Abstract
Escherichia coli is considered one of the most common agents associated with neonatal diarrhea in piglets. The aim of this work was to characterize the pathogenic and antimicrobial resistance (AMR) profiles of 122 E. coli strains isolated from pigs suffering diarrhea (n = 94) and pigs without diarrhea (n = 28) of 24 farms in Spain. Virulence factors, toxins and AMR (ESBL and colistin) genes and AMR phenotypes of E. coli isolates were analyzed. Low prevalence of pathogenic E. coli strains (26%) was found in both groups. However, ETEC and VTEC strains were more frequently isolated from diarrheic piglets. Irrespectively of diarrhea occurrence, 97.5% of the strains showed a multidrug-resistance (MDR) profile to aminopenicillins, sulfonamides and tetracyclines. It was found that 22% of E. coli was CTX-M+, with CTX-M-14 being the principal allelic variant. Remarkably, 81.5% of CTX-M+ strains were isolated from diarrheic animals and presented an extended MDR profile to aminopenicillins, quinolones and aminoglycosides. Finally, low frequencies of colistin resistance genes mcr-1 (4/122) and mcr-4 (1/122) were found. MDR E. coli strains are circulating in pig farms of Spain, representing a serious threat to animal and public health. More appropriate diagnostic approaches (genetic and AMR phenotypic analysis) should be implemented in animal health to optimize antibiotic treatments.
1. Introduction
Escherichia coli has historically been considered as one of the most common agents associated with diarrhea in suckling and post-weaned piglets [1]. There is high diversity and variants of E. coli strains integrating the normal gut microbiota, with most of them being considered not pathogenic [2]. The characterization of pathogenic E. coli strains is usually based on the presence of virulence factors [3]. In piglets, E. coli pathogenic strains can be classified into different pathotypes: enterotoxigenic (ETEC) strains releasing heat-labile (LT) and heat-stable Sta and Stb exotoxins, intimin (eae)-producing enteropathogenic (EPEC) strains and verotoxigenic (VTEC) strains producing VT1/VT2 verotoxins [4].
The routine use of antimicrobials in livestock, especially in the pig industry, for either ‘prophylaxis’ or ‘metaphylaxis’ represents a serious hazard for the selection of multidrug-resistant (MDR) Enterobacteriaceae strains [5,6]. In fact, the effectiveness of treatments against E. coli is threatened by the dramatic increase of extended-spectrum beta-lactamases (ESBL)-producing isolates worldwide [7,8,9]. A high number of ESBL have been described and designated according to the strains of bacteria or plasmid that produce them by using letters that usually evoke the name of the target antibiotic, but they are also named after substrates, their biochemical properties, location of their discovery and the name of the patient or the discovering investigators [10]. Genes encoding ESBL enzymes are usually located in plasmids and can be easily horizontally transferred among different bacterial populations. Most ESBL derive from the first enzymes discovered, TEM (named after the patient (Temoneira) providing the first sample) and SHV (sulfhydryl reagent variable), which confer resistance to beta-lactams such as ampicillin or amoxicillin. The most recent ESBLs are derived from the CTX-M (active on cefotaxime-Munich) enzymes, which confer resistance to third- and fourth-generation cephalosporins [10,11]. Cephalosporins constitute one of the largest families of antimicrobials widely used in both human and veterinary medicine [12]. On the other hand, in veterinary medicine, colistin sulfate is indicated for the treatment of Enterobacteriaceae infections and has been widely used as a preventive mass-medication of colibacillosis in piglets [13]. Although colistin is one of the antimicrobial agents with the lowest rates of resistance, the emergence of the mcr-1 (mobile colistin resistant) plasmid in the Enterobacteriaceae population has resulted in the appearance of strains with acquired resistance to this antibiotic [14,15]. In order to stop the increase of colistin-resistant strains, the European Medicines Agency established in 2016 that all EU members should restrict and reduce the use of colistin in animals for treating infections with a target level of 5 mg/PCU, where PCU refers to the ‘population correction unit’ and takes into account the animal population as well as the estimated weight of each particular animal at the time of treatment with antimicrobials. [16].
During recent decades, the importance of E. coli in porcine neonatal diarrhea seems to have decreased [17], possibly due to the successful vaccination plans implemented in the pig farms [18]. Therefore, antimicrobial treatment might not be required in most cases of diarrhea, especially in those where the role of the E. coli is not well defined. However, the systematic mass-medication of piglets suffering from diarrheic processes is still common in many cases.
The aim of the present work was to characterize the virulence factors and to determine the genotypic and phenotypic antimicrobial resistance patterns of E. coli isolates from cases of neonatal diarrhea in conventional pig farms of Catalonia (Spain). Moreover, in order to assess the clinical relevance of all isolates, samples from non-diarrheic pen-mates were also analyzed and compared to the clinical cases.
2. Materials and Methods
2.1. Microbiological Testing
A total of 122 E. coli isolates obtained from 24 conventional Spanish farms suffering neonatal diarrhea outbreaks during 2017 and 2018 were recovered from a previous study [19]. All the studied farms were located in Catalonia (NE of Spain), the Spanish region with the greatest number of production farms and one of the highest pig-density (242 animals/km2) regions in Europe. The sampling procedure included 5–10 samples from diarrheic animals and 3–5 samples from apparently healthy pen-mates for each farm. One gram of fecal sample was obtained directly from the animals using rectal swabs and submitted for diagnostic testing to the Laboratori Veterinari de Diagnosi de Malalties Infeccioses of the Universitat Autònoma de Barcelona (Spain). Besides E. coli, all samples were tested for a panel of enteric infectious agents which comprised C. perfringens types A and C toxins (Cpα, Cpβ, Cpβ2); C. difficile toxins (TcdA, TcdB); rotavirus A (RVA), B (RVB) and C (RVC); porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV), and these general results were described elsewhere [19].
For E. coli isolation, stool samples were cultured in standard Blood Agar (BD GmbH, Germany) and MacConkey Agar (Oxoid, UK) and aerobically incubated at 37 °C for 24 h. A total of 122 pure isolates were collected: 94 from diarrheic samples of 1-week-old diseased piglets and 28 from feces of apparently healthy pen-mates. All strains were confirmed as E. coli using conventional biochemical tests. Only E. coli strains obtained as pure cultures from a direct seeding of feces were used for the characterization, since the lack of microbial diversity was considered abnormal.
2.2. Characterization of E. coli Virulence Factors and Toxins
The presence of E. coli toxins (Sta, Stb, LT, EAST1, VT1 and VT2), fimbrial adhesins (F4, F5, F6, F18 and F41) and non-fimbrial adhesin eae genes was analyzed by qualitative PCR, as described by Toledo et al. (2012) with slight modifications [20]. Briefly, PCR assays were carried out using the Biotaq DNA polymerase kit (Biotaq, Ecogen) using previously described primers [19]. The final 25 µL mixture consisted of: 1× PCR Buffer, 0.8 mM of the dNTP mix, 3 mM of MgCl2, 1 mM of each primer and 1 U of Taq polymerase. As template, 2.5 µL of the DNA sample was used. The PCR program consisted of 5 min at 94 °C, followed by 30 cycles of 1 min at 94 °C, 1 min of annealing at 57 °C and 1 min of extension at 72 °C, and a final extension step of 7 min at 72 °C. Positive and negative (ultrapure water) controls were included in each run and particular care was taken to prevent carry-over contamination.
Reference E. coli strains used as positive controls were FV12048 (F4+ LT+ EAST1+), FV9722 (F5+ F41+), FV7633 (F6+), FV12047 (F18+ Sta+ Stb+) and O157-84 (eae+ VT1+ VT2+) were kindly donated by Dr. Blanco (E. coli Reference Laboratory, Santiago de Compostela, Spain).
2.3. Antimicrobial Resistance Phenotyping Analyses
Antimicrobial susceptibility of E. coli isolates was tested using the disk diffusion method [21]. Briefly, one to four colonies were suspended in 5 mL of distilled sterile water to achieve a turbidity of 0.5 in the McFarland scale. The dilution was then seeded onto Mueller–Hinton (Oxoid, UK) plates. Each isolate was tested for the following antimicrobial groups, using commercial disks: aminopenicillins (ampicillin, amoxicillin and amoxicillin/clavulanic acid); cephalosporins (ceftiofur, cephalexin, cefquinome, ceftriaxone); quinolones (ciprofloxacin, enrofloxacin, flumequine); aminoglycosides (gentamicin, neomycin, streptomycin, apramycin); tetracyclines (tetracycline, doxycycline); sulfonamides (sulfonamide, sulfamethoxazole/trimethoprim); florfenicol; colistin; lincospectin. Concentration of the commercial disks and breakpoints for each antimicrobial are shown in Table 1.

Table 1.
Concentrations and breakpoints of the antibiotic disks used for the disk diffusion method.
Additionally, minimum inhibitory concentration (MIC) test was performed in order to evaluate antimicrobial susceptibility to colistin, using the broth microdilution method in 96-well plates [22]. E. coli ATCC 25,922 was used as the quality control strain. Briefly, the tested colistin concentrations ranged from 0.25 to 8 μg/mL. The tested strains were considered resistant when their MIC value was higher than the wild type cut-off value (MIC > 2μg/mL) [23].
2.4. Genetic Characterization of Antimicrobial Resistance Genes
Molecular diagnosis of AMR genes was performed for the ESBL genes blaSHV, blaCTX-M, blaCMY1, blaCMY2 and blaTEM; carbapenemase blaOXA and mcr-1 to mcr-5 colistin-resistance genes [25]. PCR conditions were homogenized for all the reactions as follows: 5 min at 94 °C, followed by 25 cycles of 1 min at 94 °C, 1 min of annealing at 55 °C and 1 min of extension at 72 °C, and a final extension step of 7 min at 72 °C. Amplified PCR products were Sanger sequenced for verification at the Genomic and Bioinformatics Service of the Universitat Autònoma de Barcelona (Barcelona, Spain). Sequences and chromatograms were manually explored to trim bad-quality bases using the BioEdit software. The obtained sequences were aligned using Clustal Omega and blasted against those described at the National Center for Biotechnology Information (NCBI) database.
2.5. Statistical Analysis
Chi-squared or Fisher exact tests were used for comparison between proportions when appropriate. Results were considered statistically significant when p < 0.05.
3. Results
3.1. Microbiological Identification and Characterization of E. coli Isolates
Most of the E. coli isolates presented a very low prevalence of virulence factors and toxins (<5%). with the exception of EAST1, which was present in more than 70% of the strains (Table 2). Moreover, 74% of the strains were negative for all toxins and intimin (eae) genes. As such, it was not possible to classify them into any pathotype.

Table 2.
Comparison of virulence factors and pathogenic E. coli prevalence in diarrheic (n = 94) and non-diarrheic (n = 28) pig groups.
Table 3 shows the number of isolates presenting virulence factors and toxins grouped by pathotype and their total frequency in diarrheic and not diarrheic cases. The most common pathotypes within the classified strains were EPEC (n = 17) and ETEC (n = 15), with the frequency of EPEC being two times higher (21% vs. 10.6%) in non-diarrheic piglets. As expected, the frequency of ETEC was higher in animals presenting diarrhea than in those without clinical signs (15% vs. 3.5%). The presence of VTEC (4.3%) was only detected in cases of diarrhea. Similarly, E. coli strains positive for F4, LT, Sta, VT1 and VT2 genes were only isolated from clinical cases.

Table 3.
Description of the profiles of virulence factor production and classification of the toxigenic strains into pathotypes of E. coli strains.
3.2. Antimicrobial Resistance Phenotyping and Genotyping
According to the results obtained by the disk diffusion method, the overall prevalence of AMR in E. coli isolates was very high for most of the nine tested antimicrobial classes. The highest rates of resistance were found for sulfonamides (87.3%), tetracyclines (87.3%) and aminopenicillins (80.1%), followed by quinolones (49%), cephalosporins (32.9%) and aminoglycosides (27.9%). By contrast, low resistance levels were observed for colistin (6.5%) and apramycin (5.9%). Regarding the MIC results, 6 out of 122 (5%) were resistant to colistin (MIC > 2 μg/mL): 5 of them had an MIC value of 4 μg/mL, and one isolate had an MIC of 8 μg/mL. Comparing the colistin results obtained by disk diffusion and the MIC, only one strain was considered resistant to colistin by both methods.
Most of the strains, 97.5% (119/122), presented a multidrug-resistance (MDR) profile (≥3 antimicrobial categories), and 67.2% of them offered resistance to six antibiotic classes. Moreover, 30% of the strains (37/122) showed an extensive drug-resistance (XDR) profile (>7 antimicrobial classes). Two strains presented resistance to all the tested antimicrobials except for colistin.
The comparison study between E. coli isolates from diarrheic and non-diarrheic samples showed statistical differences in the proportion of AMR E. coli isolates (Figure 1). Remarkably, piglets presenting diarrhea had higher prevalence of E. coli strains resistant to quinolones and aminoglycosides (gentamicin and neomycin) compared to the non-diarrheic isolates (Fisher exact test p < 0.05).
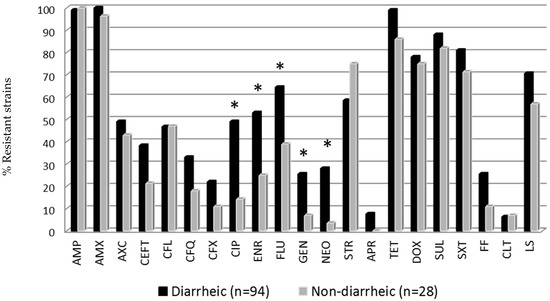
Figure 1.
Antimicrobial resistance of E. coli strains from diarrheic and non-diarrheic groups. * Statistical differences between diarrheic and non-diarrheic animals (Fisher exact test p < 0.05). Antibiotic abbreviations are shown in Table 1.
The most prevalent ESBL gene was the blaCTX-M found in 22% (27/122) of the total samples, and 81.5% (22/27) of them were isolated from diarrheic animals. All the CTX-M+ strains except one were successfully sequenced and could be classified into the following blaCTX-M genotypes: CTX-M-14 (38.5%), CTX-M-1 (19.2%), CTX-M-15 (19.2%), CTX-M-32 (11.5%), CTX-M-27 (7.7%) and CTX-M-3 (3.8%).
On the other hand, E. coli harboring CTX-M+ genes presented a wider resistance pattern than those CTX-M-negative strains (CTX-M–). Specifically, 81.5% of the CTX-M+ strains showed resistance to more than 10 antimicrobials, while in the case of CTX-M– the percentage was 37.9%. Statistical differences between CTX-M+ and CTX-M– strains were found for amoxicillin/clavulanic acid, cephalosporins, quinolones and gentamicin drugs (Figure 2). By contrast, colistin resistance genes were detected in a low proportion of cases: four strains harbored the mcr-1 gene (3%), and one strain was positive to mcr-4. Only one of the mcr-1+ strains, isolated from a diarrheic animal, exhibited phenotypic resistance to colistin in the MIC test (8 μg/mL).
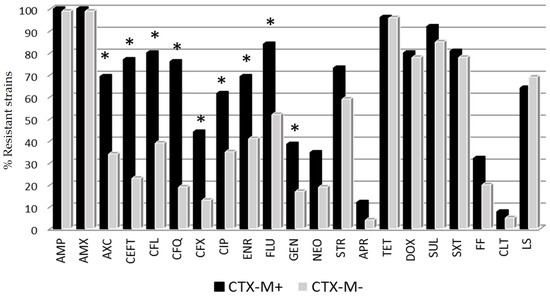
Figure 2.
Percentage of E. coli resistant strains to different antimicrobial categories according to CTX-M gene expression. * Statistical differences between CTX-M+ (positive) and CTX-M– (negative) strains (Fisher exact test p < 0.05). Antibiotic abbreviations are shown in Table 1.
4. Discussion
The aim of this work was to characterize the E. coli strains involved in porcine neonatal diarrhea outbreaks by genotyping and phenotyping isolates grown as pure cultures from both clinical cases of diarrheic and non-diarrheic healthy pen-mates.
Surprisingly, the general prevalence of strains encoding genes for virulence factors, as well as the presence of the typical E. coli pathotypes, was low in our pig population studied. Merely 26% (32/122) of the strains could be classified into a specific pathotype. EPEC was the most frequent one, representing 14% of the total isolates, followed by ETEC (12%) and VTEC (3%). The presence of EPEC is usually reported as a causative agent of diarrhea, principally in post-weaned but also in suckling pigs, but in this study, the presence of this pathotype was found more prevalent in non-diarrheic piglets (21.4%) than in the diarrheic ones (11.8%). Similar results have been reported in previous works in agreement with the results presented in the present study. Mainly, EPEC prevalence ranging from 3% to 26% in diarrheic animals and 9% to 17% in non-diarrheic animals has been described [26,27,28]. Finally, EAST1 gene was highly detected in both diarrheic (70%) and non-diarrheic groups (86%) but again with no relationship with the clinical onset. The EAST1 toxin is produced by several E. coli pathotypes [29], and although initially it was believed to be involved in the pathogenesis of diarrhea, most authors have concluded that this toxin may not be a virulence determinant, because it has been commonly isolated from healthy animals as well [30,31].
On the contrary, ETEC and VTEC were more frequently isolated from diarrheic samples, suggesting a possible association between these pathotypes and the emergence of disease in 1-week-old piglets.
The AMR phenotypes of the E. coli isolates showed a wide resistance to sulfonamides, tetracyclines and aminopenicillins in both diarrheic and non-diarrheic piglets. High percentages of antimicrobial resistance for amoxicillin and ceftiofur have been previously reported in pig farms from Catalonia [32]. However, other studies conducted in other regions of Spain [33] or in Ireland [34] reported lower levels of resistance to tetracycline, sulfamethoxazole/trimethoprim, ampicillin and streptomycin. These differences could be explained by the fact that Catalonia is one of the areas with greater density of pigs in Europe and therefore is likely subjected to a high consumption of antibiotics.
On the other hand, an interesting finding of this study was that E. coli isolates from diarrheic cases additionally presented extended AMR to quinolones and aminoglycosides (gentamicin and neomycin) compared to those from healthy pigs. These findings agree with other studies performed in Ontario [35] and in Denmark [36], which found consistently higher frequencies of resistance in diarrheic and ETEC isolates, respectively.
The genotyping study showed that the E. coli strains containing the blaCTX-M gene presented resistance to third- and fourth-generation cephalosporins, amoxicillin/clavulanic acid, quinolones and aminoglycosides. The relationship between ESBLs and quinolone resistance mechanisms has been documented by several authors [37,38,39], and it has been suggested that ESBL genes and plasmid-mediated quinolone resistance are located within the same plasmid [40]. Furthermore, strains harboring the blaCTX-M gene presented higher values of phenotypical resistance than those without this gene, mainly to all the tested beta-lactams, quinolones and gentamicin.
Regarding the genotyping of the CTX-M+ strains, all the allelic variants described in this study have previously been reported in pigs, with CTX-M-14 also being the most prevalent [41,42]
In the case of colistin, resistance was assessed using two different methods, the disk diffusion method and the MIC test, finding low concordance between both techniques. According to several authors, the disk diffusion method does not seem to be a reliable method for testing the efficacy of colistin due to the poor diffusion of polymyxins in agar. Therefore, the use of other tests, such as MIC, is more adequate for testing the susceptibility of colistin [43].
In general, the prevalence of porcine colistin-resistant E. coli strains reported in Europe is low: 0.9% in Poland [44], 1.5% in France [45], 2.9% in Germany [46] and 3.7% in Switzerland [47]. These results are in concordance with the 5% of prevalence obtained in this study using the MIC test. Nevertheless, García et al. (2018) have reported an extremely high prevalence of colistin resistance (77%) in the northwest area of Spain [48]. Variations in animal health management, biosecurity and sanitation procedures in pig production between different regions of the same country could explain this different colistin-resistance prevalence.
Likewise, the prevalence of E. coli isolates harboring the mcr-1 gene was low (3%) in this study, in agreement with results reported in Germany [46,49] and Belgium [50] with prevalence below 10%. Nevertheless, prevalences of 26% and 88% have been published in studies conducted in the north-west area of Spain [48] and in France [51], respectively. Only one E. coli isolate of this study presented the mcr-4 gene; however, the frequency of this gene in the Spanish pig farms is variable, ranging from 21% [8] to 73% [48] depending of the region. Regarding other mcr genes, none of the isolates of this study were positive for mcr-2, mcr-3 or mcr-5. The mcr-2 gene has been detected in porcine and bovine E. coli isolates from Belgium [50], mcr-3 in E. coli isolates from pigs in UK [52] and mcr-5 in 3.4% of isolates in Spain [48].
In summary, E. coli CTX-M+ strains isolated from diseased piglets showed antimicrobial multi-resistance to β-lactams, tetracyclines, sulfonamides and lincospectin extended to quinolones and aminoglycosides. This high prevalence of E. coli MDR strains in the pig population can represent a serious threat for animal and human health [53].
5. Conclusions
More appropriate diagnostic approaches, including genetic and phenotypic analysis of AMR profiles, should be implemented in animal health to optimize the use of antibiotics for treating diseased animals when necessary and with an effective antimicrobial agent, as the presence of highly resistant strains harboring ESBL and mcr genes circulating in pig farms can represent a potential threat to animal and human health.
Author Contributions
Conceptualization, A.V., L.D. and M.M.; Data curation, A.V., L.A., C.S., M.T. and N.R.; Formal analysis, A.V. and L.A.; Investigation, A.V., L.A., L.D. and M.M.; Methodology, A.V., L.A., C.S., M.T. and N.R.; Supervision, L.D. and M.M.; Writing-original draft, A.V., L.D. and M.M.; Writing-review & editing, A.V., L.A., L.D. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
Anna Vidal was supported by Universitat Autònoma de Barcelona (PIF-UAB 2015). Laia Aguirre was supported by a Collaboration scholarship from the Ministerio de Educación, Cultura y Deporte from the Spanish government (2017–2018). This research received no external funding.
Acknowledgments
Our grateful thanks to the participating veterinarians affording samples from the piglet diarrhea outbreaks. Reference E. coli strains were kindly donated by Jorge Blanco from the Laboratorio de Referencia de Escherichia coli (LREC), Santiago de Compostela, Spain.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Fairbrother, J.M.; Nadeau, E. Colibacillosis. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; pp. 807–835. [Google Scholar]
- DebRoy, C.; Maddox, C.W. Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev. 2001, 2, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Mainil, J. Escherichia coli virulence factors. Vet. Immunopathol. 2013, 152, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Robins-Browne, R.M.; Holt, K.E.; Ingle, D.J.; Hocking, D.M.; Yang, J.; Tauschek, M. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front. Cell. Infect. Microbiol. 2016, 6, 141. [Google Scholar] [CrossRef]
- Angulo, F.J.; Nargund, V.N.; Chiller, T.C. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Burow, E.; Simoneit, C.; Tenhagen, B.A.; Käsbohrer, A. Oral antimicrobials increase antimicrobial resistance. Prev. Vet. Med. 2014, 113, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Canton, R.; Gniadkowski, M.; Nordmann, P.; Rossolini, G.M.; Arlet, G.; Ayala, J.; Coque, T.M.; Kern-Zdanowicz, I.; Luzzaro, F.; et al. CTX-M: Changing the face of ESBL in Europe. J. Antimicrob. Chemother. 2007, 59, 165–174. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzoti, G.; Magistrali, C.F. Novel plasmid-mediated mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017, 22, 30589. [Google Scholar] [CrossRef]
- Kibret, M.; Abera, B. Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr. Health Sci. 2011, 11, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. β-Lactamase Nomenclature. Antimicrob. Agents Chemother. 2006, 50, 1123–1129. [Google Scholar] [CrossRef]
- Randall, L.P.; Lemma, F.; Rogers, J.P.; Cheney, T.E.A.; Powell, L.F.; Teale, C.J. Prevalence of extended-spectrum-β-lactamase-producing Escherichia coli from pigs at slaughter in the UK in 2013. J. Antimicrob. Chemother. 2014, 69, 2947–2950. [Google Scholar] [CrossRef][Green Version]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in pig production: Chemistry, mechanisms of antibacterial action, microbial resistance emergence and one health perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef] [PubMed]
- Boyen, F.; Vangroenweghe, F.; Butaye, P.; De Graef, E.; Castryck, F.; Heylen, P.; Vanrobaeys, M.; Haesebrouck, F. Disk prediffusion is a reliable method for testing colistin susceptibility in porcine E. coli strains. Vet. Microbiol. 2010, 144, 359–362. [Google Scholar] [CrossRef]
- Schwarz, S.; Johnson, A.P. Transferable resistance to colistin: A new but old threat. J. Antimicrob. Chemother. 2016, 71, 2066–2070. [Google Scholar] [CrossRef]
- EMA. Updated Advice on the Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health; EMA/CVMP/CHMP/231573/2016; European Medicines Agency: London, UK, 2016. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/updated-advice-use-colistin-products-animals-within-european-union-development-resistance-possible_en-0.pdf (accessed on 20 April 2020).
- Chan, G.; Farzan, A.; DeLay, J.; McEwen, B.; Prescott, J.F.; Friendship, R.M. A retrospective study on the etiological diagnoses of diarrhea in neonatal piglets in Ontario, Canada, between 2001 and 2010. Can. J. Vet. Res. 2013, 77, 254–260. [Google Scholar]
- Kongsted, H.; Pedersen, K.; Hjulsager, C.K.; Larsen, L.E.; Pedersen, K.S.; Jorsal, E.J.; Baekbo, P. Diarrhoea in neonatal piglets: A case control study on microbiological findings. Porc. Health Manag. 2018, 4, 17. [Google Scholar] [CrossRef]
- Vidal, A.; Martín-Valls, G.E.; Tello, M.; Mateu, E.; Martín, M.; Darwich, L. Prevalence of enteric pathogens in diarrheic and non-diarrheic samples from pig farms with neonatal diarrhea in the North East of Spain. Vet. Microbiol. 2019, 237, 108419. [Google Scholar] [CrossRef]
- Toledo, A.; Gómez, D.; Cruz, C.; Carreón, R.; López, J.; Giono, S.; Castro, A.M. Prevalence of virulence genes in Escherichia coli strains isolated from piglets in the suckling and weaning period in Mexico. J. Med. Microbiol. 2012, 61 Pt 1, 148–156. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- EUCAST. MIC and Zone Distributions and ECOFFs. Available online: http://www.eucast.org/mic_distributions_and_ecoffs/ (accessed on 16 March 2020).
- CA-SFM (Comité de L’Antibiogramme de la Société Française de Microbiologie). Recommandations Vétérinaires 2019. Available online: https://www.sfm-microbiologie.org/?s=CASFM_VET2019 (accessed on 14 April 2020).
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; López, F.; Molina-López, R.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef]
- Martins, M.F.; Martinez-Rossi, N.M.; Ferreira, A.; Brocchi, M.; Yano, T.; Castro, A.F.; Silveira, W.D. Pathogenic characteristics of Escherichia coli strains isolated from newborn piglets with diarrhea in Brazil. Vet. Microbiol. 2000, 76, 51–59. [Google Scholar] [CrossRef]
- Ngeleka, M.; Pritchard, J.; Appleyard, G.; Middleton, D.M.; Fairbrother, J.M. Isolation and association of Escherichia coli AIDA-I/STb, rather than EAST1 pathotype, with diarrhea in piglets and antibiotic sensitivity of isolates. J. Vet. Diagn. Investig. 2003, 15, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Vu-Khac, H.; Holoda, E.; Pilipcinec, E.; Blanco, M.; Blanco, J.E.; Dahbi, G.; Mora, A.; López, C.; González, E.A.; Blanco, J. Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhea in Slovakia. Vet. J. 2007, 174, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Savarino, S.J.; McVeigh, A.; Watson, J.; Cravioto, A.; Molina, J.; Echeverria, P.; Bhan, M.K.; Levine, M.M.; Fasano, A. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 1996, 173, 1019–1022. [Google Scholar] [CrossRef]
- Ruan, X.; Crupper, S.S.; Schultz, B.D.; Robertson, D.C.; Zhang, Z. Escherichia coli expressing EAST1 toxin did not cause an increase of cAMP or cGMP levels in cells, and no diarrhea in 5-day old gnotobiotic piglets. PLoS ONE 2012, 7, e43203. [Google Scholar] [CrossRef]
- Zajacova, Z.S.; Faldyna, M.; Kulich, P.; Kummer, V.; Maskova, J.; Alexa, P. Experimental infection of gnotobiotic piglets with Escherichia coli strains positive for EAST1 and AIDA. Vet. Immunol. Immunopathol. 2013, 152, 176–182. [Google Scholar] [CrossRef]
- Mateu, E.; Martín, M. Antimicrobial resistance in enteric porcine Escherichia coli strains in Spain. Vet. Rec. 2000, 146, 703–705. [Google Scholar] [CrossRef]
- Sáenz, Y.; Zarazaga, M.; Briñas, L.; Lantero, M.; Ruiz-Larrea, F.; Torres, C. Antibiotic resistance in Escherichia coli isolates obtained from animals, foods and humans in Spain. Int. J. Antimicrob. Agents 2001, 18, 353–358. [Google Scholar] [CrossRef]
- Gibbons, J.F.; Boland, F.; Egan, J.; Fanning, S.; Markey, B.K.; Leonard, F.C. Antimicrobial resistance of faecal Escherichia coli isolates from pig farms with different durations of In-feed antimicrobials use. Zoonoses Public Health 2016, 63, 241–250. [Google Scholar] [CrossRef]
- Boerlin, P.; Travis, R.; Gyles, C.L.; Reid-Smith, R.; Janecko, N.; Lim, H.; Nicholson, V.; McEwen, S.A.; Friendship, R.; Archambault, M. Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 2005, 71, 6753–6761. [Google Scholar] [CrossRef]
- Rosager, W.N.; Peter, N.J.; Erik Lind, J.S.; Svend, H.; Matthew, D.; Steen, P.K. Comparison of antimicrobial resistance in E. coli isolated from rectal and floor samples in pens with diarrhoeic nursery pigs in Denmark. Prev. Vet. Med. 2017, 147, 42–49. [Google Scholar] [CrossRef]
- Kim, M.H.; Lee, H.J.; Park, K.S.; Suh, J.T. Molecular characteristics of extended spectrum β-lactamases in Escherichia coli and Klebsiella pneumoniae and the prevalence of qnr in extended spectrum β-lactamase isolates in a tertiary care hospital in Korea. Yonsei Med. J. 2010, 51, 768–774. [Google Scholar] [CrossRef]
- Wang, Y.; He, T.; Han, J.; Wang, J.; Foley, S.L.; Yang, G.; Wan, S.; Shen, J.; Wu, C. Prevalence of ESBLs and PMQR genes in fecal Escherichia coli isolated from the non-human primates in six zoos in China. Vet. Microbiol. 2012, 159, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.E.; Sun, J.; Li, L.; Deng, H.; Liu, B.T.; Fang, L.X.; Liao, X.P.; Liu, Y.H. IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front. Microbiol. 2015, 6, 964. [Google Scholar] [CrossRef] [PubMed]
- Azargun, R.; Sadeghi, M.R.; Soroush Barhaghi, M.H.; Samadi Kafil, H.; Yeganeh, F.; Ahangar Oskouee, M.; Ghotaslou, R. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect. Drug Resist. 2018, 11, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 1, 117–123. [Google Scholar] [CrossRef]
- Zheng, H.; Zeng, Z.; Chen, S.; Liu, Y.; Yao, Q.; Deng, Y.; Chen, X.; Lv, L.; Zhuo, C.; Chen, Z.; et al. Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 2012, 39, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Landman, D.; Georgescu, C.; Martin, A.; Quale, J. Polymyxins revisited. Clin. Microbiol. Rev. 2008, 21, 449–465. [Google Scholar] [CrossRef]
- Wasyl, D.; Hoszowski, A.; Zajac, M.; Szulowski, K. Antimicrobial resistance in commensal Escherichia coli isolated from animals at slaughter. Front. Microbiol. 2013, 4, 221. [Google Scholar] [CrossRef]
- Perrin-Guyomard, A.; Bruneau, M.; Houée, P.; Deleurme, K.; Legrandois, P.; Poirier, C.; Soumet, C.; Sanders, P. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill. 2016, 21, 30135. [Google Scholar] [CrossRef]
- Irrgang, A.; Roschanski, N.; Tenhagen, B.A.; Grobbel, M.; Skladnikiewicz-Zierner, T.; Thomas, K.; Roesler, U.; Käsbohrer, A. Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010–2015. PLoS ONE 2016, 11, e0159863. [Google Scholar] [CrossRef]
- Stannarius, C.; Bürgi, E.; Regula, G.; Zychowska, M.A.; Zweifel, C.; Stephan, R. Antimicrobial resistance in Escherichia coli strains isolated from Swiss weaned pigs and sows. Schweiz. Arch. Tierheilkd. 2009, 151, 119–125. [Google Scholar] [CrossRef][Green Version]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-occurence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef]
- Roschanski, N.; Falgenhauer, L.; Grobbel, M.; Guenther, S.; Kreienbrock, L.; Imirzalioglu, C.; Roesler, U. Retrospective survey of mcr-1 and mcr-2 in German pig-fattening farms, 2011–2012. Int. J. Antimicrob. Agents 2017, 50, 266–271. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016, 21, 30280. [Google Scholar] [CrossRef]
- Delannoy, S.; Le Devendec, L.; Jouy, E.; Fach, P.; Drider, D.; Kempf, I. Characterization of colistin-resistant Escherichia coli isolated from diseased pigs in France. Front. Microbiol. 2017, 8, 2278. [Google Scholar] [CrossRef]
- Duggett, N.A.; Randall, L.P.; Horton, R.A.; Lemma, F.; Kirchner, M.; Nunez-Garcia, J.; Brena, C.; Williamson, S.M.; Teale, C.; Anjum, M.F. Molecular epidemiology of isolates with multiple mcr plasmids from a pig farm in Great Britain: The effects of colistin withdrawal in the short and long term. J. Antimicrob. Chemother. 2018, 73, 3025–3033. [Google Scholar] [CrossRef]
- Morales, A.S.; Fragoso de Araújo, J.; de Moura Gomes, V.T.; Reis Costa, A.T.; dos Prazeres Rodrigues, D.; Porfida Ferreira, T.S.; de Lima Filsner, P.H.; Felizardo, M.R.; Micke Moreno, A. Colistin resistance in Escherichia coli and Salmonella enterica strains isolated from swine in Brazil. Sci. World J. 2012, 2012, 109795. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).