mechanisms and Resistance; Wiley: Hoboken, NJ, USA, 2013; Volume 1, ISBN 978-3-527-33305-9. [Google Scholar]
- Upmanyu, N.; Malviya, V.N. Antibiotics: Mechanisms of action and modern challenges. Microorg. Sustain. Environ. Health 2020, 1, 367–382. [Google Scholar] [CrossRef]
- Ueda, K.; Beppu, T. Antibiotics in microbial coculture. J. Antibiot. 2016, 70, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Clardy, J.; Fischbach, M.A.; Currie, C.R. The natural history of antibiotics. Curr. Biol. 2009, 19, R437. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Moody, S.C. Microbial co-culture: Harnessing intermicrobial signaling for the production of novel antimicrobials. Future Microbiol. 2014, 9, 575–578. [Google Scholar] [CrossRef]
- Zhang, C.; Straight, P.D. Antibiotic discovery through microbial interactions. Curr. Opin. Microbiol. 2019, 51, 64–71. [Google Scholar] [CrossRef]
- Lyddiard, D.; Jones, G.L.; Greatrex, B.W. Keeping it simple: Lessons from the golden era of antibiotic discovery. FEMS Microbiol. Lett. 2016, 363, 84. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Zhgun, A.A. Fungal BGCs for Production of Secondary Metabolites: Main Types, Central Roles in Strain Improvement, and Regulation According to the Piano Principle. Int. J. Mol. Sci. 2023, 24, 11184. [Google Scholar] [CrossRef] [PubMed]
- Felnagle, E.A.; Jackson, E.E.; Chan, Y.A.; Podevels, A.M.; Berti, A.D.; McMahon, M.D.; Thomas, M.G. Nonribosomal Peptide Synthetases Involved in the Production of Medically Relevant Natural Products. Mol. Pharm. 2008, 5, 191. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Chen, H.; Keating, T.A.; Hubbard, B.K.; Losey, H.C.; Luo, L.; Marshall, C.G.; Miller, D.A.; Patel, H.M. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 2001, 5, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, J.D.; Chang, C.Y. Terpene synthases in disguise: Enzymology, structure, and opportunities of non-canonical terpene synthases. Nat. Prod. Rep. 2020, 37, 425–463. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.H.; Tang, Y. Navigating the fungal polyketide chemical space: From genes to molecules. J. Org. Chem. 2012, 77, 9933–9953. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, P.; Hur, J.S.; Yun, H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb. Cell Fact. 2016, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- Ames, B.D.; Nguyen, C.; Bruegger, J.; Smith, P.; Xu, W.; Ma, S.; Wong, E.; Wong, S. Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 11144–11149. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, W.G.; Matsuda, Y. Branching and converging pathways in fungal natural product biosynthesis. Fungal Biol. Biotechnol. 2022, 9, 6. [Google Scholar] [CrossRef]
- Avalos, J.; Limón, M.C. Fungal Secondary Metabolism. Encyclopedia 2021, 2, 1–13. [Google Scholar] [CrossRef]
- Sheehan, J.C.; Buhle, E.L.; Corby, E.J.; Laubach, G.D.; Ryan, J.J. The total synthesis of a 5-phenyl penicillin: Methyl 5-phenyl-(2-carbomethoxyethyl)-penicillinate. J. Am. Chem. Soc. 1950, 72, 3828–3829. [Google Scholar] [CrossRef]
- Wright, P.M.; Seiple, I.B.; Myers, A.G. The Evolving Role of Chemical Synthesis in Antibacterial Drug Discovery. Angew. Chem. Int. Ed. Engl. 2014, 53, 8840. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, K. Total synthesis of the big four antibiotics and related antibiotics. J. Antibiot. 2013, 66, 107–129. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Chakravarty, I.; Ojha, S.; Kundu, K. Design and Development of Antibiotic Fermentation Using Different Processing Strategies: Challenges and Perspectives. In Applied Microbiology and Bioengineering: An Interdisciplinary Approach; Academic Press: Cambridge, MA, USA, 2019; Volume 1, pp. 163–183. [Google Scholar]
- Fedorenko, V.; Genilloud, O.; Horbal, L.; Marcone, G.L.; Marinelli, F.; Paitan, Y.; Ron, E.Z. Antibacterial Discovery and Development: From Gene to Product and Back. Biomed Res. Int. 2015, 2015, 591349. [Google Scholar] [CrossRef] [PubMed]
- Tevyashova, A.N. Recent Trends in Synthesis of Chloramphenicol New Derivatives. Antibiotics 2021, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, J.; Bartz, Q.R.; Smith, R.M.; Joslyn, D.A.; Burkholder, P.R. Chloromycetin, a New Antibiotic From a Soil Actinomycete. Science 1947, 106, 417. [Google Scholar] [CrossRef] [PubMed]
- Boruwa, J.; Borah, J.C.; Gogoi, S.; Barua, N.C. A short asymmetric total synthesis of chloramphenicol using a selectively protected 1,2-diol. Tetrahedron Lett. 2005, 46, 1743–1746. [Google Scholar] [CrossRef]
- Salo, O.V.; Ries, M.; Medema, M.H.; Lankhorst, P.P.; Vreeken, R.J.; Bovenberg, R.A.L.; Driessen, A.J.M. Genomic mutational analysis of the impact of the classical strain improvement program on β-lactam producing Penicillium chrysogenum. BMC Genom. 2015, 16, 937. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Dumina, M.V.; Voinova, T.M.; Dzhavakhiya, V.V.; Eldarov, M.A. Role of acetyl-CoA Synthetase and LovE Regulator Protein of Polyketide Biosynthesis in Lovastatin Production by Wild-Type and Overproducing Aspergillus terreus Strains. Appl. Biochem. Microbiol. 2018, 54, 188–197. [Google Scholar] [CrossRef]
- Dumina, M.V.; Zhgun, A.A.; Novak, M.I.; Domratcheva, A.G.; Petukhov, D.V.; Dzhavakhiya, V.V.; Eldarov, M.A.; Bartoshevitch, I.E. Comparative gene expression profiling reveals key changes in expression levels of cephalosporin C biosynthesis and transport genes between low and high-producing strains of Acremonium chrysogenum. World J. Microbiol. Biotechnol. 2014, 30, 2933–2941. [Google Scholar] [CrossRef]
- Van Den Berg, M.A. Impact of the penicillium chrysogenum genome on industrial production of metabolites. Appl. Microbiol. Biotechnol. 2011, 92, 45–53. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Ivanova, M.A.; Domracheva, A.G.; Novak, M.I.; Elidarov, M.A.; Skryabin, K.G.; Bartoshevich, Y.E. Genetic transformation of the mycelium fungi Acremonium chrysogenum. Appl. Biochem. Microbiol. 2008, 44, 600–607. [Google Scholar] [CrossRef]
- Abe, Y.; Baba, S.; Suzuki, T.; Ono, C.; Iwamoto, K.; Hosobuchi, M. Molecular basis of ML-236B production in the high-producing mutant No. 41520 of Penicillium citrinum. J. Gen. Appl. Microbiol. 2004, 50, 169–176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Domratcheva, A.G.; Zhgun, A.A.; Novak, N.V.; Dzhavakhiya, V.V. The Influence of Chemical Mutagenesis on the Properties of the Cyclosporine a High-Producer Strain Tolypocladium inflatum VKM F-3630D. Appl. Biochem. Microbiol. 2018, 54, 53–57. [Google Scholar] [CrossRef]
- Bartoshevich, Y.; Novak, M.; Domratcheva, A.; Skrybin, K. Method of Cephalosporin C Biosynthesis by Using New Acremonium Chrysogenum Strain RNCM NO F-4081D. Patent. RU2426793C2, 20 August 2011. [Google Scholar]
- Zhgun, A.A.; Nuraeva, G.K.; Volkov, I.A. High-yielding lovastatin producer aspergillus terreus shows increased resistance to inhibitors of polyamine biosynthesis. Appl. Sci. 2020, 10, 8290. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef] [PubMed]
- Gould, K. Antibiotics: From prehistory to the present day. J. Antimicrob. Chemother. 2016, 71, 572–575. [Google Scholar] [CrossRef]
- Podolsky, S.H. The evolving response to antibiotic resistance (1945–2018). Palgrave Commun. 2018, 4, 124. [Google Scholar] [CrossRef]
- Fernandez, R.; Paz, L.I.; Rosato, R.R.; Rosato, A.E. Ceftaroline Is Active against Heteroresistant Methicillin-Resistant Staphylococcus aureus Clinical Strains despite Associated Mutational Mechanisms and Intermediate Levels of Resistance. Antimicrob. Agents Chemother. 2014, 58, 5736. [Google Scholar] [CrossRef]
- Volpato, G.; Rodrigues, R.C.; Fernandez-Lafuente, R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: Drawbacks and perspectives. Curr. Med. Chem. 2010, 17, 3855–3873. [Google Scholar] [CrossRef]
- Srirangan, K.; Orr, V.; Akawi, L.; Westbrook, A.; Moo-Young, M.; Chou, C.P. Biotechnological advances on penicillin G acylase: Pharmaceutical implications, unique expression mechanism and production strategies. Biotechnol. Adv. 2013, 31, 1319–1332. [Google Scholar] [CrossRef] [PubMed]
- Eldarov, M.A.; Sklyarenko, A.V.; Mardanov, A.V.; Beletsky, A.V.; Zhgun, A.A.; Dumina, M.V.; Medvedeva, N.V.; Satarova, D.E.; Ravin, N.V.; Yarockii, S.V. Cephalosporin-acid synthetase of Escherichia coli strain VKPM B-10182: Genomic context, gene identification, producer strain production. Appl. Biochem. Microbiol. 2015, 51, 505–510. [Google Scholar] [CrossRef]
- Leisner, J.J. The Diverse Search for Synthetic, Semisynthetic and Natural Product Antibiotics from the 1940s and up to 1960 Exemplified by a Small Pharmaceutical Player. Front. Microbiol. 2020, 11, 529905. [Google Scholar] [CrossRef] [PubMed]
- Tevyashova, A.N.; Shapovalova, K.S. Potential for the Development of a New Generation of Aminoglycoside Antibiotics. Pharm. Chem. J. 2021, 55, 860–875. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.P.; Balasubramanian, A.; Ramalingam, K. Cephalosporins: An imperative antibiotic over the generations. Int. J. Res. Pharm. Sci. 2020, 11, 623–630. [Google Scholar] [CrossRef][Green Version]
- da Cunha, B.R.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef]
- Blondeau, J.M. Fluoroquinolones: Mechanism of action, classification, and development of resistance. Surv. Ophthalmol. 2004, 49, S73–S78. [Google Scholar] [CrossRef]
- Bañuls, A.-L.; Nguyen, T.V.A.; Nguyen, Q.H.; Nguyen, T.N.A.; Tran, H.H.; Godreuil, S. Antimicrobial resistance: The 70-year arms race between humans and bacteria. In Ecology and Evolution of Infectious Diseases: Pathogen Control and Public Health Management in Low-Income Countries; Roche, B., Ed.; Oxford University Press: Oxford, UK, 2018; Volume 1, pp. 77–90. [Google Scholar]
- Iskandar, K.; Murugaiyan, J.; Halat, D.H.; Hage, S.E.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089. [Google Scholar] [CrossRef]
- Donadio, S.; Maffioli, S.; Monciardini, P.; Sosio, M.; Jabes, D. Antibiotic discovery in the twenty-first century: Current trends and future perspectives. J. Antibiot. 2010, 63, 423–430. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Kautsar, S.A.; Medema, M.H.; Linington, R.G. Microbial natural product databases: Moving forward in the multi-omics era. Nat. Prod. Rep. 2021, 38, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Shim, H. Three Innovations of Next-Generation Antibiotics: Evolvability, Specificity, and Non-Immunogenicity. Antibiotics 2023, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Dannert, C. NextGen microbial natural products discovery. Microb. Biotechnol. 2015, 8, 26–28. [Google Scholar] [CrossRef]
- Prescott, T.A.K.; Hill, R.; Mas-Claret, E.; Gaya, E.; Burns, E. Fungal Drug Discovery for Chronic Disease: History, New Discoveries and New Approaches. Biomolecules 2023, 13, 986. [Google Scholar] [CrossRef]
- Baker, K.V.; Takano, E.; Breitling, R. The “Three Cs” of Novel Antibiotic Discovery and Production through Synthetic Biology: Biosynthetic Gene Clusters, Heterologous Chassis, and Synthetic Microbial Consortia. Adv. Biosyst. 2018, 2, 1800064. [Google Scholar] [CrossRef]
- Cook, M.A.; Wright, G.D. The past, present, and future of antibiotics. Sci. Transl. Med. 2022, 14, eabo7793. [Google Scholar] [CrossRef]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Normansell, I.D. Strain improvement in antibiotic-producing microorganisms. J. Chem. Technol. Biotechnol. 1982, 32, 296–303. [Google Scholar] [CrossRef]
- Lal, R.; Khanna, R.; Kaur, H.; Khanna, M.; Dhingra, N.; Lal, S.; Gartemann, K.H.; Eichenlaub, R.; Ghosh, P.K. Engineering antibiotic producers to overcome the limitations of classical strain improvement programs. Crit. Rev. Microbiol. 1996, 22, 201–255. [Google Scholar] [CrossRef]
- Lal, R.; Khanna, M.; Kaur, H.; Srivastava, N.; Tripathi, K.K.; Lal, S. Rifamycins: Strain improvement program. Crit. Rev. Microbiol. 1995, 21, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Zhgun, A.A. Random Mutagenesis of Filamentous Fungi Stains for High-Yield Production of Secondary Metabolites: The Role of Polyamines. In Genotoxicity and Mutagenicity—Mechanisms and Test Methods, Chapter 2; Soloneski, S., Larramendy, M.L., Eds.; IntechOpen: London, UK, 2021; pp. 25–41. ISBN 978-1-83880-041-3. [Google Scholar]
- Sawant, A.M.; Vamkudoth, K.R. Biosynthetic process and strain improvement approaches for industrial penicillin production. Biotechnol. Lett. 2022, 44, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Crook, N.; Alper, H.S. Classical Strain Improvement. In Engineering Complex Phenotypes in Industrial Strains; Patnaik, R., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2012; Volume 1, pp. 1–33. ISBN 9780470610756. [Google Scholar]
- García-Estrada, C.; Martín, J.F.; Cueto, L.; Barreiro, C. Omics Approaches Applied to Penicillium chrysogenum and Penicillin Production: Revealing the Secrets of Improved Productivity. Genes 2020, 11, 712. [Google Scholar] [CrossRef] [PubMed]
- Divakar, K.; Wijayawardene, N.N.; Boonyuen, N.; Ranaweera, C.B.; De Zoysa, H.K.S.; Padmathilake, R.E.; Nifla, F.; Dai, D.-Q.; Liu, Y.; Suwannarach, N.; et al. OMICS and Other Advanced Technologies in Mycological Applications. J. Fungi 2023, 9, 688. [Google Scholar] [CrossRef]
- Song, P.; Yuan, K.; Qin, T.; Zhang, K.; Ji, X.; Ren, L.; Guan, R.; Wen, J.; Huang, H. Metabolomics profiling reveals the mechanism of increased pneumocandin B0 production by comparing mutant and parent strains. J. Ind. Microbiol. Biotechnol. 2018, 45, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Aris, P.; Yan, L.; Wei, Y.; Chang, Y.; Shi, B.; Xia, X. Conservation of griseofulvin genes in the gsf gene cluster among fungal genomes. G3 Genes|Genomes|Genet. 2022, 12, jkab39. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, R. Engineering complex phenotypes in industrial strains. Biotechnol. Prog. 2008, 24, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xie, L.; Gong, G.; Zhang, W.; Zhu, B.; Hu, Y. De novo comparative transcriptome analysis of Acremonium chrysogenum: High-yield and wild-type strains of cephalosporin C producer. PLoS ONE 2014, 9, e104542. [Google Scholar] [CrossRef]
- Terfehr, D.; Dahlmann, T.A.; Kück, U. Transcriptome analysis of the two unrelated fungal β-lactam producers Acremonium chrysogenum and Penicillium chrysogenum: Velvet-regulated genes are major targets during conventional strain improvement programs. BMC Genom. 2017, 18, 272. [Google Scholar] [CrossRef]
- Barreiro, C.; Martín, J.F.; García-Estrada, C. Proteomics shows new faces for the old penicillin producer Penicillium chrysogenum. J. Biomed. Biotechnol. 2012, 2012, 105109. [Google Scholar] [CrossRef]
- Dumina, M.V.; Zhgun, A.A.; Domracheva, A.G.; Novak, M.I.; El’darov, M.A. Chromosomal polymorphism of Acremonium chrysogenum strains producing cephalosporin C. Russ. J. Genet. 2012, 48, 778–784. [Google Scholar] [CrossRef]
- Martín, J.; García-Estrada, C.; Kosalková, K.; Ullán, R.V.; Albillos, S.M.; Martín, J.-F. The inducers 1,3-diaminopropane and spermidine produce a drastic increase in the expression of the penicillin biosynthetic genes for prolonged time, mediated by the LaeA regulator. Fungal Genet. Biol. 2012, 49, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhgun, A.A.; Nuraeva, G.K.; Dumina, M.V.; Voinova, T.M.; Dzhavakhiya, V.V.; Eldarov, M.A. 1,3-Diaminopropane and Spermidine Upregulate Lovastatin Production and Expression of Lovastatin Biosynthetic Genes in Aspergillus terreus via LaeA Regulation. Appl. Biochem. Microbiol. 2019, 55, 243–254. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Eldarov, M.A. Polyamines Upregulate Cephalosporin C Production and Expression of β-Lactam Biosynthetic Genes in High-Yielding Acremonium chrysogenum Strain. Molecules 2021, 26, 6636. [Google Scholar] [CrossRef] [PubMed]
- Hyvönen, M.T.; Keinänen, T.A.; Nuraeva, G.K.; Yanvarev, D.V.; Khomutov, M.; Khurs, E.N.; Kochetkov, S.N.; Vepsäläinen, J.; Zhgun, A.A.; Khomutov, A.R. Hydroxylamine analogue of agmatine: Magic bullet for arginine decarboxylase. Biomolecules 2020, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Crismaru, C.G.; Salo, O.; Bovenberg, R.A.L.; Driessena, A.J.M. Impact of Classical Strain Improvement of Penicillium rubens on Amino Acid Metabolism during β-Lactam Production. Appl. Environ. Microbiol. 2020, 86, e01561-19. [Google Scholar] [CrossRef] [PubMed]
- Hook, D.J. Production of antibiotics by fermentation. In Basic Biotechnology, 3rd ed.; Ratledge, C., Kristiansen, B., Eds.; Cambridge University Press: Cambridge, UK, 2006; Volume 1, pp. 433–456. ISBN 9780511802409. [Google Scholar]
- Olsufyeva, E.N.; Yankovskaya, V.S. Main trends in the design of semi-synthetic antibiotics of a new generation. Russ. Chem. Rev. 2020, 89, 339. [Google Scholar] [CrossRef]
- Bruggink, A. Synthesis of β-Lactam Antibiotics, 1st ed.; Bruggink, A., Roy, P.D., Eds.; Springer: Dordrecht, The Netherlands, 2001; Volume 1. [Google Scholar]
- Russell, A.D. Types of Antibiotics and Synthetic Antimicrobial Agents. In Hugo and Russell’s Pharmaceutical Microbiology, 7th ed.; Denyer, S.P., Hodges, N.A., Gorman, S.P., Eds.; John Wiley & Sons, Ltd.: Malden, MA, USA, 2007; Volume 1, pp. 152–186. ISBN 9780470988329. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef]
- Hou, L.W.; Giraldo, A.; Groenewald, J.W.; Rämä, T.; Summerbell, R.C.; Huang, G.Z.; Cai, L.; Crous, P.W. Redisposition of acremonium-like fungi in Hypocreales. Stud. Mycol. 2023, 105, 23–203. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Z.; Liu, W.; Ke, X.; Tian, X.; Chu, J. Cephalosporin C biosynthesis and fermentation in Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 2022, 106, 6413–6426. [Google Scholar] [CrossRef]
- Boger, D.L.; Kim, S.H.; Mori, Y.; Weng, J.H.; Rogel, O.; Castle, S.L.; McAtee, J.J. First and second generation-total synthesis of the teicoplanin aglycon. J. Am. Chem. Soc. 2001, 123, 1862–1871. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, A.S.; Alani, B.G.; Ibrahim, I.T.; Mahdi, A.S.; Alani, B.G.; Ibrahim, I.T. Oxazolidinones Antibiotics, Chemistry, Applications and Role in COVID-19 Treatment. Pharmacol. Pharm. 2023, 14, 19–32. [Google Scholar] [CrossRef]
- Correia, J.; Borges, A.; Simões, M.; Simões, L.C. Beyond Penicillin: The Potential of Filamentous Fungi for Drug Discovery in the Age of Antibiotic Resistance. Antibiotics 2023, 12, 1250. [Google Scholar] [CrossRef] [PubMed]
- Powers-Fletcher, M.V.; Kendall, B.A.; Griffin, A.T.; Hanson, K.E. Filamentous Fungi. Microbiol. Spectr. 2016, 4, 1–29. [Google Scholar] [CrossRef]
- Blackwell, M. The fungi: 1, 2, 3... 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.A.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C.; Day, N.P.J.; et al. Global antibiotic consumption and usage in humans, 2000–2018: A spatial modelling study. Lancet Planet. Health 2021, 5, e893–e904. [Google Scholar] [CrossRef] [PubMed]
- Dicker, D.; Nguyen, G.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; et al. Global, regional, and national age-sex-specific mortality and life expectancy, 1950-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1684–1735. [Google Scholar] [CrossRef]
- Guzmán-Chávez, F.; Zwahlen, R.D.; Bovenberg, R.A.L.; Driessen, A.J.M. Engineering of the filamentous fungus penicillium chrysogenumas cell factory for natural products. Front. Microbiol. 2018, 9, 2768. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Choudhry, H.; Asseri, A.H.; Elfaky, M.A.; Mohamed, S.G.A.; Mohamed, G.A. Stachybotrys chartarum—A Hidden Treasure: Secondary Metabolites, Bioactivities, and Biotechnological Relevance. J. Fungi 2022, 8, 504. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Sirwi, A.; Eid, B.G.; Mohamed, S.G.A.; Mohamed, G.A. Bright Side of Fusarium oxysporum: Secondary Metabolites Bioactivities and Industrial Relevance in Biotechnology and Nanotechnology. J. Fungi 2021, 7, 943. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Regulation of secondary metabolism in fungi. Pure Appl. Chem. 1986, 58, 219–226. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Steiniger, C.; Cairns, T.C.; Wisecaver, J.H.; Lind, A.; Pohl, C.; Regner, C.; Rokas, A.; Meyer, V. Beyond the biosynthetic gene cluster paradigm: Genome-wide co-expression networks connect clustered and unclustered transcription factors to secondary metabolic pathways. bioRxiv 2020. bioRxiv2020.04.15.040477. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Duban, M.; Cociancich, S.; Leclère, V. Nonribosomal Peptide Synthesis Definitely Working out of the Rules. Microorganisms 2022, 10, 577. [Google Scholar] [CrossRef] [PubMed]
- Cummings, M.; Breitling, R.; Takano, E. Steps towards the synthetic biology of polyketide biosynthesis. Fems Microbiol. Lett. 2014, 351, 116. [Google Scholar] [CrossRef]
- Yuan, Y.; Cheng, S.; Bian, G.; Yan, P.; Ma, Z.; Dai, W.; Chen, R.; Fu, S.; Huang, H.; Chi, H.; et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi. Nat. Catal. 2022, 5, 277–287. [Google Scholar] [CrossRef]
- Kjærbølling, I.; Mortensen, U.H.; Vesth, T.; Andersen, M.R. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal Genet. Biol. 2019, 130, 107–121. [Google Scholar] [CrossRef]
- Youssef, A.-K.; Anwar, S.; Ghareeb, D.A.; Hur, J.Y.; Jeong, E.; Kim, Y.C.; Lee, S.R. Strategies for Natural Product Discovery by Unlocking Cryptic Biosynthetic Gene Clusters in Fungi. Separations 2023, 10, 333. [Google Scholar] [CrossRef]
- antiSMASH Fungal Version. Available online: https://fungismash.secondarymetabolites.org/#!/start (accessed on 29 November 2023).
- Terlouw, B.R.; Blin, K.; Navarro-Muñoz, J.C.; Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; et al. MIBiG 3.0: A community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023, 51, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Mózsik, L.; Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Transcriptional Activation of Biosynthetic Gene Clusters in Filamentous Fungi. Front. Bioeng. Biotechnol. 2022, 10, 901037. [Google Scholar] [CrossRef] [PubMed]
- Pillay, L.C.; Nekati, L.; Makhwitine, P.J.; Ndlovu, S.I. Epigenetic Activation of Silent Biosynthetic Gene Clusters in Endophytic Fungi Using Small Molecular Modifiers. Front. Microbiol. 2022, 13, 815008. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ran, H.; Fan, J.; Keller, N.P.; Liu, Z.; Wu, F.; Yin, W.B. Fungal-fungal cocultivation leads to widespread secondary metabolite alteration requiring the partial loss-of-function VeA1 protein. Sci. Adv. 2022, 8, 6094. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Gacek, A.; Strauss, J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 2012, 95, 1389–1404. [Google Scholar] [CrossRef] [PubMed]
- Pfannenstiel, B.T.; Keller, N.P. On top of biosynthetic gene clusters: How epigenetic machinery influences secondary metabolism in fungi. Biotechnol. Adv. 2019, 37, 107345. [Google Scholar] [CrossRef]
- Spagnolo, F.; Trujillo, M.; Dennehy, J.J. Why Do Antibiotics Exist? MBio 2021, 12, e01966-21. [Google Scholar] [CrossRef]
- O’Brien, E.M.; Morgan, B.J.; Mulrooney, C.A.; Carroll, P.J.; Kozlowski, M.C. Perylenequinone Natural Products: Total Synthesis of Hypocrellin A. J. Org. Chem. 2010, 75, 57. [Google Scholar] [CrossRef]
- Dikshit, R.; Tallapragada, P. Development and screening of mutants from Monascus sanguineus for secondary metabolites production. Beni-Suef Univ. J. Basic Appl. Sci. 2018, 7, 235–240. [Google Scholar] [CrossRef]
- Seidl, V.; Seibel, C.; Kubicek, C.P.; Schmoll, M. Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. USA 2009, 106, 13909–13914. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Sugui, J.A. Sexual reproduction in Aspergillus species of medical or economical importance: Why so fastidious? Trends Microbiol. 2009, 17, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Böhm, J.; Hoff, B.; O’Gorman, C.M.; Wolfers, S.; Klix, V.; Binger, D.; Zadra, I.; Kürnsteiner, H.; Pöggeler, S.; Dyer, P.S.; et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA 2013, 110, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Krijgsheld, P.; Bleichrodt, R.; van Veluw, G.J.; Wang, F.; Müller, W.H.; Dijksterhuis, J.; Wösten, H.A.B. Development in aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Clutterbuck, A.J. Parasexual recombination in fungi. J. Genet. 1996, 75, 281–286. [Google Scholar] [CrossRef]
- Bodie, E.A.; Armstrong, G.L.; Dunn-Coleman, N.S. Strain improvement of chymosin-producing strains of Aspergillus niger var. awamori using parasexual recombination. Enzyme Microb. Technol. 1994, 16, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Sarangbin, S.; Morikawa, S.; Kirimura, K.; Usami, S. Formation of autodiploid strains in Aspergillus niger and their application to citric acid production from starch. J. Ferment. Bioeng. 1994, 77, 474–478. [Google Scholar] [CrossRef]
- McLean, K.J.; Hans, M.; Meijrink, B.; van Scheppingen, W.B.; Vollebregt, A.; Tee, K.L.; van der Laan, J.-M.; Leys, D.; Munro, A.W.; van den Berg, M.A. Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum. Proc. Natl. Acad. Sci. USA 2015, 112, 2847–2852. [Google Scholar] [CrossRef]
- Anyaogu, D.C.; Mortensen, U.H. Heterologous production of fungal secondary metabolites in Aspergilli. Front. Microbiol. 2015, 6, 77. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; de Vries, R.P.; Garrigues, S. Strategies for the Development of Industrial Fungal Producing Strains. J. Fungi 2023, 9, 834. [Google Scholar] [CrossRef]
- Nevalainen, K.M.H. Strain improvement in filamentous fungi-an overview. Appl. Mycol. Biotechnol. 2001, 1, 289–304. [Google Scholar] [CrossRef]
- Wang, Q.; Zhong, C.; Xiao, H. Genetic Engineering of Filamentous Fungi for Efficient Protein Expression and Secretion. Front. Bioeng. Biotechnol. 2020, 8, 526947. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.J.; Hu, S.; Wang, B.T.; Jin, L. Advances in Genetic Engineering Technology and Its Application in the Industrial Fungus Aspergillus oryzae. Front. Microbiol. 2021, 12, 644404. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A.B. Filamentous fungi for the production of enzymes, chemicals and materials. Curr. Opin. Biotechnol. 2019, 59, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Dzhavakhija, V.G.; Voinova, T.M.; Vavilova, N.A.; Santsevich, N.I.; Vinokurova, N.G.; Kadomtseva, V.M.; Dzhavakhija, V.V.; Mishin, A.G. Fungus Strain Aspergillus Terreus 44–62 as Producer of Lovastatin, Industrial Method for Isolation of Lovastatin and Method for Lactoninization of. Statins. Patent RU 2261901C2, 10 October 2003. [Google Scholar]
- Smith, A.W.; Collis, K.; Ramsden, M.; Fox, H.M.; Peberdy, J.F. Chromosome rearrangements in improved cephalosporin C-producing strains of Acremonium chrysogenum. Curr. Genet. 1991, 19, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Zolan, M.E. Chromosome-length polymorphism in fungi. Microbiol. Rev. 1995, 59, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Kbvresearch.com. Available online: https://www.kbvresearch.com/antibiotics-market/ (accessed on 17 November 2023).
- Latinoamericana, R.; Campos Muñiz, C.; Cuadra Zelaya, T.E.; Esquivel, G.R.; Fernández, F.J. Penicillin and cephalosporin production: A historical perspective. Rev. Latinoam. Microbiol. 2007, 49, 88–98. [Google Scholar]
- Pandey, N.; Cascella, M. Beta-Lactam Antibiotics, 1st ed.; StatPearls Publishing: St. Petersburg, FL, USA, 2023; Volume 1. [Google Scholar]
- Binod, P.; Sindhu, R.; Pandey, A. Production of antibiotics by filamentous fungi. In Current Developments in Biotechnology and Bioengineering: Filamentous Fungi Biorefinery; Taherzadeh, M.J., Ferreira, J.A., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; Volume 1, pp. 477–496. ISBN 9780323918725. [Google Scholar]
- De Rosa, M.; Verdino, A.; Soriente, A.; Marabotti, A. The Odd Couple(s): An Overview of Beta-Lactam Antibiotics Bearing More Than One Pharmacophoric Group. Int. J. Mol. Sci. 2021, 22, 617. [Google Scholar] [CrossRef]
- Zhgun, A.; Dumina, M.; Valiakhmetov, A.; Eldarov, M. The critical role of plasma membrane H+-ATPase activity in cephalosporin C biosynthesis of Acremonium chrysogenum. PLoS ONE 2020, 15, e0238452. [Google Scholar] [CrossRef]
- Martín, J.F. Insight into the Genome of Diverse Penicillium chrysogenum Strains: Specific Genes, Cluster Duplications and DNA Fragment Translocations. Int. J. Mol. Sci. 2020, 21, 3936. [Google Scholar] [CrossRef]
- Why the Most Dreaded Injection is Called the “Peanut Butter” Shot. Available online: https://www.military.com/off-duty/2020/02/10/why-most-dreaded-injection-called-peanut-butter-shot.html (accessed on 13 September 2023).
- BICILLIN L-A-Penicillin g Benzathine Injection, Suspension. Available online: https://labeling.pfizer.com/showlabeling.aspx?id=691 (accessed on 13 September 2023).
- Elander, R.P. Strain Improvement and Preservation of β-Lactam-Producing Microorganisms. In Antibiotics: Containing the Beta-Lactam Structure; Demain, A.L., Solomon, N.A., Eds.; Springer: Berlin, Heidelberg, 1983; Volume 1, pp. 97–146. [Google Scholar]
- Laich, F.; Fierro, F.; Martín, J.F. Production of Penicillin by Fungi Growing on Food Products: Identification of a Complete Penicillin Gene Cluster in Penicillium griseofulvum and a Truncated Cluster in Penicillium verrucosum. Appl. Environ. Microbiol. 2002, 68, 1211. [Google Scholar] [CrossRef] [PubMed]
- Aharonowitz, Y.; Cohen, G.; Martin, J.F. Penicillin and cephalosporin biosynthetic genes: Structure, organization, regulation, and evolution. Annu. Rev. Microbiol. 1992, 46, 461–495. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Liras, P. Transfer of Secondary Metabolite Gene Clusters: Assembly and Reorganization of the Β-Lactam Gene Cluster from Bacteria to Fungi and Arthropods. In Horizontal Gene Transfer: Breaking Borders between Living Kingdoms; Villa, T.G., Viñas, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 1, pp. 337–359. ISBN 9783030218621. [Google Scholar]
- Bud, R. Innovators, deep fermentation and antibiotics: Promoting applied science before and after the Second World War. Dynamis 2011, 31, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.S.; Bovenberg, R.A.L.; Driessen, A.J.M. Biosynthetic concepts for the production of β-lactam antibiotics in Penicillium chrysogenum. Biotechnol. J. 2012, 7, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Newbert, R.W.; Barton, B.; Greaves, P.; Harper, J.; Turner, G. Analysis of a commercially improved Penicillium chrysogenum strain series: Involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J. Ind. Microbiol. Biotechnol. 1997, 19, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Penicillium Strain Named State Microbe of Illinois. Available online: https://www.ars.usda.gov/news-events/news/research-news/2021/penicillium-strain-named-state-microbe-of-illinois/ (accessed on 13 September 2023).
- Backus, M.P.; Stauffer, J.F. The Production and Selection of a Family of Strains in Penicillium chrysogenum. Mycologia 1955, 47, 429–463. [Google Scholar] [CrossRef]
- Perlman, D. Some mycological aspects of penicillin production. Bot. Rev. 1950, 16, 449–523. [Google Scholar] [CrossRef]
- Elander, R.P. Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 2003, 61, 385–392. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Liu, C.; Hang, H.; Chu, J.; Guo, M.; Zhuang, Y. Method for Increasing Penicillin Fermentation Production Unit. China Patent CN113388658A, 14 September 2021. [Google Scholar]
- Jørgensen, H.; Nielsen, J.; Villadsen, J.; Møllgaard, H. Analysis of penicillin V biosynthesis during fed-batch cultivations with a high-yielding strain of Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 1995, 43, 123–130. [Google Scholar] [CrossRef]
- Duan, S.; Yuan, G.; Zhao, Y.; Ni, W.; Luo, H.; Shi, Z.; Liu, F. Simulation of computational fluid dynamics and comparison of cephalosporin C fermentation performance with different impeller combinations. Korean J. Chem. Eng. 2013, 30, 1097–1104. [Google Scholar] [CrossRef]
- Yang, Y.; Xia, J.; Li, J.; Chu, J.; Li, L.; Wang, Y.; Zhuang, Y.; Zhang, S. A novel impeller configuration to improve fungal physiology performance and energy conservation for cephalosporin C production. J. Biotechnol. 2012, 161, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Godtfredsen, W.O.; Jahnsen, S.; Lorck, H.; Roholt, K.; Tybring, L. Fusidic acid: A new antibiotic. Nature 1962, 193, 987. [Google Scholar] [CrossRef] [PubMed]
- Verbist, L. The antimicrobial activity of fusidic acid. J. Antimicrob. Chemother. 1990, 25, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.W.; Ge, X.Y.; Huang, Y.; Chai, X.T.; Zhang, L.; Zhang, Y.X.; Deng, L.N.; Liu, C.Q.; Xu, H.; Gao, J. High-yield strain of fusidic acid obtained by atmospheric and room temperature plasma mutagenesis and the transcriptional changes involved in improving its production in fungus Fusidium coccineum. J. Appl. Microbiol. 2021, 130, 405–415. [Google Scholar] [CrossRef]
- Venkata Dasu, V.; Panda, T.; Chidambaram, M. Determination of significant parameters for improved griseofulvin production in a batch bioreactor by Taguchi’s method. Process Biochem. 2003, 38, 877–880. [Google Scholar] [CrossRef]
- K1yoshi, U.; Abe, S. Production of Griseofulvin by Some Strains of the Genus Penicillum. J. Antibiot. Ser. A 1961, 14, 215–220. [Google Scholar] [CrossRef]
- Nigam, P.; Singh, D. METABOLIC PATHWAYS|Production of Secondary Metabolites—Fungi. Encycl. Food Microbiol. 1999, 2, 1319–1328. [Google Scholar] [CrossRef]
- Song, P.; Zhang, K.; Zhang, S.; Huang, B.Q.; Ji, X.J.; Ren, L.J.; Gao, S.; Wen, J.P.; Huang, H. Enhancement of Pneumocandin B0 Production in Glarea lozoyensis by Low-Temperature Adaptive Laboratory Evolution. Front. Microbiol. 2018, 9, 2788. [Google Scholar] [CrossRef]
- Hu, Z.C.; Li, W.J.; Zou, S.P.; Niu, K.; Zheng, Y.G. Mutagenesis of echinocandin B overproducing Aspergillus nidulans capable of using starch as main carbon source. Prep. Biochem. Biotechnol. 2020, 50, 745–752. [Google Scholar] [CrossRef]
- Men, P.; Zhou, Y.; Xie, L.; Zhang, X.; Zhang, W.; Huang, X.; Lu, X. Improving the production of the micafungin precursor FR901379 in an industrial production strain. Microb. Cell Fact. 2023, 22, 44. [Google Scholar] [CrossRef]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus Glob. Mycol. J. 2011, 2, 87. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.J.; Doña, I.; Fernández, T.D.; Bogas, G. Penicillin. In Cutaneous Drug Hypersensitivity: Clinical Features, Mechanisms, Diagnosis, and Management; Bircher, A.J., Maibach, H.I., Brockow, K., Barbaud, A., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2022; Volume 1, pp. 169–176. ISBN 9783030827434. [Google Scholar]
- Ball, A.P.; Gray, J.A.; Murdoch, J.M. The Natural Penicillins—Benzylpenicillin (Penicillin G) and Phenoxymethylpenicillin (Penicillin V). In Antibacterial Drugs Today; Springer: Dordrecht, The Netherlands, 1978; Volume 1, pp. 6–18. [Google Scholar]
- Perron, Y.G.; Minor, W.F.; Holdrege, C.T.; Gottstein, W.J.; Godfrey, J.C.; Crast, L.B.; Babel, R.B.; Cheney, L.C. Derivatives of 6-Aminopenicillanic Acid. I. Partially Synthetic Penicillins Prepared from α-Aryloxyalkanoic Acids. J. Am. Chem. Soc. 1960, 82, 3934–3938. [Google Scholar] [CrossRef]
- Güngör, Ö.Ö.N.; Gürkan, P.; Özçelik, B.; Oyardi, Ö. Synthesis and antimicrobial activities of new higher amino acid Schiff base derivatives of 6-aminopenicillanic acid and 7-aminocephalosporanic acid. J. Mol. Struct. 2016, 1106, 181–191. [Google Scholar] [CrossRef]
- Cheptea, C.; Zara, A.; Dimitriu, D.G.; Sunel, V.; Dorohoi, D.O.; Cigu, T.A. New Semisynthetic Penicillins Obtained by Coupling of the 6-Aminopenicillanic Acid with 5-Mercapto-1,2,4-triazoles-3,4-disubstituted. Int. J. Mol. Sci. 2023, 24, 24. [Google Scholar] [CrossRef] [PubMed]
- Bruggink, A.; Roy, P.D. Industrial Synthesis of Semisynthetic Antibiotics. In Synthesis of β-Lactam Antibiotics; Bruggink, A., Ed.; Springer: Dordrecht, The Netherlands, 2001; Volume 1, pp. 12–54. [Google Scholar]
- Möller, J.; Niehoff, J.; Dors, M.; Hiddessen, R.; Schügerl, K. Influence of medium composition on penicillin V production in a stirred tank reactor. J. Biotechnol. 1992, 25, 245–259. [Google Scholar] [CrossRef]
- Chang, L.T.; McGrory, E.L.; Elander, R.P.; Hook, D.J. Decreased production of para-hydroxypenicillin V in penicillin V fermentations. J. Ind. Microbiol. 1991, 7, 175–179. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Velasco, J.; Fernandez, F.J.; Martín, J.F. The cefG gene of Cephalosporium acremonium is linked to the cefEF gene and encodes a deacetylcephalosporin C acetyltransferase closely related to homoserine O-acetyltransferase. J. Bacteriol. 1992, 174, 3056. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Fierro, F.; Casqueiro, J.; Martín, J.F. Gene organization and plasticity of the beta-lactam genes in different filamentous fungi. Antonie Leeuwenhoek 1999, 75, 81–94. [Google Scholar] [CrossRef]
- Newton, G.G.; Abraham, E.P. Isolation of cephalosporin C, a penicillin-like antibiotic containing D-alpha-aminoadipic acid. Biochem. J. 1956, 62, 651–658. [Google Scholar] [CrossRef]
- Acremonium chrysogenum (Thirumalachar et Sukapure) Gams—11550|ATCC. Available online: https://www.atcc.org/products/11550 (accessed on 22 September 2023).
- Brakhage, A.A. (Ed.) Molecular Biotechnolgy of Fungal Beta-Lactam Antibiotics and Related Peptide Synthetases. In Advances in Biochemical Engineering/Biotechnology, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 88, ISBN 978-3-540-22032-9. [Google Scholar]
- García-Estrada, C.; Martín, J.-F. Penicillins and Cephalosporins. In Comprehensive Biotechnology; Moo-Young, M., Ed.; Academic Press: Burlington, VT, USA, 2011; Volume 1, pp. 255–268. ISBN 978-0-08-088504-9. [Google Scholar]
- Ehl’darov, M.A.; Redo, V.A.; Zhgun, A.A.; Zejnalov, O.A.; Skrjabin, K.G. RU 231 68 C1 pETTvDAO2 RECOMBINANT PLASMID PROVIDING SYNTHESIS OF YEAST Trigonopsis variabilis D-AMINO ACID OXYDASE (DAO) IN Escherichia coli CELLS AND RECOMBINANT Escherichia coli STRAIN C41(DE3)/pETTvDAO2 AS PRODUCER OF DAO. Patent RU23168C1, 17 March 2006. [Google Scholar]
- Lee, S.K.; Park, S.W.; Kim, Y.I.; Chung, K.H.; Hong, S.I.; Kim, S.W. Immobilization of GL-7-ACA Acylase for the Production of 7-ACA. Korean J. Chem. Eng. 2002, 19, 261–266. [Google Scholar] [CrossRef]
- Deaguero, A.L.; Blum, J.K.; Bommarius, A.S. Biocatalytic synthesis of β-lactam antibiotics. In Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology; John Wiley Sons, Inc.: Hoboken, NJ, USA, 2010; pp. 1–32. [Google Scholar]
- Eldarov, M.A.; Sklyarenko, A.V.; Dumina, M.; Medvedeva, N.V.; Jgoun, A.A.; Satarova, J.E.; Sidorenko, A.I.; Emperian, A.S.; Yarotsky, S.V. Recombinant cephalosporin-acid synthesase: Optimisation of expression in E. coli cells, immobilisation and application for biocatalytic cefazolin synthesis. Biomed. Khim. 2015, 61, 646–651. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Falagas, M.E.; Grammatikos, A.P.; Michalopoulos, A. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev. Anti. Infect. Ther. 2008, 6, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Beekman, A.M.; Barrow, R.A.; Beekman, A.M.; Barrow, R.A. Fungal Metabolites as Pharmaceuticals. Aust. J. Chem. 2014, 67, 827–843. [Google Scholar] [CrossRef]
- Kavanagh, F.; And, A.H.; Robbins, W.J. Antibiotic Substances from Basidiomycetes: VIII. Pleurotus Multilus (Fr.) Sacc. and Pleurotus Passeckerianus Pilat*. Proc. Natl. Acad. Sci. USA 1951, 37, 570. [Google Scholar] [CrossRef] [PubMed]
- Hartley, A.J.; De Mattos-Shipley, K.; Collins, C.M.; Kilaru, S.; Foster, G.D.; Bailey, A.M. Investigating pleuromutilin-producing Clitopilus species and related basidiomycetes. FEMS Microbiol. Lett. 2009, 297, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Watkins, R.R.; File, T.M. Lefamulin: A Novel Semisynthetic Pleuromutilin Antibiotic for Community-acquired Bacterial Pneumonia. Clin. Infect. Dis. 2020, 71, 2757–2762. [Google Scholar] [CrossRef] [PubMed]
- Szymański, M.; Chmielewska, S.; Czyżewska, U.; Malinowska, M.; Tylicki, A. Echinocandins—Structure, mechanism of action and use in antifungal therapy. J. Enzyme Inhib. Med. Chem. 2022, 37, 876. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; Hassan, K.; Kemkuignou, B.M.; Čmoková, A.; Surup, F.; Kuhnert, E.; Paomephan, P.; Cheng, T.; de Hoog, S.; et al. Ten decadal advances in fungal biology leading towards human well-being. Fungal Divers. 2022, 116, 547–614. [Google Scholar] [CrossRef]
- European Medicines Agency—Human Medicines—Fusafungine Containing Medicinal Products for Oromucosal and Nasal Use. Available online: https://web.archive.org/web/20180620220021/http://www.ema.europa.eu/ema//index.jsp?curl=pages%2Fmedicines%2Fhuman%2Freferrals%2FFusafungine_for_oromucosal_and_nasal_use%2Fhuman_referral_prac_000054.jsp&mid=WC0b01ac05805c516f (accessed on 26 September 2023).
- Mast, Y.; Stegmann, E. Actinomycetes: The Antibiotics Producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef]
- Taurino, C.; Frattini, L.; Marcone, G.L.; Gastaldo, L.; Marinelli, F. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Fact. 2011, 10, 82. [Google Scholar] [CrossRef]
- Stapley, E.O.; Jackson, M.; Hernandez, S.; Zimmerman, S.B.; Currie, S.A.; Mochales, S.; Mata, J.M.; Woodruff, H.B.; Hendlin, D. Cephamycins, a New Family of β-Lactam Antibiotics I. Production by Actinomycetes, Including Streptomyces lactamdurans sp. Antimicrob. Agents Chemother. 1972, 2, 122. [Google Scholar] [CrossRef] [PubMed]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.P.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Badshah, S.L.; Muska, M.; Ahmad, N.; Khan, K. The Current Case of Quinolones: Synthetic Approaches and Antibacterial Activity. Molecules 2016, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Ovung, A.; Bhattacharyya, J. Sulfonamide drugs: Structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021, 13, 259. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.M.; Hu, D.; Gao, H.; Kushiro, T.; Awakawa, T.; Chen, G.D.; Wang, C.X.; Abe, I.; Yao, X.S. Biosynthesis of helvolic acid and identification of an unusual C-4-demethylation process distinct from sterol biosynthesis. Nat. Commun. 2017, 8, 1644. [Google Scholar] [CrossRef] [PubMed]
- Bills, G.F.; Platas, G.; Gams, W. Conspecificity of the cerulenin and helvolic acid producing ‘Cephalosporium caerulens’, and the hypocrealean fungus Sarocladium oryzae. Mycol. Res. 2004, 108, 1291–1300. [Google Scholar] [CrossRef]
- Chou, T.S.; Eisenbraun, E.J.; Rapala, R.T. The chemistry of cephalosporin P1. Tetrahedron Lett. 1967, 8, 409–414. [Google Scholar] [CrossRef]
- Sy-Cordero, A.A.; Pearce, C.J.; Oberlies, N.H. Revisiting the enniatins: A review of their isolation, biosynthesis, structure determination and biological activities. J. Antibiot. 2012, 65, 541–549. [Google Scholar] [CrossRef]
- Pelaez, F.; Cabello, A.; Platas, G.; Díez, M.T.; Del Val, A.G.; Basilio, A.; Martán, I.; Vicente, F.; Bills, G.F.; Giacobbe, R.A.; et al. The discovery of enfumafungin, a novel antifungal compound produced by an endophytic Hormonema species biological activity and taxonomy of the producing organisms. Syst. Appl. Microbiol. 2000, 23, 333–343. [Google Scholar] [CrossRef]
- Ali, H.; Ries, M.I.; Lankhorst, P.P.; Van Der Hoeven, R.A.M.; Schouten, O.L.; Noga, M.; Hankemeier, T.; Van Peij, N.N.M.E.; Bovenberg, R.A.L.; Vreeken, R.J.; et al. A non-canonical NRPS is involved in the synthesis of fungisporin and related hydrophobic cyclic tetrapeptides in Penicillium chrysogenum. PLoS ONE 2014, 9, e98212. [Google Scholar] [CrossRef]
- Saleh, I.; Goktepe, I. The characteristics, occurrence, and toxicological effects of patulin. Food Chem. Toxicol. 2019, 129, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Saniewski, M.; Horbowicz, M.; Kanlayanarat, S. The biological activities of troponoids and their use in agriculture a review. J. Hortic. Res. 2014, 22, 5–19. [Google Scholar] [CrossRef]
- Vicente, M.F.; Cabello, A.; Platas, G.; Basilio, A.; Díez, M.T.; Dreikorn, S.; Giacobbe, R.A.; Onishi, J.C.; Meinz, M.; Kurtz, M.B.; et al. Antimicrobial activity of ergokonin A from Trichoderma longibrachiatum. J. Appl. Microbiol. 2001, 91, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Prompanya, C.; Dethoup, T.; Bessa, L.J.; Pinto, M.M.M.; Gales, L.; Costa, P.M.; Silva, A.M.S.; Kijjoa, A. New Isocoumarin Derivatives and Meroterpenoids from the Marine Sponge-Associated Fungus Aspergillus similanensis sp. nov. KUFA 0013. Mar. Drugs 2014, 12, 5160. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.H.; Cacho, R.; Tang, Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 2010, 17, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, J.; Song, Z.; Zhu, G.; Liu, M.; Dai, H.; Hsiang, T.; Liu, X.; Zhang, L.; Quinn, R.J.; et al. Genome-based mining of new antimicrobial meroterpenoids from the phytopathogenic fungus Bipolaris sorokiniana strain 11134. Appl. Microbiol. Biotechnol. 2020, 104, 3835–3846. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Kim, W.S.; Fang, A.; Demain, A.L. Carbon and nitrogen source nutrition of fumagillin biosynthesis by Aspergillus fumigatus. Curr. Microbiol. 2003, 46, 275–279. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, L.; Li, X.; Lv, R.; Cao, W.; Gao, W.; Liu, J.; Xie, Z.; Liu, H. Effective production of kojic acid in engineered Aspergillus niger. Microb. Cell Fact. 2023, 22, 40. [Google Scholar] [CrossRef]
- Aris, P.; Wei, Y.; Mohamadzadeh, M.; Xia, X. Griseofulvin: An Updated Overview of Old and Current Knowledge. Molecular 2022, 27, 7034. [Google Scholar] [CrossRef]
- Oxford, A.E.; Raistrick, H.; Simonart, P. Studies in the biochemistry of micro-organisms: Griseofulvin, C(17)H(17)O(6)Cl, a metabolic product of Penicillium griseo-fulvum Dierckx. Biochem. J. 1939, 33, 240–248. [Google Scholar] [CrossRef]
- Venkata Dasu, V.; Panda, T. Studies on production of griseofulvin. Bioprocess Eng. 1999, 21, 489–495. [Google Scholar] [CrossRef]
- Drug and Biologic Approval and IND Activity Reports > NME Drug and New Biologic Approvals in 2007. Available online: https://wayback.archive-it.org/7993/20170406061415/https://www.fda.gov/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/DrugandBiologicApprovalReports/ucm081690.htm (accessed on 26 September 2023).
- Jones, R.N.; Fritsche, T.R.; Sader, H.S.; Ross, J.E. Activity of Retapamulin (SB-275833), a Novel Pleuromutilin, against Selected Resistant Gram-Positive Cocci. Antimicrob. Agents Chemother. 2006, 50, 2583. [Google Scholar] [CrossRef] [PubMed]
- Robert Daum, R.S.; Kar, S.; Kirkpatrick Retapamulin, P. Retapamulin. Nat. Rev. Drug Discov. 2007, 6, 865–866. [Google Scholar] [CrossRef]
- Dillon, C.; Guarascio, A.J.; Covvey, J.R. Lefamulin: A promising new pleuromutilin antibiotic in the pipeline. Expert Rev. Anti. Infect. Ther. 2019, 17, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Charles, C.V. A Review of Newly Approved Antibiotic Treatment for Community-Acquired Bacterial Pneumonia: Lefamulin. Sr. Care Pharm. 2020, 35, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yu, B.; Liu, H.M. New drug approvals for 2019: Synthesis and clinical applications. Eur. J. Med. Chem. 2020, 205, 112667. [Google Scholar] [CrossRef]
- Waites, K.B.; Crabb, D.M.; Duffy, L.B.; Jensen, J.S.; Liu, Y.; Paukner, S. In Vitro Activities of Lefamulin and Other Antimicrobial Agents against Macrolide-Susceptible and Macrolide-Resistant Mycoplasma pneumoniae from the United States, Europe, and China. Antimicrob. Agents Chemother. 2017, 61, 10-1128. [Google Scholar] [CrossRef]
- Emri, T.; Majoros, L.; Tóth, V.; Pócsi, I. Echinocandins: Production and applications. Appl. Microbiol. Biotechnol. 2013, 97, 3267–3284. [Google Scholar] [CrossRef]
- Denning, D.W. Echinocandins: A new class of antifungal. J. Antimicrob. Chemother. 2002, 49, 889–891. [Google Scholar] [CrossRef]
- Debono, M.; Gordee, R.S. Antibiotics that inhibit fungal cell wall development. Annu. Rev. Microbiol. 1994, 48, 471–497. [Google Scholar] [CrossRef]
- Yue, Q.; Chen, L.; Zhang, X.; Li, K.; Sun, J.; Liu, X.; An, Z.; Bills, G.F. Evolution of Chemical Diversity in Echinocandin Lipopeptide Antifungal Metabolites. Eukaryot. Cell 2015, 14, 698–718. [Google Scholar] [CrossRef] [PubMed]
- Pharmacotherapy Update|New Antifungal Agents Additions to the Existing Armamentarium (Part 1). Available online: https://www.clevelandclinicmeded.com/medicalpubs/pharmacy/mayjune2003/antifungal.htm (accessed on 25 September 2023).
- Benz, F.; Knüsel, F.; Nüesch, J.; Treichler, H.; Voser, W.; Nyfeler, R.; Keller-Schierlein, W. Metabolites of microorganisms. 143. Echinocandin B, a novel polypeptide-antibiotic from Aspergillus nidulans var. echinulatus: Isolation and structural components. Helv. Chim. Acta 1974, 57, 2459–2477. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Boyken, L.; Hollis, R.J.; Messer, S.A.; Tendolkar, S.; Diekema, D.J. In Vitro Activities of Anidulafungin against More than 2,500 Clinical Isolates of Candida spp., Including 315 Isolates Resistant to Fluconazole. J. Clin. Microbiol. 2005, 43, 5425. [Google Scholar] [CrossRef] [PubMed]
- Zida, A.; Bamba, S.; Yacouba, A.; Ouedraogo-Traore, R.; Guiguemdé, R.T. Anti-Candida albicans natural products, sources of new antifungal drugs: A review. J. Mycol. Med. 2017, 27, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bouffard, F.A.; Zambias, R.A.; Dropinski, J.F.; Balkovec, J.M.; Hammond, M.L.; Abruzzo, G.K.; Bartizal, K.F.; Marrinan, J.A.; Kurtz, M.B.; McFadden, D.C.; et al. Synthesis and Antifungal Activity of Novel Cationic Pneumocandin Bo Derivatives. J. Med. Chem. 1994, 37, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Masurekar, P.S.; Fountoulakis, J.M.; Hallada, T.C.; Sosa, M.S.; Kaplan, L. Pneumocandins from Zalerion arboricola. II. Modification of product spectrum by mutation and medium manipulation. J. Antibiot. 1992, 45, 1867–1874. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tkacz, J.S.; Giacobbe, R.A.; Monaghan, R.L. Improvement in the titer of echinocandin-type antibiotics: A magnesium-limited medium supporting the biphasic production of pneumocandins A0 and B0. J. Ind. Microbiol. 1993, 11, 95–103. [Google Scholar] [CrossRef]
- Qin, T.; Song, P.; Wang, X.; Ji, X.; Ren, L.; Huang, H. Protoplast mutant selection of Glarea Lozoyensis and statistical optimization of medium for pneumocandin B0 yield-up. Biosci. Biotechnol. Biochem. 2016, 80, 2241–2246. [Google Scholar] [CrossRef]
- Fujie, A. Discovery of micafungin (FK463): A novel antifungal drug derived from a natural product lead. Pure Appl. Chem. 2007, 79, 603–614. [Google Scholar] [CrossRef]
- Ueda, S.; Sakamoto, K.; Oohata, N.; Tsuboi, M.; Yamashita, M.; Hino, M.; Yamada, M.; Hashimoto, S. Screening and characterization of microorganisms with FR901379 acylase activity. J. Antibiot. 2010, 63, 65–70. [Google Scholar] [CrossRef]
- Kanda, M.; Tsuboi, M.; Sakamoto, K.; Shimizu, S.; Yamashita, M.; Honda, H. Improvement of FR901379 production by mutant selection and medium optimization. J. Biosci. Bioeng. 2009, 107, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Yamamoto, E.; Hayashi, A.; Yabutani, T.; Yamashita, M.; Honda, H. Scale-up fermentation of echinocandin type antibiotic FR901379. J. Biosci. Bioeng. 2010, 109, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Men, P.; Wang, M.; Li, J.; Geng, C.; Huang, X.; Lu, X. Establishing an Efficient Genetic Manipulation System for Sulfated Echinocandin Producing Fungus Coleophoma empetri. Front. Microbiol. 2021, 12, 734780. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.E.; Smith, S.K.; Onishi, J.C.; Meinz, M.; Kurtz, M.; Giacobbe, R.A.; Wilson, K.E.; Liesch, J.; Zink, D.; Horn, W.; et al. Isolation and structural determination of enfumafungin, a triterpene glycoside antifungal agent that is a specific inhibitor of glucan synthesis. J. Am. Chem. Soc. 2000, 122, 4882–4886. [Google Scholar] [CrossRef]
- Kuhnert, E.; Li, Y.; Lan, N.; Yue, Q.; Chen, L.; Cox, R.J.; An, Z.; Yokoyama, K.; Bills, G.F. Enfumafungin synthase represents a novel lineage of fungal triterpene cyclases. Environ. Microbiol. 2018, 20, 3325. [Google Scholar] [CrossRef]
- Jiménez-Ortigosa, C.; Perez, W.B.; Angulo, D.; Borroto-Esoda, K.; Perlin, D.S. De Novo Acquisition of Resistance to SCY-078 in Candida glabrata Involves FKS Mutations That both Overlap and Are Distinct from Those Conferring Echinocandin Resistance. Antimicrob. Agents Chemother. 2017, 61, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Rhomberg, P.R.; Borroto-Esoda, K.; Castanheira, M. Differential Activity of the Oral Glucan Synthase Inhibitor SCY-078 against Wild-Type and Echinocandin-Resistant Strains of Candida Species. Antimicrob. Agents Chemother. 2017, 61, 10-1128. [Google Scholar] [CrossRef]
- SCYNEXIS Announces FDA Approval of BREXAFEMME® (Ibrexafungerp Tablets) as the First and Only Oral Non-Azole Treatment for Vaginal Yeast Infections: SCYNEXIS, Inc. (SCYX). Available online: https://web.archive.org/web/20211231040808/https://www.scynexis.com/news-media/press-releases/detail/240/scynexis-announces-fda-approval-of-brexafemme (accessed on 9 October 2023).
- Olleik, H.; Nicoletti, C.; Lafond, M.; Courvoisier-Dezord, E.; Xue, P.; Hijazi, A.; Baydoun, E.; Perrier, J.; Maresca, M. Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins 2019, 11, 514. [Google Scholar] [CrossRef]
- Gäumann, E.; Roth, S.; Ettlinger, L.; Plattner, P.A.; Nager, U. Enniatin, a new antibiotic that works against mycobacteria. Experientia 1947, 3, 202–203. [Google Scholar] [CrossRef]
- Osman, M.F. A clinical assessment of locabiotal in the treatment of infections of the nose and throat. J. Int. Med. Res. 1977, 5, 139–146. [Google Scholar] [CrossRef]
- Hornbogen, T.; Glinski, M.; Zocher, R. Biosynthesis of Depsipeptide Mycotoxins in Fusarium. Eur. J. Plant Pathol. 2002, 108, 713–718. [Google Scholar] [CrossRef]
- Zocher, R.; Keller, U.; Kleinkauf, H. Enniatin synthetase, a novel type of multifunctional enzyme catalyzing depsipeptide synthesis in Fusarium oxysporum. Biochemistry 1982, 21, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Zocher, R.; Haese, A. Enniatin Production by Fusarium Strains and Its Effect on Potato Tuber Tissue. Appl. Environ. Microbiol. 1996, 62, 393. [Google Scholar] [CrossRef]
- Hedenmalm, K.; Kurz, X.; Morales, D. Effect of withdrawal of fusafungine from the market on prescribing of antibiotics and other alternative treatments in Germany: A pharmacovigilance impact study. Eur. J. Clin. Pharmacol. 2019, 75, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Debbab, A.; Proksch, P. Fifty years of drug discovery from fungi. Fungal Divers. 2011, 50, 3–19. [Google Scholar] [CrossRef]
- Pela´ez, F. Biological Activities of Fungal Metabolites. In Handbook of Industrial Mycology; An, Z., Ed.; CRC Press: Boca Raton, FL, USA, 2004; Volume 1, pp. 68–111. ISBN 9780429224560. [Google Scholar]
- Schueffler, A.; Anke, T. Fungal natural products in research and development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef]
- Demain, A.L.; Martens, E. Production of valuable compounds by molds and yeasts. J. Antibiot. 2016, 70, 347–360. [Google Scholar] [CrossRef]
- Butler, M.S.; Henderson, I.R.; Capon, R.J.; Blaskovich, M.A.T. Antibiotics in the clinical pipeline as of December 2022. J. Antibiot. 2023, 76, 431–473. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, M.; Chen, Y.; Ren, H.; Zhang, H.; Yu, C.; Lu, J.; You, L.; Yu, J.; Liang, H.; et al. Pharmacokinetics of benapenem for injection in subjects with mild to moderate renal impairment. Eur. J. Clin. Pharmacol. 2022, 78, 1079–1086. [Google Scholar] [CrossRef]
- Iterum Therapeutics Receives Complete Response Letter from U.S. Food and Drug Administration for Oral Sulopenem: Iterum Therapeutics plc (ITRM). Available online: https://www.iterumtx.com/news/press-releases/detail/73/iterum-therapeutics-receives-complete-response-letter-from (accessed on 30 November 2023).
- Minamimura, M.; Taniyama, Y.; Inoue, E.; Mitsuhashi, S. In vitro antibacterial activity and beta-lactamase stability of CP-70,429 a new penem antibiotic. Antimicrob. Agents Chemother. 1993, 37, 1547. [Google Scholar] [CrossRef]
- Doern, G.V.; Pierce, G.; Brueggemann, A.B. In vitro activity of sanfetrinem (GV104326), a new trinem antimicrobial agent, versus Streptococcus pneumonia, Haemophilus influenzae, and Moraxella catarrhalis. Diagn. Microbiol. Infect. Dis. 1996, 26, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Sanfetrinem|Working Group for New TB Drugs. Available online: https://www.newtbdrugs.org/pipeline/compound/sanfetrinem (accessed on 30 November 2023).
- Brian, P.W.; Curtis, P.J.; Hemming, H.G.; Norris, G.L.F. Wortmannin, an antibiotic produced by Penicillium wortmanni. Trans. Br. Mycol. Soc. 1957, 40, 365-IN3. [Google Scholar] [CrossRef]
- Karve, S.; Werner, M.E.; Sukumar, R.; Cummings, N.D.; Copp, J.A.; Wang, E.C.; Li, C.; Sethi, M.; Chen, R.C.; Pacold, M.E.; et al. Revival of the abandoned therapeutic wortmannin by nanoparticle drug delivery. Proc. Natl. Acad. Sci. USA 2012, 109, 8230–8235. [Google Scholar] [CrossRef] [PubMed]
- Pitz, M.W.; Eisenhauer, E.A.; MacNeil, M.V.; Thiessen, B.; Easaw, J.C.; Macdonald, D.R.; Eisenstat, D.D.; Kakumanu, A.S.; Salim, M.; Chalchal, H.; et al. Phase II study of PX-866 in recurrent glioblastoma. Neuro. Oncol. 2015, 17, 1270. [Google Scholar] [CrossRef] [PubMed]
- Heiss, J.D.; Argersinger, D.P.; Theodore, W.H.; Butman, J.A.; Sato, S.; Khan, O.I. Convection-Enhanced Delivery of Muscimol in Patients with Drug-Resistant Epilepsy. Neurosurgery 2019, 85, E4. [Google Scholar] [CrossRef]
- Davis, A.K.; Barrett, F.S.; May, D.G.; Cosimano, M.P.; Sepeda, N.D.; Johnson, M.W.; Finan, P.H.; Griffiths, R.R. Effects of Psilocybin-Assisted Therapy on Major Depressive Disorder: A Randomized Clinical Trial. JAMA Psychiatry 2021, 78, 1. [Google Scholar] [CrossRef]
- Goodwin, G.M.; Aaronson, S.T.; Alvarez, O.; Arden, P.C.; Baker, A.; Bennett, J.C.; Bird, C.; Blom, R.E.; Brennan, C.; Brusch, D.; et al. Single-Dose Psilocybin for a Treatment-Resistant Episode of Major Depression. N. Engl. J. Med. 2022, 387, 1637–1648. [Google Scholar] [CrossRef]
- Lee, T.H.; Lee, C.K.; Tsou, W.L.; Liu, S.Y.; Kuo, M.T.; Wen, W.C. A new cytotoxic agent from solid-state fermented mycelium of Antrodia camphorata. Planta Med. 2007, 73, 1412–1415. [Google Scholar] [CrossRef]
- Ho, C.L.; Wang, J.L.; Lee, C.C.; Cheng, H.Y.; Wen, W.C.; Cheng, H.H.Y.; Chen, M.C.M. Antroquinonol blocks Ras and Rho signaling via the inhibition of protein isoprenyltransferase activity in cancer cells. Biomed. Pharmacother. 2014, 68, 1007–1014. [Google Scholar] [CrossRef]
- Beheshtirouy, S.; Khani, E.; Khiali, S.; Entezari-Maleki, T. Investigational antiviral drugs for the treatment of COVID-19 patients. Arch. Virol. 2022, 167, 751–805. [Google Scholar] [CrossRef]
- Makhwitine, J.P.; Kumalo, H.M.; Ndlovu, S.I.; Mkhwanazi, N.P. Epigenetic Induction of Secondary Metabolites Production in Endophytic Fungi Penicillium chrysogenum and GC-MS Analysis of Crude Metabolites with Anti-HIV-1 Activity. Microorganisms 2023, 11, 1404. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; García-Estrada, C.; Rumbero, A.; Recio, E.; Albillos, S.M.; Ullán, R.V.; Martín, J.-F. Characterization of an autoinducer of penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 2011, 77, 5688–5696. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Li, M.; Hu, Z.; Shao, Y.; Ying, J.; Zhang, H. The Potential Use of Fungal Co-Culture Strategy for Discovery of New Secondary Metabolites. Microorganisms 2023, 11, 464. [Google Scholar] [CrossRef]
- Ozcengiz, G.; Demain, A.L. Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol. Adv. 2013, 31, 287–311. [Google Scholar] [CrossRef] [PubMed]
- Khomutov, A.R.; Dzhavakhiya, V.G.; Voinova, T.M.; Ermolinskii, B.S.; Khomutov, R.M. Aminooxy analogue of putrescine inhibits polyketide biosynthetic pathway of natural products. Bioorganicheskaya Khimiya 1989, 15, 706–709. [Google Scholar]
- Thines, E.; Daußmann, T.; Semar, M.; Sterner, O.; Anke, H. Fungal Melanin Biosynthesis Inhibitors: Introduction of a Test System Based on the Production of Dihydroxynaphthalene (DHN) Melanin in Agar Cultures. Z. Fur Naturforsch.-Sect. C J. Biosci. 1995, 50, 813–819. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Eldarov, M.A. Spermidine and 1,3-Diaminopropane Have Opposite Effects on the Final Stage of Cephalosporin C Biosynthesis in High-Yielding Acremonium chrysogenum Strain. Int. J. Mol. Sci. 2022, 23, 14625. [Google Scholar] [CrossRef]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6, 299. [Google Scholar] [CrossRef]
- Elhamouly, N.A.; Hewedy, O.A.; Zaitoon, A.; Miraples, A.; Elshorbagy, O.T.; Hussien, S.; El-Tahan, A.; Peng, D. The hidden power of secondary metabolites in plant-fungi interactions and sustainable phytoremediation. Front. Plant Sci. 2022, 13, 1044896. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; Real-Santillán, R.O.; López-Carmona, D.; García-Gómez, G.; Galicia-Gallardo, A.P.; Alfaro-Cuevas, R.; González-Esquivel, C.E.; Najera-Rincón, M.B.; Adame-Garnica, S.G.; et al. In a belowground multitrophic interaction, Trichoderma harzianum induces maize root herbivore tolerance against Phyllophaga vetula. Pest Manag. Sci. 2021, 77, 3952–3963. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed]
- Bagheri-Gavkosh, S.; Bigdeli, M.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Inhibitory effects of Ephedra major Host on Aspergillus parasiticus growth and aflatoxin production. Mycopathologia 2009, 168, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Jermnak, U.; Yoshinari, T.; Sugiyama, Y.; Tsuyuki, R.; Nagasawa, H.; Sakuda, S. Isolation of methyl syringate as a specific aflatoxin production inhibitor from the essential oil of Betula alba and aflatoxin production inhibitory activities of its related compounds. Int. J. Food Microbiol. 2012, 153, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Eng, F.; Marin, J.E.; Zienkiewicz, K.; Gutiérrez-Rojas, M.; Favela-Torres, E.; Feussner, I. Jasmonic acid biosynthesis by fungi: Derivatives, first evidence on biochemical pathways and culture conditions for production. PeerJ 2021, 9, e10873. [Google Scholar] [CrossRef] [PubMed]
- Deepthi, S.; Satheeshkumar, K. Cell line selection combined with jasmonic acid elicitation enhance camptothecin production in cell suspension cultures of Ophiorrhiza mungos L. Appl. Microbiol. Biotechnol. 2017, 101, 545–558. [Google Scholar] [CrossRef]
- Yukimune, Y.; Tabata, H.; Higashi, Y.; Hara, Y. Methyl jasmonate-induced overproduction of paclitaxel and baccatin III in Taxus cell suspension cultures. Nat. Biotechnol. 1996, 14, 1129–1132. [Google Scholar] [CrossRef]
- Pande, A.; Mun, B.G.; Lee, D.S.; Khan, M.; Lee, G.M.; Hussain, A.; Yun, B.W. NO Network for Plant–Microbe Communication Underground: A Review. Front. Plant Sci. 2021, 12, 658679. [Google Scholar] [CrossRef]
- Jedelská, T.; Luhová, L.; Petřivalský, M. Nitric oxide signalling in plant interactions with pathogenic fungi and oomycetes. J. Exp. Bot. 2021, 72, 848–863. [Google Scholar] [CrossRef]
- Zhao, Y.; Lim, J.; Xu, J.; Yu, J.H.; Zheng, W. Nitric oxide as a developmental and metabolic signal in filamentous fungi. Mol. Microbiol. 2020, 113, 872–882. [Google Scholar] [CrossRef]
- Baidya, S.; Cary, J.W.; Grayburn, W.S.; Calvo, A.M. Role of Nitric Oxide and Flavohemoglobin Homolog Genes in Aspergillus nidulans Sexual Development and Mycotoxin Production. Appl. Environ. Microbiol. 2011, 77, 5524. [Google Scholar] [CrossRef]
- Khalil, Z.G.; Kalansuriya, P.; Capon, R.J. Lipopolysaccharide (LPS) stimulation of fungal secondary metabolism. Mycology 2014, 5, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Li, X.P.; Wang, Y.; Wang, J.W. Nitric oxide donor sodium nitroprusside-induced transcriptional changes and hypocrellin biosynthesis of Shiraia sp. S9. Microb. Cell Fact. 2021, 20, 92. [Google Scholar] [CrossRef] [PubMed]
- Khiralla, A.; Mohammed, A.O.; Yagi, S. Fungal perylenequinones. Mycol. Prog. 2022, 21, 38. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Sun, C.X.; Wang, J.W. Enhanced Production of Hypocrellin A in Submerged Cultures of Shiraia bambusicola by Red Light. Photochem. Photobiol. 2019, 95, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Estey, E.P.; Brown, K.; Diwu, Z.; Liu, J.; Lown, J.W.; Miller, G.G.; Moore, R.B.; Tulip, J.; McPhee, M.S. Hypocrellins as photosensitizers for photodynamic therapy: A screening evaluation and pharmacokinetic study. Cancer Chemother. Pharmacol. 1996, 37, 343–350. [Google Scholar] [CrossRef]
- Higgins, S.A.; Schadt, C.W.; Matheny, P.B.; Löffler, F.E. Phylogenomics Reveal the Dynamic Evolution of Fungal Nitric Oxide Reductases and Their Relationship to Secondary Metabolism. Genome Biol. Evol. 2018, 10, 2474–2489. [Google Scholar] [CrossRef]
- Meinwald, J. Understanding the chemistry of chemical communication: Are we there yet? Proc. Natl. Acad. Sci. USA 2003, 100, 14514–14516. [Google Scholar] [CrossRef]
- Cohen, S.S. A Guide to the Polyamines, 1st ed.; Oxford University Press: New York, NY, USA, 1998; Volume 1, ISBN -10. [Google Scholar]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Fukuma, I.; Cohen, S.S. Polyamines in bacteriophage R17 and its RNA. J. Virol. 1975, 16, 222. [Google Scholar] [CrossRef]
- Gibson, W.; Roizman, B. Compartmentalization of Spermine and Spermidine in the Herpes Simplex Virion. Proc. Natl. Acad. Sci. USA 1971, 68, 2818. [Google Scholar] [CrossRef]
- Mounce, B.C.; Poirier, E.Z.; Passoni, G.; Simon-Loriere, E.; Cesaro, T.; Prot, M.; Stapleford, K.A.; Moratorio, G.; Sakuntabhai, A.; Levraud, J.P.; et al. Interferon-Induced Spermidine-Spermine Acetyltransferase and Polyamine Depletion Restrict Zika and Chikungunya Viruses. Cell Host Microbe 2016, 20, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in aging and disease. Aging 2011, 3, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Perez-Leal, O.; Merali, S. Regulation of polyamine metabolism by translational control. Amino Acids 2012, 42, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Panagiotidis, C.A.; Shu-Ching, H.; Canellakis, E.S. Post-translational and transcriptional regulation of polyamine biosynthesis in Escherichia coli. Int. J. Biochem. 1994, 26, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Palanimurugan, R.; Scheel, H.; Hofmann, K.; Dohmen, R.J. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004, 23, 4857–4867. [Google Scholar] [CrossRef] [PubMed]
- Wimalasekera, R.; Tebartz, F.; Scherer, G.F.E. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 2011, 181, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Tun, N.N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Segal Floh, E.I.; Scherer, G.F.E. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 2006, 47, 346–354. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Herbert, N.; Arst, J. Regulation of Gene Expression by Ambient pH in Filamentous Fungi and Yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 426. [Google Scholar] [CrossRef]
- Jain, S.; Keller, N. Insights to fungal biology through LaeA sleuthing. Fungal Biol. Rev. 2013, 27, 51–59. [Google Scholar] [CrossRef]
- García-Estrada, C.; Barreiro, C.; Jami, M.-S.; Martín-González, J.; Martín, J.-F. The inducers 1,3-diaminopropane and spermidine cause the reprogramming of metabolism in Penicillium chrysogenum, leading to multiple vesicles and penicillin overproduction. J. Proteomics 2013, 85, 129–159. [Google Scholar] [CrossRef]
- Zhgun, A.A.; Nuraeva, G.K.; Eldarov, M.A. The Role of LaeA and LovE Regulators in Lovastatin Biosynthesis with Exogenous Polyamines in Aspergillus terreus. Appl. Biochem. Microbiol. 2019, 55, 639–648. [Google Scholar] [CrossRef]
- Murray Stewart, T.; Dunston, T.T.; Woster, P.M.; Casero, R.A. Polyamine catabolism and oxidative damage. J. Biol. Chem. 2018, 293, 18736–18745. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Su, G.-C.; Huang, W.-Y.; Ko, M.-Y.; Yeh, H.-Y.; Chang, G.-D.; Lin, S.-J.; Chi, P. Promotion of homology-directed DNA repair by polyamines. Nat. Commun. 2019, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235. [Google Scholar] [CrossRef]
- Valdés-Santiago, L.; Ruiz-Herrera, J. Stress and polyamine metabolism in fungi. Front. Chem. 2013, 1, 42. [Google Scholar] [CrossRef]
- Rato, C.; Amirova, S.R.; Bates, D.G.; Stansfield, I.; Wallace, H.M. Translational recoding as a feedback controller: Systems approaches reveal polyamine-specific effects on the antizyme ribosomal frameshift. Nucleic Acids Res. 2011, 39, 4587–4597. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Thön, M.; Spröte, P.; Scharf, D.H.; Al-Abdallah, Q.; Wolke, S.M.; Hortschansky, P. Aspects on evolution of fungal beta-lactam biosynthesis gene clusters and recruitment of trans-acting factors. Phytochemistry 2009, 70, 1801–1811. [Google Scholar] [CrossRef]
- Smith, D.J.; Bull, J.H.; Edwards, J.; Turner, G. Amplification of the isopenicillin N synthetase gene in a strain of Penicillium chrysogenum producing high levels of penicillin. MGG Mol. Gen. Genet. 1989, 216, 492–497. [Google Scholar] [CrossRef]
- Barredo, J.L.; Díez, B.; Alvarez, E.; Martín, J.F. Large amplification of a 35-kb DNA fragment carrying two penicillin biosynthetic genes in high penicillin producing strains of Penicillium chrysogenum. Curr. Genet. 1989, 16, 453–459. [Google Scholar] [CrossRef]
- Fierro, F.; Barredo, J.L.; Díez, B.; Gutierrez, S.; Fernández, F.J.; Martín, J.F. The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc. Natl. Acad. Sci. USA 1995, 92, 6200–6204. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Eldarov, M.A.; Sklyarenko, A.V.; Dumina, M.V.; Beletsky, A.V.; Yarotsky, S.V.; Ravin, N. V Draft Genome Sequence of Escherichia coli Strain VKPM B-10182, Producing the Enzyme for Synthesis of Cephalosporin Acids. Genome Announc. 2014, 2, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Nijland, J.G.; Ebbendorf, B.; Woszczynska, M.; Boer, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Nonlinear biosynthetic gene cluster dose effect on penicillin production by Penicillium chrysogenum. Appl. Environ. Microbiol. 2010, 76, 7109–7115. [Google Scholar] [CrossRef] [PubMed]
- Seidl, V.; Seiboth, B. Trichoderma reesei: Genetic approaches to improving strain efficiency. Biofuels 2010, 1, 343–354. [Google Scholar] [CrossRef]
- Ziemons, S.; Koutsantas, K.; Becker, K.; Dahlmann, T.; Kück, U. Penicillin production in industrial strain Penicillium chrysogenum P2niaD18 is not dependent on the copy number of biosynthesis genes. BMC Biotechnol. 2017, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lv, Y.; Li, X.; Lin, Y.; Deng, H.; Pan, L. Profiling of secondary metabolite gene clusters regulated by LaeA in Aspergillus niger FGSC A1279 based on genome sequencing and transcriptome analysis. Res. Microbiol. 2017, 169, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Valiakhmetov, A.I.; Trilisenko, L.V.; Vagabov, V.M.; Bartoshevich, I.E.; Kulaev, I.S.; Novak, M.I.; Domracheva, A.G.; El’darov, M.A.; Skriabin, K.G. The concentration dynamics of inorganic polyphosphates during the cephalosporin C synthesis by Acremonium chrysogenum. Prikl. Biokhim. Mikrobiol. 2010, 46, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Burgstaller, W. Transport of Small Ions and Molecules through the Plasma Membrane of Filamentous Fungi. Crit. Rev. Microbiol. 1997, 23, 1–46. [Google Scholar] [CrossRef]
- Gradmann, D.; Hansen, U.-P.; Long, W.S.; Slayman, C.L.; Warncke, J. Current-voltage relationships for the plasma membrane and its principal electrogenic pump inNeurospora crassa: I. Steady-state conditions. J. Membr. Biol. 1978, 39, 333–367. [Google Scholar] [CrossRef]
- Kallow, W.; Von Döhren, H.; Kleinkauf, H. Penicillin biosynthesis: Energy requirement for tripeptide precursor formation by delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Acremonium chrysogenum. Biochemistry 1998, 37, 5947–5952. [Google Scholar] [CrossRef]
- Jami, M.-S.; Barreiro, C.; García-Estrada, C.; Martín, J.-F. Proteome Analysis of the Penicillin Producer Penicillium chrysogenum. Mol. Cell. Proteomics 2010, 9, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Alam, M.T.; Breitling, R.; Takano, E. The future of industrial antibiotic production: From random mutagenesis to synthetic biology. Bioeng. Bugs 2011, 2, 230–233. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).