Abstract
A growing number of recent studies have revealed that non-coding RNAs play a wide variety of roles beyond expectation. A lot of non-coding RNAs have been shown to function by forming intracellular structures either in the nucleus or the cytoplasm. In the fission yeast Schizosaccharomyces pombe, a non-coding RNA termed meiRNA has been shown to play multiple vital roles in the course of meiosis. meiRNA is tethered to its genetic locus after transcription and forms a peculiar intranuclear dot structure. It ensures stable expression of meiotic genes in cooperation with an RNA-binding protein Mei2. Chromosome-associated meiRNA also facilitates recognition of homologous chromosome loci and induces robust pairing. In this review, the quarter-century history of meiRNA, from its identification to functional characterization, will be outlined.
1. A Long Non-Coding RNA Essential for the Progression of Meiosis in Fission Yeast
It is nearly impossible to avoid issues relating to non-coding RNAs in biology nowadays. Non-coding RNAs play crucial roles in a wide range of biological processes. In particular, an increasing number of non-coding RNAs have been shown to be active in gene expression regulation. Participation of non-coding RNAs in gene expression makes great contribution to enable fine-tuned regulation to adapt to environmental changes. Furthermore, it has been demonstrated that non-coding RNAs function in diverse cellular contexts beyond gene expression [1,2].
In the fission yeast Schizosaccharomyces pombe, a unicellular eukaryotic model organism, a long non-coding RNA termed meiRNA acts as an essential factor for meiosis. S. pombe cells grow preferentially in the haploid state. When starved for nutrients, S. pombe cells conjugate with cells of the opposite mating type and form zygotes. Resultant diploid zygotes then undergo meiosis and eventually form spores that are tolerant to stress conditions including starvation [3]. meiRNA was initially identified a quarter-century ago in the screening for high copy suppressors of a conditional meiotic mutant, mei2, which was defective in the initiation of the first meiotic division (meiosis I) [4]. Removal of meiRNA from S. pombe cells by deleting the sme2 gene that encodes meiRNA also results in meiotic cell cycle arrest before meiosis I, indicating an essential role of meiRNA in the induction of meiosis I. meiRNA was originally reported to be about 0.5 kb polyadenylated transcripts [4]. meiRNA has lately been shown to have two types of isoforms, meiRNA-S (about 0.5 kb) and meiRNA-L (about 1.5 kb) (Figure 1) [5]. Whereas meiRNA-L is shown to have full activity to carry forward meiosis, the function, if any, of meiRNA-S remains unknown [6,7].
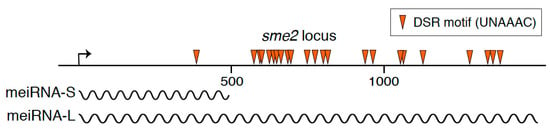
Figure 1.
A schematic diagram of the sme2 locus on chromosome II, which encodes meiRNA. Wavy lines indicate two isoforms of the meiRNA transcripts, namely meiRNA-S and meiRNA-L. Arrowheads indicate the determinant of selective removal (DSR) motif, UNAAAC, which is recognized by Mmi1.
2. Binding Partner of meiRNA
Over-production of meiRNA suppresses the meiotic defect of the mei2 mutant. The mei2 gene encodes an RRM-type RNA binding protein. This evokes the possibility of meiRNA directly interacting with Mei2 protein. It has been actually demonstrated that Mei2 binds to the 5′ region of meiRNA-L, corresponding to meiRNA-S [4,6]. meiRNA also binds to another RNA-binding protein, Mmi1, through the 3′ region, as mentioned below. Interaction of meiRNA with Mei2 and Mmi1 has also been confirmed by several genome-wide high-throughput approaches [8,9,10,11].
Mei2 is a key player in the initiation and progression of meiosis in S. pombe [12]. Mei2 is inactivated by phosphorylation in mitotically growing cells [13]. Forced expression of an unphosphorylated active form of Mei2 leads to induction of the whole process of meiosis even in haploid cells under nutrient rich conditions, indicating that Mei2 is a molecular switch to turn on meiosis [13]. Mei2 is also removed from mitotically growing cells through the ubiquitin-proteasome pathway [14,15]. An intriguing and unanswered question is how Mei2 induces meiosis.
After inducing meiosis, Mei2 has another function of inducing meiosis I. This was demonstrated using a temperature-sensitive mei2 mutant that is able to initiate meiosis, but does not undergo meiosis I [4]. The following observations led to the conclusion that Mei2 and meiRNA play an essential role in meiosis I as an RNA-protein complex: The meiosis I defect in the mei2 mutant is suppressed by over-expression of meiRNA; cells lacking meiRNA cannot enter meiosis I; meiRNA directly associates with Mei2 both in vivo and in vitro [4].
3. Mei2 Dot
Microscopic observations revealed that the Mei2–meiRNA complex functions by forming a nuclear dot structure, known as Mei2 dot [13,16]. In cells lacking meiRNA or in cells expressing a mutant form of Mei2 that has lost its ability to bind to meiRNA, the Mei2 dot is not formed, indicating that interaction between Mei2 and meiRNA is crucial for the assembly of the Mei2 dot [16].
During the premeiotic S phase and meiotic prophase, the S. pombe nucleus undergoes oscillatory movement between the two poles of the cell [17]. This dynamic nuclear movement is called “horsetail” movement due to its shape, and it facilitates homologous chromosome pairing, which is essential for proper meiotic chromosome segregation. During the horsetail movement, the nucleus is led by a spindle pole body (SPB), a centrosome equivalent structure, to which all the telomeres are clustered, forming a bouquet chromosome arrangement [17,18]. The Mei2 dot was observed at a fixed position near the SPB in the oscillating horsetail nucleus, suggesting its association with a specific region of the chromosome (Figure 2a) [13,16]. Another study using chromosome translocation has revealed that the Mei2 dot associates with the sme2 locus on chromosome II, which encodes meiRNA [19]. meiRNA has also been demonstrated to localize to its genetic locus, sme2 (Figure 2b) [6,16,19]. Mei2 is a shuttling protein between the nucleus and cytoplasm [20]. From these observations, it is hypothesized that meiRNA is tethered to the sme2 locus, encoding itself, after transcription by an unclear mechanism, and then catches the shuttling Mei2 through direct interaction, to form the Mei2 dot.
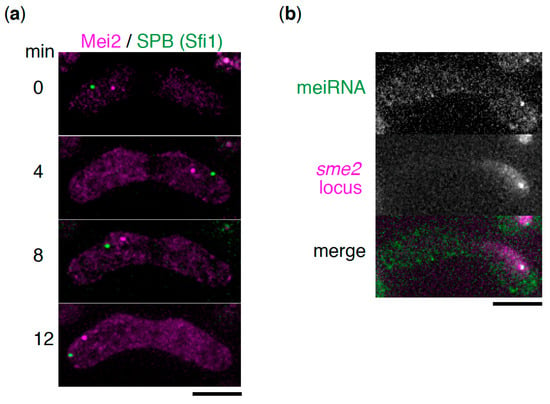
Figure 2.
meiRNA forms a chromosome-associated dot structure in cooperation with its binding partner, Mei2. (a) Time-lapse images of Mei2 (magenta) and the spindle pole body (SPB) (Sfi1, green) during the horsetail movement. Scale bar: 5 µm. (b) Localization of meiRNA. meiRNA was visualized by the MS2 system, in which MS2-loop-tagged meiRNA and MS2-YFP were expressed (green) [6]. The sme2 locus was marked by inserting lacO repeats and expressing 4xmCherry-LacI-NLS (magenta). Scale bar: 5 µm.
4. Molecular Function of Mei2 Dot
The next natural question is about the molecular function of the Mei2 dot. To get a clear answer, we have had to wait for identification and characterization of another key player, Mmi1.
The first step was a high-copy number suppressor screening for the meiotic arrest of cells lacking meiRNA [21]. Although no common feature was found among the isolated suppressors, some of them were expressed exclusively during meiosis. These included mei4, ssm4, rec8, and spo5, all of which encode factors required for proper meiosis progression. Intriguingly, mei4 transcripts did not accumulate in mitotically growing cells, even when they were forced to express from a strong constitutive promoter [22]. A specific region on mei4 transcripts was shown to be responsible for their elimination from mitotic cells. When this region was fused to a reporter gene such as GFP, the resultant chimeric gene was expressed exclusively in meiotic cells. Similar regions responsible for meiosis-specific expression were found in ssm4, rec8, and spo5. These cis-acting regions were designated as determinant of selective removal (DSR). DSR activity was demonstrated to be exhibited by the enrichment of the hexanucleotide UNAAC motif [5,8]. DSR-carrying transcripts are not limited to the four genes mentioned above, and it is assumed that dozens of transcripts have DSR.
Mmi1, a key factor for DSR-dependent meiotic transcript elimination, was isolated by genetic screening [22]. Mmi1 is a YTH-type RNA binding protein and interacts with DSR directly, by recognizing the UNAAAC motif [5,22]. While YTH-RNA-binding proteins generally recognize N6-methyladenosine (m6A)-containing RNAs [23], Mmi1 binds to its target RNAs in a unique manner and does not recognize m6A [24,25,26]. When DSR-carrying transcripts are recognized by Mmi1, RNA degradation by the nuclear exosome, a highly conserved 3′-5′ exonuclease complex, is induced [22,27]. This nuclear exosome-mediated RNA degradation requires polyadenylation by a canonical poly(A) polymerase [27]. Mmi1 also suppresses expression of a subset of its target genes by inducing facultative heterochromatin assembly at these loci [8,28,29]. Moreover, Mmi1 regulates transcriptional termination of its target genes [10,30,31] and prevents nuclear export of its target transcripts [32]. Mmi1 associates with a nuclear protein complex called MTREC (Mtl1-Red1 core) composed of a zinc-finger protein, Red1, and an Mtr4-like helicase, Mtl1 [33,34]. MTREC is suggested to be a counterpart of PAXT (poly(A) tail exosome targeting), which plays a crucial role in the selective elimination of polyadenylated transcripts in the nucleus of human cells [35]. MTREC is essential for Mmi1-mediated RNA degradation and facultative heterochromatin assembly [33,34]. Mmi1 also associates with a conserved multiprotein complex, Ccr4-Not [36,37,38,39]. It is suggested that Not4/Mot2 ubiquitin ligase subunit of the Ccr4-Not complex is recruited to Mei2 by Mmi1 and suppresses expression of DSR-containing transcripts including meiRNA by limiting the Mei2 accumulation in mitotically growing cells [39]. These Mmi1-mediated multi-layered regulations on meiotic genes may reflect the risk of their mistimed expression, which has been demonstrated to cause chromosome segregation errors [40].
Mmi1 is crucial for mitotic growth, but becomes an obstacle for meiosis, since it suppresses expression of meiotic genes. Thus, the Mmi1-DSR system should be blocked during meiosis. In this regard, the function of the Mei2 dot was found out to be inhibition of Mmi1 [22]. In mitotically growing cells, Mmi1 forms several nuclear foci. Many factors involved in the Mmi1-DSR system, including the nuclear exosome, localize to the Mmi1 nuclear foci [27,34,38,41,42,43]. DSR transcripts also localize to the Mmi1 foci [32]. When cells initiate meiosis, the Mmi1 foci converge on a single focus [22]. Importantly, the Mmi1 convergence point coincides with the Mei2 dot. Furthermore, Mmi1 does not converge in mei2 or sme2 deletion mutant cells both of which fail to form the Mei2 dot. From these observations, it has been thought that Mmi1 may be sequestered and inactivated by the Mei2 dot. Physical interaction of Mmi1 with Mei2 and with meiRNA supports this hypothesis [6,22]. Moreover, consistent with the hypothesis, the meiotic arrest of cells lacking the Mei2 dot could be suppressed by a hypomorphic mmi1 mutation [22]. As mentioned before, the meiosis arrest phenotype in cells lacking meiRNA is suppressed by overexpression of DSR-carrying transcripts [5,21]. Based on the hypothesis, this is rational because excess target transcripts may sequester Mmi1.
meiRNA carries numerous copies of the DSR motif in its 3′ region (Figure 1) and binds to Mmi1 through the 3′ region [6]. Moreover, meiRNA is degraded by the Mmi1-mediated degradation system in mitotically growing cells and is stably expressed only in meiotic cells [5,6]. Expression of meiRNA is upregulated by a transcription factor, Ste11, upon nutrient starvation [4,44]. From these observations, it is suggested that meiRNA serves as a decoy to lure Mmi1. meiRNA is indeed able to reduce the activity of Mmi1, while complete inactivation of Mmi1 might require Mei2 [6]. In mitotically growing cells, overexpression of meiRNA induces ectopic expression of Mmi1-target DSR transcripts in mmi1 mutant cells but not in wild-type cells. Meanwhile, in mutant cells lacking the 5′ region of meiRNA, to which Mei2 binds, DSR transcripts are expressed upon nutrient starvation, although lower than in wild-type cells. The mutant cells lacking the 5′ region are able to undergo meiosis [6], suggesting that the Mmi1 function may be repressed through an unknown mechanism in the course of meiosis and that inactivation of Mmi1 may be accomplished by meiRNA, independent of the binding of Mei2.
5. Contribution of meiRNA in Homologous Chromosome Pairing
It has been demonstrated that meiRNA plays a distinct role other than meiotic gene expression regulation [7]. In vivo live-cell imaging has revealed that the two sme2 loci on homologous chromosomes, which encode meiRNA, pair robustly in the early stage of meiotic prophase. When translocated to another site, the sme2 locus induces robust pairing there. Furthermore, the sme2 locus is able to facilitate ectopic pairing between nonhomologous chromosome sites when both sites carry the locus. The robust pairing at the sme2 locus depends on transcription of meiRNA. Dampening of meiRNA transcription at one sme2 locus of two homologous chromosomes impairs the robust early pairing. From these observations, it is thought that meiRNA stays at its genetic locus and triggers recognition and interaction of the meiRNA-associated site on homologous chromosomes through unknown mechanisms. A 5′ region of meiRNA, corresponding to meiRNA-S, is dispensable for induction of robust pairing. meiRNA binds to Mei2 through this 5′ region [4,6], indicating that the association of Mei2 with meiRNA is not necessary for the robust pairing [7]. Chromosome tethering of meiRNA at the sme2 locus requires Mmi1, to which meiRNA binds through the 3′ region [6]. However, involvement of Mmi1 in chromosome pairing remains to be proven.
6. Outlook
Multiple roles of meiRNA during meiosis have been demonstrated, namely, the regulation of meiotic gene expression and induction of homologous chromosome pairing. meiRNA might be one of the most characterized non-coding RNAs, but there remain unanswered questions. For instance, it is unclear how meiRNA is tethered to its genetic locus. It has been shown that Mmi1 is essential for chromosomal tethering of meiRNA [6], but the precise mechanisms are yet to be clarified. Further studies are also required to understand the molecular mechanisms of meiRNA-mediated pairing. Moreover, yet another intriguing question is whether RNA-driven pairing is observed at other loci. Several non-coding RNAs have been demonstrated to be upregulated during meiosis [45], although their molecular function is unknown. Future studies will tell us how meiRNA functions, in greater detail, and shed light on the generality of such non-coding RNA-mediated regulations.
Funding
This research received no external funding.
Acknowledgments
The author thanks Yuichi Shichino for preparing Figure 2b and Yoko Otsubo for helpful comments.
Conflicts of Interest
The author declares no conflict of interest.
References
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Shichino, Y.; Yamamoto, M. The long non-coding RNA world in yeasts. Biochim. Biophys. Acta 2016, 1859, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Sakuno, T.; Watanabe, Y.; Yamamoto, M. Analysis of Schizosaccharomyces pombe meiosis. Cold Spring Harb. Protoc. 2017, 2017, pdb top079855. [Google Scholar] [CrossRef]
- Watanabe, Y.; Yamamoto, M.S. pombe mei2 encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 1994, 78, 487–498. [Google Scholar] [CrossRef]
- Yamashita, A.; Shichino, Y.; Tanaka, H.; Hiriart, E.; Touat-Todeschini, L.; Vavasseur, A.; Ding, D.Q.; Hiraoka, Y.; Verdel, A.; Yamamoto, M. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Bio. 2012, 2, 120014. [Google Scholar] [CrossRef] [PubMed]
- Shichino, Y.; Yamashita, A.; Yamamoto, M. Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Bio. 2014, 4, 140022. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.Q.; Okamasa, K.; Yamane, M.; Tsutsumi, C.; Haraguchi, T.; Yamamoto, M.; Hiraoka, Y. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science 2012, 336, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Hiriart, E.; Vavasseur, A.; Touat-Todeschini, L.; Yamashita, A.; Gilquin, B.; Lambert, E.; Perot, J.; Shichino, Y.; Nazaret, N.; Boyault, C.; et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012, 31, 2296–2308. [Google Scholar] [CrossRef]
- Kilchert, C.; Wittmann, S.; Passoni, M.; Shah, S.; Granneman, S.; Vasiljeva, L. Regulation of mRNA levels by decay-promoting introns that recruit the exosome specificity factor Mmi1. Cell Rep. 2015, 13, 2504–2515. [Google Scholar] [CrossRef]
- Touat-Todeschini, L.; Shichino, Y.; Dangin, M.; Thierry-Mieg, N.; Gilquin, B.; Hiriart, E.; Sachidanandam, R.; Lambert, E.; Brettschneider, J.; Reuter, M.; et al. Selective termination of lncRNA transcription promotes heterochromatin silencing and cell differentiation. EMBO J. 2017, 36, 2626–2641. [Google Scholar] [CrossRef]
- Mukherjee, K.; Futcher, B.; Leatherwood, J. mmi1 and rep2 mRNAs are novel RNA targets of the Mei2 RNA-binding protein during early meiosis in Schizosaccharomyces pombe. Open Bio. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M. The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast. P. Jpn. Acad. B-Phys. 2010, 86, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Yabana, S.; Chikashige, Y.; Hiraoka, Y.; Yamamoto, M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 1997, 386, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, K.; Katayama, S.; Dhut, S.; Sato, M.; Watanabe, Y.; Yamamoto, M.; Toda, T. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 2001, 1, 389–399. [Google Scholar] [CrossRef]
- Otsubo, Y.; Yamashita, A.; Ohno, H.; Yamamoto, M.S. pombe TORC1 activates the ubiquitin-proteasomal degradation of the meiotic regulator Mei2 in cooperation with Pat1 kinase. J. Cell Sci. 2014, 127, 2639–2646. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Watanabe, Y.; Nukina, N.; Yamamoto, M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 1998, 95, 115–123. [Google Scholar] [CrossRef]
- Chikashige, Y.; Ding, D.Q.; Funabiki, H.; Haraguchi, T.; Mashiko, S.; Yanagida, M.; Hiraoka, Y. Telomere-led premeiotic chromosome movement in fission yeast. Science 1994, 264, 270–273. [Google Scholar] [CrossRef]
- Chikashige, Y.; Ding, D.Q.; Imai, Y.; Yamamoto, A.; Haraguchi, T.; Hiraoka, Y. Meiotic nuclear reorganization: Switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 1997, 16, 193–202. [Google Scholar] [CrossRef]
- Shimada, T.; Yamashita, A.; Yamamoto, M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 2003, 14, 2461–2469. [Google Scholar] [CrossRef]
- Sato, M.; Shinozaki-Yabana, S.; Yamashita, A.; Watanabe, Y.; Yamamoto, M. The fission yeast meiotic regulator Mei2p undergoes nucleocytoplasmic shuttling. FEBS Lett. 2001, 499, 251–255. [Google Scholar] [CrossRef]
- Yamashita, A.; Watanabe, Y.; Yamamoto, M. Microtubule-associated coiled-coil protein Ssm4 is involved in the meiotic development in fission yeast. Genes Cells 1997, 2, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Harigaya, Y.; Tanaka, H.; Yamanaka, S.; Tanaka, K.; Watanabe, Y.; Tsutsumi, C.; Chikashige, Y.; Hiraoka, Y.; Yamashita, A.; Yamamoto, M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 2006, 442, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Hazra, D.; Chapat, C.; Graille, M. m(6)A mRNA destiny: Chained to the rhYTHm by the YTH-containing proteins. Genes 2019, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Sanchez, A.M.; Goldgur, Y.; Shuman, S.; Schwer, B. Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast. RNA 2016, 22, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, Y.; Bao, H.; Jiang, Y.; Xu, C.; Wu, J.; Shi, Y. A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res. 2016, 44, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Xu, J.; Su, S.; Liu, H.; Gan, J.; Ma, J. Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1. Biochem. Biophys. Res. Commun. 2017, 491, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S.; Yamashita, A.; Harigaya, Y.; Iwata, R.; Yamamoto, M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010, 29, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Zofall, M.; Yamanaka, S.; Reyes-Turcu, F.E.; Zhang, K.; Rubin, C.; Grewal, S.I. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 2012, 335, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, S.; Asano, T.; Kanoh, J.; Ishikawa, F. Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells 2013, 18, 327–339. [Google Scholar] [CrossRef]
- Chalamcharla, V.R.; Folco, H.D.; Dhakshnamoorthy, J.; Grewal, S.I. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl. Acad. Sci. USA 2015, 112, 15548–15555. [Google Scholar] [CrossRef]
- Shah, S.; Wittmann, S.; Kilchert, C.; Vasiljeva, L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014, 28, 231–244. [Google Scholar] [CrossRef]
- Shichino, Y.; Otsubo, Y.; Kimori, Y.; Yamamoto, M.; Yamashita, A. YTH-RNA-binding protein prevents deleterious expression of meiotic proteins by tethering their mRNAs to nuclear foci. eLife 2018, 7, e32155. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.N.; Chalamcharla, V.R.; Reyes-Turcu, F.; Mehta, S.; Zofall, M.; Balachandran, V.; Dhakshnamoorthy, J.; Taneja, N.; Yamanaka, S.; Zhou, M.; et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 2013, 155, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Egan, E.D.; Braun, C.R.; Gygi, S.P.; Moazed, D. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA 2014, 20, 867–881. [Google Scholar] [CrossRef]
- Meola, N.; Domanski, M.; Karadoulama, E.; Chen, Y.; Gentil, C.; Pultz, D.; Vitting-Seerup, K.; Lykke-Andersen, S.; Andersen, J.S.; Sandelin, A.; et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 2016, 64, 520–533. [Google Scholar] [CrossRef]
- Cotobal, C.; Rodriguez-Lopez, M.; Duncan, C.; Hasan, A.; Yamashita, A.; Yamamoto, M.; Bahler, J.; Mata, J. Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenet. Chromatin. 2015, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Stowell, J.A.W.; Webster, M.W.; Kogel, A.; Wolf, J.; Shelley, K.L.; Passmore, L.A. Reconstitution of targeted deadenylation by the Ccr4-Not complex and the YTH domain protein Mmi1. Cell Rep. 2016, 17, 1978–1989. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Thillainadesan, G.; Chalamcharla, V.R.; Meng, Z.; Balachandran, V.; Dhakshnamoorthy, J.; Zhou, M.; Grewal, S.I.S. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell 2016, 61, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, F.; Candelli, T.; Leon, S.; Libri, D.; Rougemaille, M. Ubiquitination-dependent control of sexual differentiation in fission yeast. eLife 2017, 6. [Google Scholar] [CrossRef]
- Folco, H.D.; Chalamcharla, V.R.; Sugiyama, T.; Thillainadesan, G.; Zofall, M.; Balachandran, V.; Dhakshnamoorthy, J.; Mizuguchi, T.; Grewal, S.I. Untimely expression of gametogenic genes in vegetative cells causes uniparental disomy. Nature 2017, 543, 126–130. [Google Scholar] [CrossRef]
- Sugiyama, T.; Sugioka-Sugiyama, R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011, 30, 1027–1039. [Google Scholar] [CrossRef]
- Yamashita, A.; Takayama, T.; Iwata, R.; Yamamoto, M. A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res. 2013, 41, 9680–9687. [Google Scholar] [CrossRef]
- Sugiyama, T.; Wanatabe, N.; Kitahata, E.; Tani, T.; Sugioka-Sugiyama, R. Red5 and three nuclear pore components are essential for efficient suppression of specific mRNAs during vegetative growth of fission yeast. Nucleic Acids Res. 2013, 41, 6674–6686. [Google Scholar] [CrossRef]
- Mata, J.; Bahler, J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 15517–15522. [Google Scholar] [CrossRef]
- Watanabe, T.; Miyashita, K.; Saito, T.T.; Yoneki, T.; Kakihara, Y.; Nabeshima, K.; Kishi, Y.A.; Shimoda, C.; Nojima, H. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001, 29, 2327–2337. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).