The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers
Abstract
1. Early History of the Growth-Arrest-Specific (GAS)-5 Transcriptional Unit: Discovery in a Screen for G0 Genes
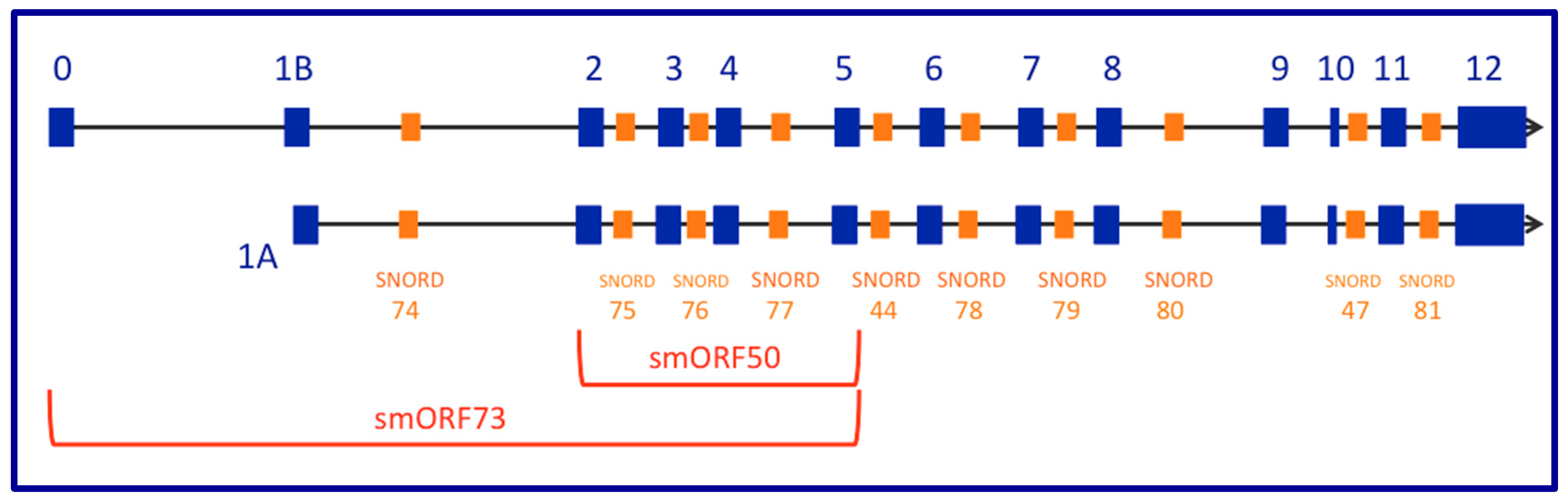
2. The GAS5 Locus Encodes a Long Non-Coding RNA (lncRNA) Gene That Contains Small Nucleolar (sno) RNAs in Its Introns While Demonstrating snoRNA-Independent Functions
3. The Evidence Supporting Orthology of Rodent and Primate GAS5 Genes

4. GAS5 Expression in Cancers
5. The Kinase mTOR, Rapamycin, and GAS5 Transcription
6. Clinical Aspects of GAS5 Function in Breast Cancer
7. Most Likely There Are Multiple Mechanisms, Distinct and Independent, through Which GAS5 Expression Impacts Cell Phenotype
8. GAS5 lncRNA as a Scaffold Seeding the Assembly of a Nucleoprotein (RNP) Complex
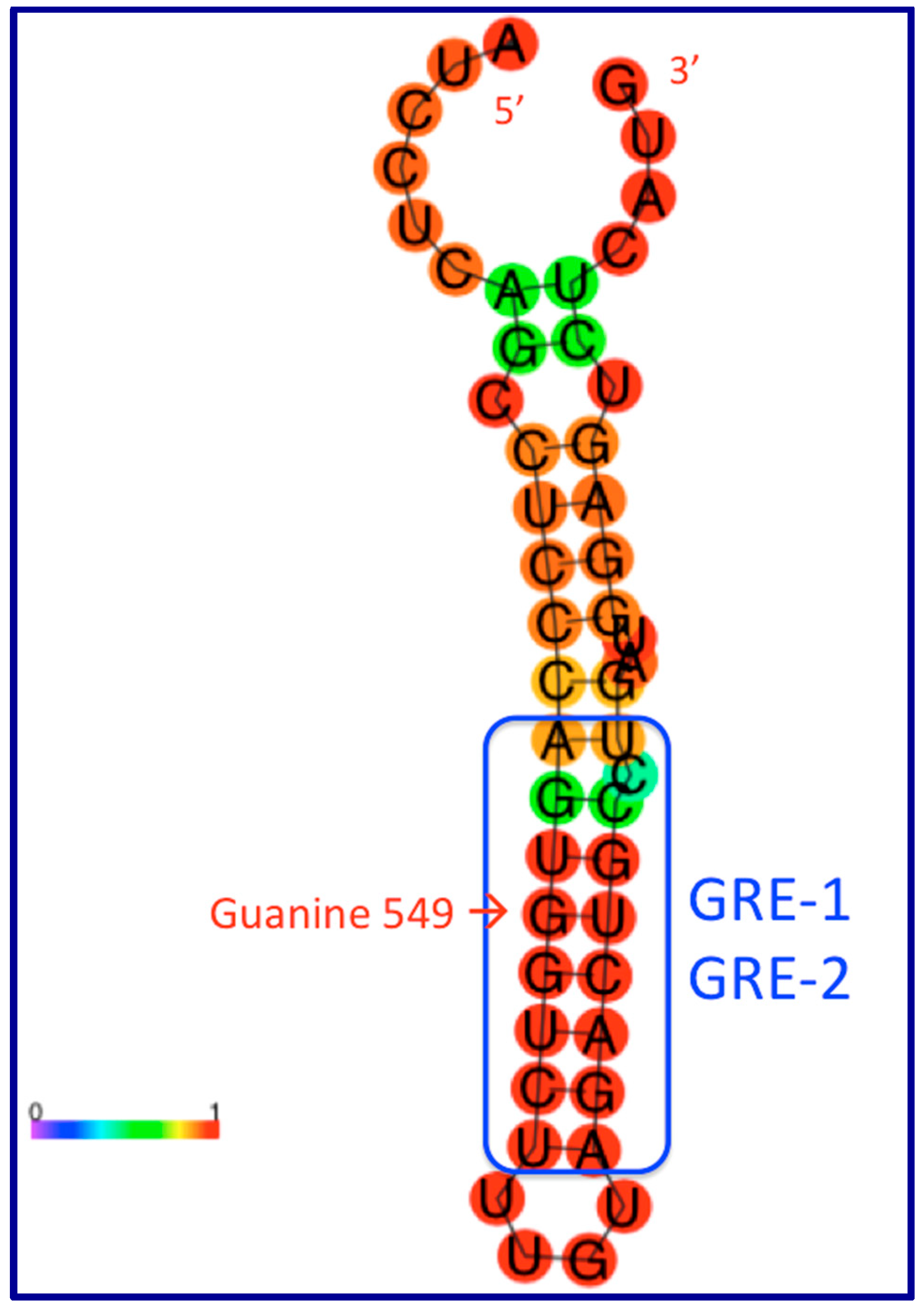
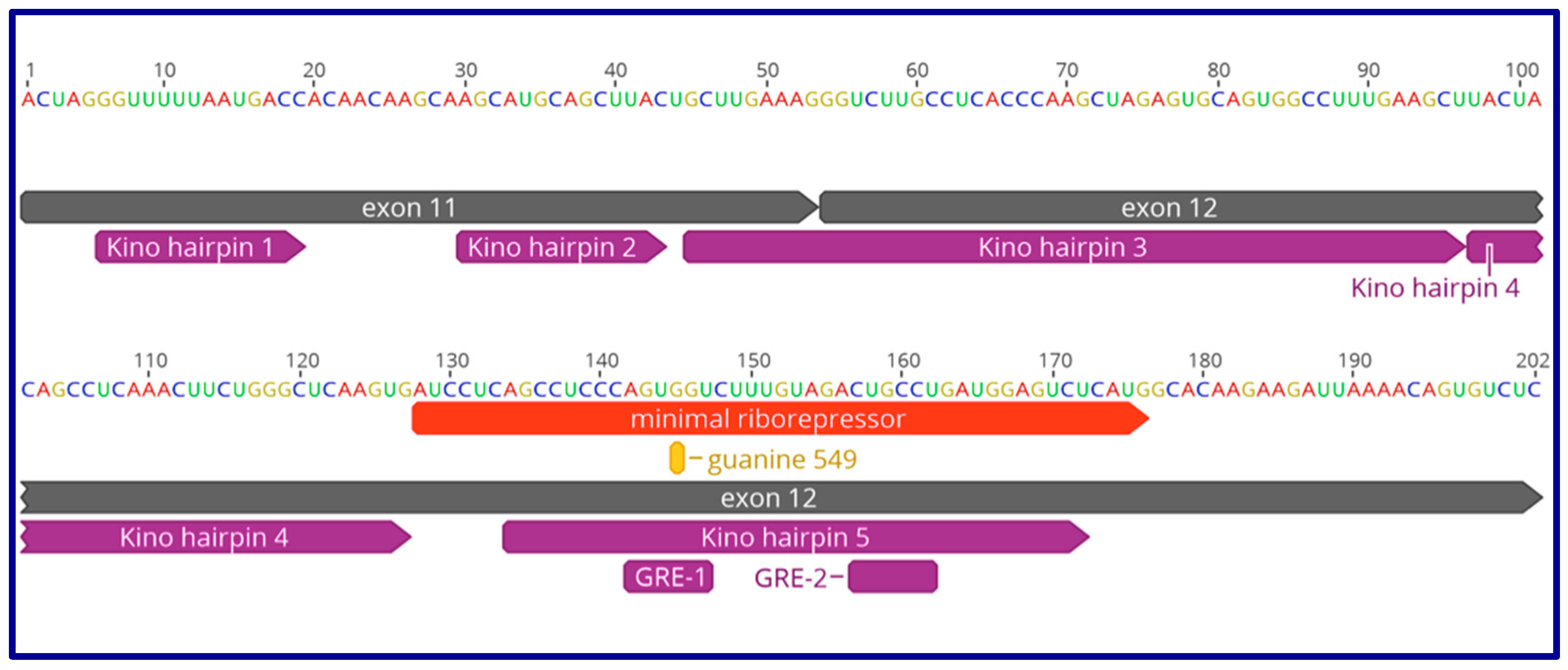
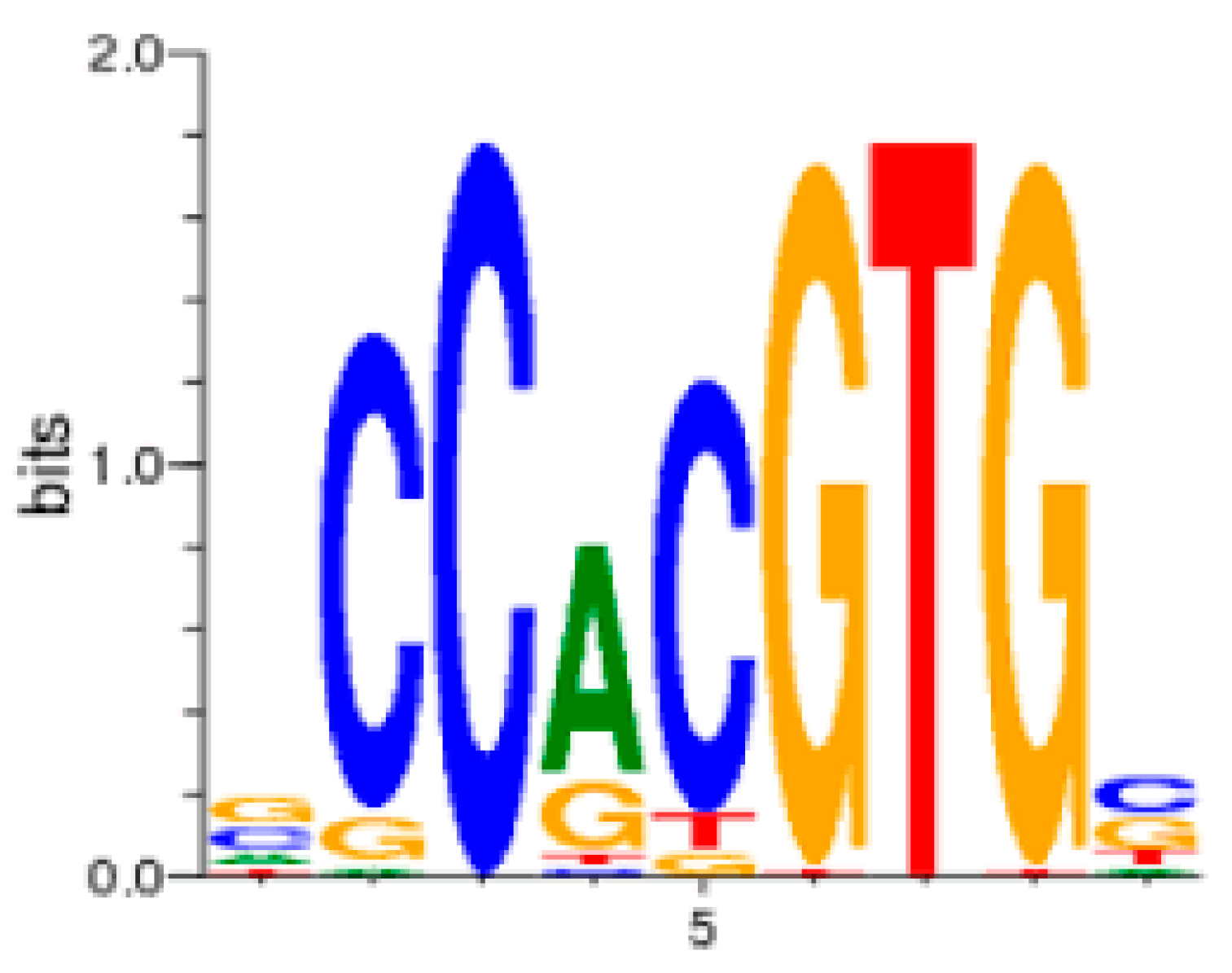
9. The Role of GAS5 lncRNA as an RNA Mimic of the Glucocorticoid Receptor (“Riborepressor”)
10. Evolution of the GAS5 Glucocorticoid Receptor RNA Mimic
11. Interaction of GAS5 Transcripts with microRNAs
12. Other Plausible Molecular Mechanisms of GAS5 Function
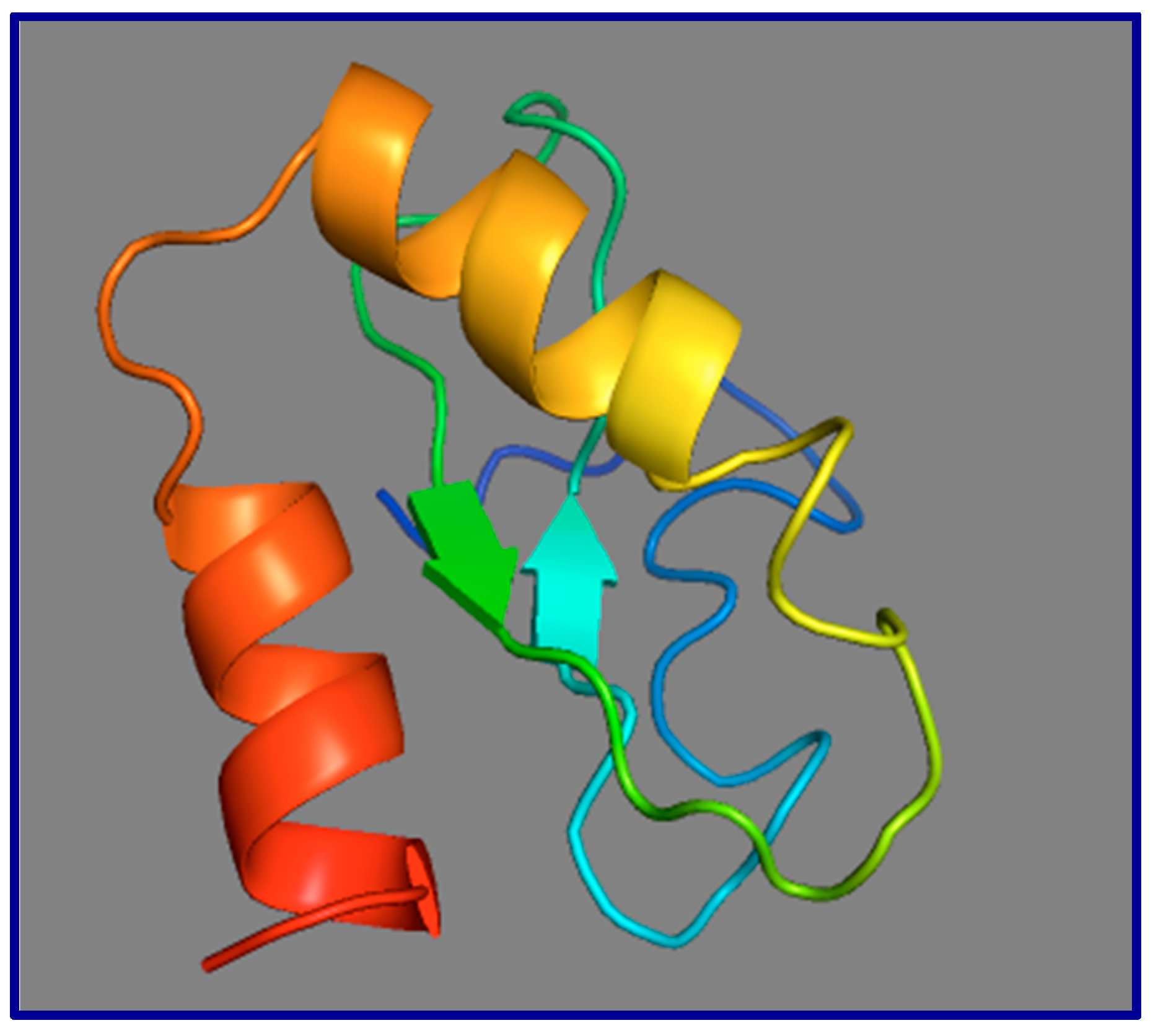
13. Small Open Reading Frames (smORFs), Whose Short-Protein Products Are Potentially Functional, Are Not Conserved between Mouse Gas5 and Human GAS5
14. Evolution of the GAS5 Gene in Primates
15. A Major Unexplored Question about GAS5 Function: Genes Responding Downstream to GAS5 RNA Level
16. Can Any Elements of the Human GAS5 Gene Operate Independently to Yield a Tumor Suppressor Phenotype?
17. Conclusions
18. Wrap-Up
Funding
Acknowledgments
Conflicts of Interest
References
- Baserga, R.; Huang, C.H.; Rossini, M.; Chang, H.; Ming, P.M. The role of nuclei and nucleoli in the control of cell proliferation. Cancer Res. 1976, 36, 4297–4300. [Google Scholar] [PubMed]
- Chiu, N.; Baserga, R. Changes in template activity and structure of nuclei from WI-38 cells in the prereplicative phase. Biochemistry 1975, 14, 3126–3132. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Baserga, R. Cytoplasmic ribonucleic acid synthesis in the prereplicative phase of isoproterenol-induced cell proliferation. Exp. Mol. Pathol. 1970, 13, 25–35. [Google Scholar] [CrossRef]
- Liebhaber, H.; Krugman, S.; Giles, J.P.; McGregor, D.M. Recovery of Cytopathic Agents from Patients with Infectious Hepatitis: Isolation and Propagation in Cultures of Human Diploid Lung Fibroblasts (Wi-38). Virology 1964, 24, 109–113. [Google Scholar] [CrossRef]
- Schneider, C.; King, R.M.; Philipson, L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988, 54, 787–793. [Google Scholar] [CrossRef]
- York, J.P.; Ren, Y.A.; Zeng, J.; Bin, Z.; Wang, F.; Chen, R.; Liu, J.; Xia, X.; Zhang, P. Growth Arrest Specific 2 (GAS2) is a Critical Mediator of Germ Cell Cyst Breakdown and Folliculogenesis in Mice. Sci. Rep. 2016, 6, 34956. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Kutter, C.; Watt, S.; Stefflova, K.; Wilson, M.D.; Goncalves, A.; Ponting, C.P.; Odom, D.T.; Marques, A.C. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012, 8, e1002841. [Google Scholar] [CrossRef]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Williams, G.T. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes 2015, 6, 484–499. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Steitz, J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998, 18, 6897–6909. [Google Scholar] [CrossRef] [PubMed]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Sizing up long non-coding RNAs: Do lncRNAs have secondary and tertiary structure? Bioarchitecture 2012, 2, 189–199. [Google Scholar] [CrossRef]
- Carninci, P.; Hayashizaki, Y. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 2007, 17, 139–144. [Google Scholar] [CrossRef]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef]
- Falaleeva, M.; Welden, J.R.; Duncan, M.J.; Stamm, S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. Bioessays 2017, 39, 1600264. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Ohira, M.; Li, Y.; Niizuma, H.; Oo, M.L.; Zhu, Y.; Ozaki, T.; Isogai, E.; Nakamura, Y.; Koda, T.; et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int. J. Oncol. 2009, 34, 931–938. [Google Scholar] [PubMed]
- Wang, Q.; Li, Q.; Zhou, P.; Deng, D.; Xue, L.; Shao, N.; Peng, Y.; Zhi, F. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed. Pharmacother. 2017, 91, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Damas, N.D.; Marcatti, M.; Come, C.; Christensen, L.L.; Nielsen, M.M.; Baumgartner, R.; Gylling, H.M.; Maglieri, G.; Rundsten, C.F.; Seemann, S.E.; et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat. Commun. 2016, 7, 13875. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.T. Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics 2009, 36, 447–454. [Google Scholar] [CrossRef]
- Valleron, W.; Laprevotte, E.; Gautier, E.F.; Quelen, C.; Demur, C.; Delabesse, E.; Agirre, X.; Prosper, F.; Kiss, T.; Brousset, P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia 2012, 26, 2052–2060. [Google Scholar] [CrossRef] [PubMed]
- Valleron, W.; Ysebaert, L.; Berquet, L.; Fataccioli, V.; Quelen, C.; Martin, A.; Parrens, M.; Lamant, L.; de Leval, L.; Gisselbrecht, C.; et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood 2012, 120, 3997–4005. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Bi, J.; Xue, X.; Zheng, L.; Zhi, K.; Hua, J.; Fang, G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012, 279, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P. Gametic imprinting in mammals. Science 1995, 270, 1610–1613. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Williams, G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res. Treat. 2014, 145, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Arshi, A.; Sharifi, F.S.; Khorramian Ghahfarokhi, M.; Faghih, Z.; Doosti, A.; Ostovari, S.; Mahmoudi Maymand, E.; Ghahramani Seno, M.M. Expression Analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in Breast Cancer Tissues from Young Women and Women over 45 Years of Age. Mol. Ther. Nucleic Acids 2018, 12, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Fei, T.; Verhaak, R.G.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Li, Y.; Li, L.; Hou, W.; You, Z. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol. Rep. 2016, 36, 3241–3250. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, W.; Jiang, J.; Bao, E.; Xu, D.; Zeng, Y.; Tao, L.; Qiu, J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS ONE 2013, 8, e73991. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Li, S.; Zhang, Q.; Wang, H.; Qin, H.; Wang, S. Long non-coding RNA GAS5 inhibits ovarian cancer cell proliferation via the control of microRNA-21 and SPRY2 expression. Exp. Ther. Med. 2018, 16, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, Y.; Tabei, M.B.; Askari, A.; Izadi, P.; Daraei, A.; Bastami, M.; Naghizadeh, M.M.; Nariman-Saleh-Fam, Z.; Mansoori, B.; Tavakkoly-Bazzaz, J. Expression levels of breast cancer-related GAS5 and LSINCT5 lncRNAs in cancer-free breast tissue: Molecular associations with age at menarche and obesity. Breast J. 2018, 24, 876–882. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Wen, Q.; Liu, Y.; Lyu, H.; Xu, X.; Wu, Q.; Liu, N.; Yin, Q.; Li, J.; Sheng, X. Long Noncoding RNA GAS5, Which Acts as a Tumor Suppressor via microRNA 21, Regulates Cisplatin Resistance Expression in Cervical Cancer. Int. J. Gynecol. Cancer 2017, 27, 1096–1108. [Google Scholar] [CrossRef]
- Pickard, M.R.; Mourtada-Maarabouni, M.; Williams, G.T. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta 2013, 1832, 1613–1623. [Google Scholar] [CrossRef]
- Li, G.; Cai, Y.; Wang, C.; Huang, M.; Chen, J. LncRNA GAS5 regulates the proliferation, migration, invasion and apoptosis of brain glioma cells through targeting GSTM3 expression. The effect of LncRNA GAS5 on glioma cells. J. Neurooncol. 2019, 143, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Luo, N.; Hu, Y.; Li, X.; Zhang, K. MiR-137 Suppresses Triple-Negative Breast Cancer Stemness and Tumorigenesis by Perturbing BCL11A-DNMT1 Interaction. Cell. Physiol. Biochem. 2018, 47, 2147–2158. [Google Scholar] [CrossRef] [PubMed]
- Bian, D.; Shi, W.; Shao, Y.; Li, P.; Song, G. Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma. Am. J. Transl. Res. 2017, 9, 1509–1520. [Google Scholar] [PubMed]
- Song, J.; Shu, H.; Zhang, L.; Xiong, J. Long noncoding RNA GAS5 inhibits angiogenesis and metastasis of colorectal cancer through the Wnt/beta-catenin signaling pathway. J. Cell. Biochem. 2019, 120, 6937–6951. [Google Scholar] [CrossRef]
- Efeyan, A.; Sabatini, D.M. mTOR and cancer: Many loops in one pathway. Curr. Opin. Cell Biol. 2010, 22, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Yacqub-Usman, K.; Pickard, M.R.; Williams, G.T. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate 2015, 75, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Williams, G.T. Role of GAS5 noncoding RNA in mediating the effects of rapamycin and its analogues on mantle cell lymphoma cells. Clin. Lymphoma Myeloma Leuk. 2014, 14, 468–473. [Google Scholar] [CrossRef]
- Heppner, G.H. Tumor heterogeneity. Cancer Res. 1984, 44, 2259–2265. [Google Scholar]
- Zong, Y.; Zhang, Y.; Sun, X.; Xu, T.; Cheng, X.; Qin, Y. miR-221/222 promote tumor growth and suppress apoptosis by targeting lncRNA GAS5 in breast cancer. Biosci. Rep. 2019, 39, BSR20181859. [Google Scholar] [CrossRef]
- Baselga, J.; Campone, M.; Piccart, M.; Burris, H.A., 3rd; Rugo, H.S.; Sahmoud, T.; Noguchi, S.; Gnant, M.; Pritchard, K.I.; Lebrun, F.; et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012, 366, 520–529. [Google Scholar] [CrossRef]
- Yardley, D.A.; Noguchi, S.; Pritchard, K.I.; Burris, H.A., 3rd; Baselga, J.; Gnant, M.; Hortobagyi, G.N.; Campone, M.; Pistilli, B.; Piccart, M.; et al. Everolimus plus exemestane in postmenopausal patients with HR(+) breast cancer: BOLERO-2 final progression-free survival analysis. Adv. Ther. 2013, 30, 870–884. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Shiekhattar, R. Long non-coding RNAs and enhancers. Curr. Opin. Genet. Dev. 2011, 21, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- Ramanathan, M.; Porter, D.F.; Khavari, P.A. Methods to study RNA-protein interactions. Nat. Methods 2019, 16, 225–234. [Google Scholar] [CrossRef]
- Raho, G.; Barone, V.; Rossi, D.; Philipson, L.; Sorrentino, V. The gas 5 gene shows four alternative splicing patterns without coding for a protein. Gene 2000, 256, 13–17. [Google Scholar] [CrossRef]
- Coccia, E.M.; Cicala, C.; Charlesworth, A.; Ciccarelli, C.; Rossi, G.B.; Philipson, L.; Sorrentino, V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell. Biol. 1992, 12, 3514–3521. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.; Jackson, S.E.; Wills, M.R.; Weissman, J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Torimura, M.; Akimitsu, N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE 2013, 8, e55684. [Google Scholar] [CrossRef] [PubMed]
- Maquat, L.E.; Tarn, W.Y.; Isken, O. The pioneer round of translation: Features and functions. Cell 2010, 142, 368–374. [Google Scholar] [CrossRef]
- Xu, X.; Lv, J.; Huang, Y.; Pu, J.; Hou, J.; Wang, L. Overexpression of lncRNA GAS5 suppresses prostatic epithelial cell proliferation by regulating COX-2 in chronic non-bacterial prostatitis. Cell Cycle 2019, 18, 923–931. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Yi, Z.; Jiang, L.; Li, Y.; Han, Q.; Yang, Y.; Zhang, Q.; Yang, Z.; Kuang, Y.; et al. LncRNA GAS5 regulates redox balance and dysregulates the cell cycle and apoptosis in malignant melanoma cells. J. Cancer Res. Clin. Oncol. 2019, 145, 637–652. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.L.; Maler, B.A.; Yamamoto, K.R. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell 1983, 33, 489–499. [Google Scholar] [CrossRef]
- Frank, F.; Okafor, C.D.; Ortlund, E.A. The first crystal structure of a DNA-free nuclear receptor DNA binding domain sheds light on DNA-driven allostery in the glucocorticoid receptor. Sci. Rep. 2018, 8, 13497. [Google Scholar] [CrossRef] [PubMed]
- Hudson, W.H.; Pickard, M.R.; de Vera, I.M.; Kuiper, E.G.; Mourtada-Maarabouni, M.; Conn, G.L.; Kojetin, D.J.; Williams, G.T.; Ortlund, E.A. Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat. Commun. 2014, 5, 5395. [Google Scholar] [CrossRef]
- Luisi, B.F.; Xu, W.X.; Otwinowski, Z.; Freedman, L.P.; Yamamoto, K.R.; Sigler, P.B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 1991, 352, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Parsonnet, N.V.; Lammer, N.C.; Holmes, Z.E.; Batey, R.T.; Wuttke, D.S. The glucocorticoid receptor DNA-binding domain recognizes RNA hairpin structures with high affinity. Nucleic Acids Res. 2019, 47, 8180–8192. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, N.A.; Busan, S.; Rice, G.M.; Nelson, J.A.; Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Ouyang, Z.; Chang, H.Y. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 2013, 8, 849–869. [Google Scholar] [CrossRef]
- Pickard, M.R.; Williams, G.T. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget 2016, 7, 10104–10116. [Google Scholar] [CrossRef]
- Kuo, T.; Lew, M.J.; Mayba, O.; Harris, C.A.; Speed, T.P.; Wang, J.C. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 11160–11165. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, L.; Wang, E.; Zhang, C.; Li, X. LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA for miR-137 to regulate the Notch1 signaling pathway. Biochem. Biophys. Res. Commun. 2018, 496, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Tian, G.; Cheung, H.H.; Wei, W.; Lee, T.L. Gas5 is an essential lncRNA regulator for self-renewal and pluripotency of mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res. Ther. 2018, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, J.; Xiong, G.; He, G.; Guan, X.; Yang, K.; Bai, Y. Negative regulation of lncRNA GAS5 by miR-196a inhibits esophageal squamous cell carcinoma growth. Biochem. Biophys. Res. Commun. 2018, 495, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Y.; Zheng, J.; Liu, X.; Chen, J.; Liu, L.; Wang, P.; Xue, Y. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim. Biophys. Acta 2017, 1864, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Shen, Z.; She, Q. Association Between the Deletion Allele of Ins/Del Polymorphism (Rs145204276) in the Promoter Region of GAS5 with the Risk of Atherosclerosis. Cell. Physiol. Biochem. 2018, 49, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jiang, J. GAS5 Regulates RECK Expression and Inhibits Invasion Potential of HCC Cells by Sponging miR-135b. Biomed. Res. Int. 2019, 2019, 2973289. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H. RNAa Induced by TATA Box-Targeting MicroRNAs. Adv. Exp. Med. Biol. 2017, 983, 91–111. [Google Scholar]
- Hu, G.; Lou, Z.; Gupta, M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS ONE 2014, 9, e107016. [Google Scholar] [CrossRef]
- Shi, Y.; Parag, S.; Patel, R.; Lui, A.; Murr, M.; Cai, J.; Patel, N.A. Stabilization of lncRNA GAS5 by a Small Molecule and Its Implications in Diabetic Adipocytes. Cell Chem. Biol. 2019, 26, 319–330. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Yang, T.; Zhang, N.; Liu, Z.; Jiang, Y. Low Long Noncoding RNA Growth Arrest-Specific Transcript 5 Expression in the Exosomes of Lung Cancer Cells Promotes Tumor Angiogenesis. J. Oncol. 2019, 2019, 2476175. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lv, Y.; Shao, C.; Chen, C.; Zhang, T.; Wei, Y.; Fan, H.; Lv, T.; Liu, H.; Song, Y. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J. Cell. Physiol. 2019, 234, 20721–20727. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Chen, B.; Calin, G.A. Exosomal lncRNAs as new players in cell-to-cell communication. Transl. Cancer Res. 2018, 7, S243–S252. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Muller, A.J.; Chatterjee, S.; Teresky, A.; Levine, A.J. The gas5 gene is disrupted by a frameshift mutation within its longest open reading frame in several inbred mouse strains and maps to murine Chromosome 1. Mamm. Genome 1998, 9, 773–774. [Google Scholar] [CrossRef]
- Stevens, N.J.; Seiffert, E.R.; O’Connor, P.M.; Roberts, E.M.; Schmitz, M.D.; Krause, C.; Gorscak, E.; Ngasala, S.; Hieronymus, T.L.; Temu, J. Palaeontological evidence for an Oligocene divergence between Old World monkeys and apes. Nature 2013, 497, 611–614. [Google Scholar] [CrossRef]
- Boursot, P.; Auffray, J.-C.; Britton-Davidian, J.; Bonhomme, F. The evolution of house mice. Annu. Rev. Ecol. Syst. 1993, 24, 119–152. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Chugunova, A.; Loseva, E.; Mazin, P.; Mitina, A.; Navalayeu, T.; Bilan, D.; Vishnyakova, P.; Marey, M.; Golovina, A.; Serebryakova, M.; et al. LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 4940–4945. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Xu, X.; Fang, Y.; Zhao, S.; Hu, J.; Xu, J.; Jia, P.; Ding, X.; Teng, J. The effect of long noncoding RNA GAS5 on apoptosis in renal ischemia/reperfusion injury. Nephrology 2018, 24, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, H.; Jin, L.; Li, G.; Hu, S.; Ning, C.; Guo, J.; Shuai, S.; Li, X.; Li, M. Long Noncoding RNA GAS5 Suppresses 3T3-L1 Cells Adipogenesis Through miR-21a-5p/PTEN Signal Pathway. DNA Cell Biol. 2018, 37, 767–777. [Google Scholar] [CrossRef] [PubMed]
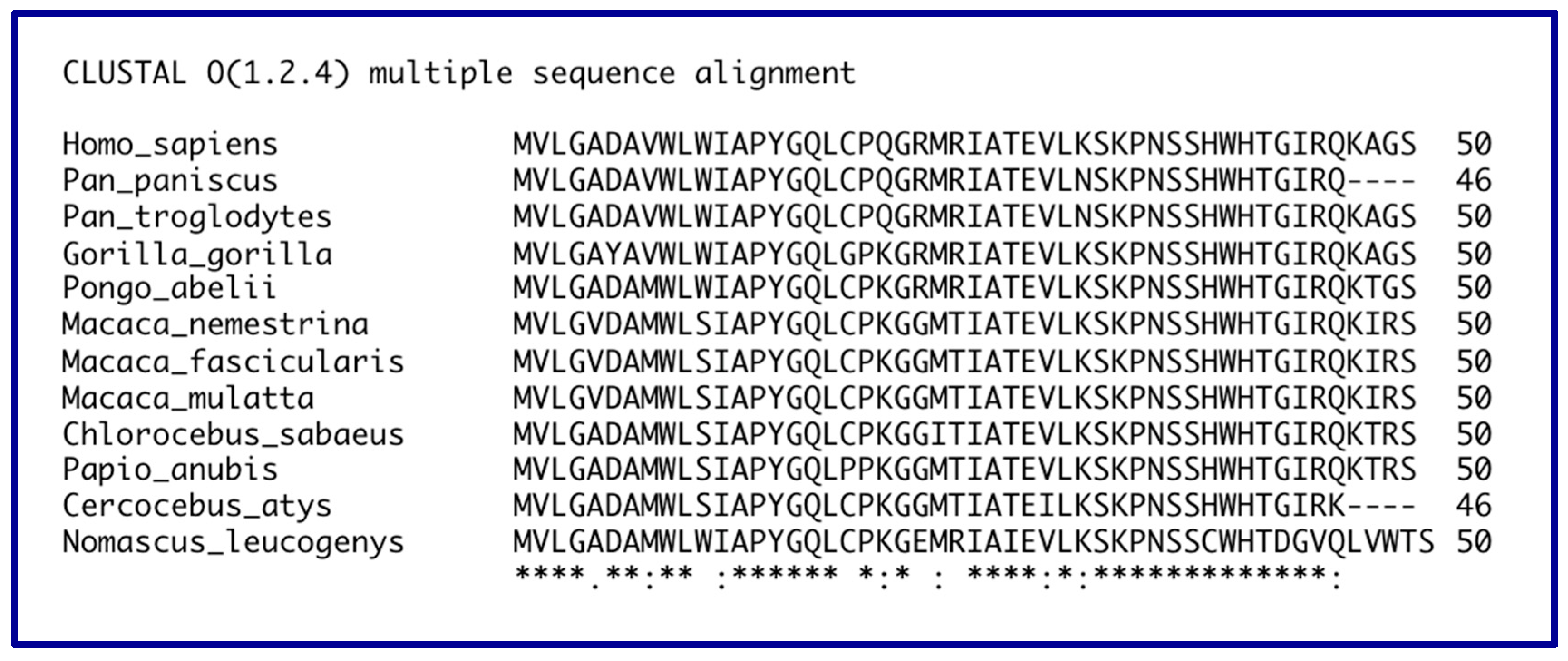

| Gene Name/Aliases | Phenotype | Chromosomal Locale * | Small ORFs | Effect of Estrogen | Intronic snoRNAs |
|---|---|---|---|---|---|
| Mouse Gas5 | Tumor suppressor | Chromosome 1 chr1:161,035,166-161,038,537 | Yes | Unknown | Nine SNORD class snoRNAs |
| Human GAS5 | Tumor suppressor | Chromosome 1q25; chr1:173,833,037-173,837,129 | Yes | Repression by ~60% | 10 SNORD class |
| Human SNHG16 ** | Oncogene | Chromosome 17q25.1; chr17:74,553,860-74,561,430 | Yes; Yu et al., [22] | Little or no effect | Three SNORD class snoRNAs |
| Human SNHG1 | Oncogene | 11q12.3; chr11:62,619,460-62,623,357 | Wang Q et al., [23] | Little or no effect | Nine SNORD class snoRNAs |
| Human SNHG5 | Oncogene | 6q14.3; chr6:86,386,725-86,388,438 | Dong et al., [25] | Very small downregulation with E2 | Two SNORD class snoRNAs |
| MicroRNA/Aliases | Site of Interaction with GAS5 lncRNA | Tumor or Cell Type | Effect Demonstrated | Reference |
|---|---|---|---|---|
| miR-221/222 | Exon | Gliomas | Unknown | Zong et al., [49] |
| miRNA-21 | Exon 5 * | Endothelial cells, ovarian carcinoma, cervical carcinoma | Reduced | Ma, et al. [36] Shen and She [86]; Wen et al. [39] |
| miRNA-135b | Exon | Hepatocellular carcinoma (HCC) | Sponging or translational inhibition | Yang and Jiang [87]. |
| miRNA-196a | GAS5 promoter ** or exon | Glioma, esophageal squamous cells | Downregulation of GAS5 RNA (Wang) | Wang et al. [83]; Zhao et al. [84] |
| miRNA-137 | Exon | Melanoma | GAS5 RNA and miRNA co-regulated | Bian et al. [43] |
| Species | Exons Spliced to Form cDNA | Predicted Amino Acid Sequence | RefSeq File |
|---|---|---|---|
| Human | Exons 2-3-4-5 * | MVLGADAVWLWIAPYGQLCPQGRMRIATEVLKSKPNSSHWHTGIRQKAGS Predicted MW 5547 daltons, predicted pI 10.02 | NR_002578 |
| Human | Exon 0-1-2-3-4-5 * | MGTRRRAGEPALRSPGTILSFRGMVLGADAVW LWIAPYGQLCPQGRMRIATEVLKSKPNSSHWHTGIRQKAGS * Predicted MW 8016, predicted pI 11.55 | NR_152521 |
| Mouse | Exon 3-4-5-6 ** | MKAYEDSSGSWITERAQCARIEDQKMKWWSLRLDRQFES * Predicted MW 4767 daltons; predicted pI 6.11 | NR_153812 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goustin, A.S.; Thepsuwan, P.; Kosir, M.A.; Lipovich, L. The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers. Non-Coding RNA 2019, 5, 46. https://doi.org/10.3390/ncrna5030046
Goustin AS, Thepsuwan P, Kosir MA, Lipovich L. The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers. Non-Coding RNA. 2019; 5(3):46. https://doi.org/10.3390/ncrna5030046
Chicago/Turabian StyleGoustin, Anton Scott, Pattaraporn Thepsuwan, Mary Ann Kosir, and Leonard Lipovich. 2019. "The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers" Non-Coding RNA 5, no. 3: 46. https://doi.org/10.3390/ncrna5030046
APA StyleGoustin, A. S., Thepsuwan, P., Kosir, M. A., & Lipovich, L. (2019). The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers. Non-Coding RNA, 5(3), 46. https://doi.org/10.3390/ncrna5030046





