Abstract
Background: Globally, Salmonella enterica is one of the leading causes of foodborne illness in humans. Food of animal origin is obligatorily tested for the presence of this pathogen. Unfortunately, in meat and meat products, this is often hampered by the presence of background microbiota, which may present as false-positive Salmonella. Methods: For the identification of Salmonella spp. from meat samples of beef, pork, and poultry, the authorized detection method is PN-EN ISO 6579-1:2017-04 with the White–Kauffmann–Le Minor scheme, two biochemical tests: API 20E and VITEK II, and a real-time PCR-based technique. Results: Out of 42 presumptive strains of Salmonella, 83.3% Salmonella enterica spp. enterica, 14.3% Citrobacter braakii, and 12.4% Proteus mirabilis were detected from 180 meat samples. Conclusions: Presumptive strains of Salmonella should be identified based on genotypic properties such as DNA-based methods. The aim of this study was the isolation and identification of Salmonella spp. from miscellaneous meat sorts: beef, pork, and poultry.
1. Introduction
Salmonella is one of the most important foodborne pathogens and a leading cause of foodborne illness in humans in the EU [1]. The source of the infection is usually contaminated food products of animal origin. Continuous surveillance of the occurrence of this pathogen in foods is required to ensure public health. Therefore, official food testing methodology according to European and national food legislation is necessary. Moreover, the crucial issues are the rapidity, efficiency, and accuracy of these methods. Traditionally, bacteriological culture methods have been used for isolation and identification of Salmonella spp. Colonies with a morphology indicative of Salmonella spp. are then selected for further studies, and identification is based on the determination of biochemical features and completed by serotyping. The specificity of meat samples makes it a challenging material for routine bacteriological testing as it contains a high number of indigenous microorganisms. Another complication may be the low Salmonella number compared to other bacteria in the sample. Enrichment culture procedures are aimed at increasing the salmonellae population but, at the same time, the number of background bacteria also increases. The reliable identification of Salmonella spp. is essential to ensure food safety. However, the presence of similarities in phenotypic characteristics between closely related bacteria may lead to incorrect identification. Bacteria belonging to the genus Citrobacter are particularly often incorrectly identified as Salmonella spp.
This study aimed to compare the identification of Salmonella spp. isolates obtained from meat samples using routine bacteriological methods and molecular biology methods.
2. Materials and Methods
2.1. Sampling
A total of 180 raw meat samples (60 beef, 60 pork, and 60 poultry) were obtained from meat processing plants cutting beef, pork, and poultry carcasses in central Poland. All samples were derived from carcass parts classified by official veterinary inspections as fit for human consumption. All samples collected as a single sample weighed at least 200 g for each type of meat (from parts of carcasses for culinary use, i.e., roast beef, entrecote, boneless ribs, neck, breast, and thigh). The meat samples were collected randomly using the aseptic technique and were packed in separate sterile bags, which were then labeled. All samples were transported to the laboratory in refrigerated containers at a temperature of 4 °C within one hour of collection.
2.2. Salmonella spp. Isolation and Identification
Salmonella spp. from all samples were isolated in accordance with PN-EN ISO 6579-1:2017-04 Microbiology of the food chain—Horizontal method for the detection, enumeration and serotyping of Salmonella—Part 1: Detection of Salmonella spp. [2].
Samples were pre-enriched: for pork and beef samples, 10 g of each sample was mixed with 90 mL buffered peptone water (BPW GRASO, Starogard, Poland), and 25 g of each poultry sample was mixed with 225 mL BPW at a temperature of 25 °C (±3 °C) in sterile stomacher bags (Whirl-Pak, NAsco, Madison, WI, USA), placed in a stomacher and crushed for 2 min. The selective proliferation of Salmonella spp. was carried out using modified semi-solid Rappaport-Vassiliadis (MSRV) agar (GRASO, Starogard, Poland) and Muller–Kauffmann tetrathionate-novobiocin (MKTTn) broth (GRASO, Starogard, Poland). Two selective enrichment media, xylose lysine deoxycholate agar (XLD; GRASO, Starogard, Poland) and brilliant green agar (BGA; OXOID, Hampshire, United Kingdom), were used. Salmonella suspect colonies were transferred to a non-selective nutrient agar (GRASO, Starogard, Poland) to obtain the pure culture for further testing and a semi-solid medium by Garda for testing flagellar antigens. Serotyping was performed by slide agglutination with commercial H poly antisera for verification of the genus Salmonella enterica (IBSS Biomed, Kraków, Poland), O group antisera to determine O group (IBSS Biomed, Kraków, Poland), and H phase and H factor antisera to determine H phase and H factor (IBSS Biomed, Kraków, Poland, Bio-Rad, Hercules, CA, USA), according to the White–Kauffmann–Le Minor scheme.
2.2.1. Biochemical Strain Identification
Colonies showing morphology typical for Salmonella spp. on selective agars were subjected to biochemical identification using two commercially available tests: API 20E (BioMérieux, Craponne, France) and a VITEK2 COMPACT automated system for bacterial identification. VITEK® 2 GN cards (BioMérieux, Craponne, France) with reference strains for E. coli ATCC 25922, Salmonella Typhimurium ATCC 14028, Salmonella Enteritidis ATCC 1307,6 and Pseudomonas aeruginosa ATCC 27853 served as a quality check. Both tests were used according to the manufacturer’s instructions.
2.2.2. Confirmation of Salmonella Identification with Molecular Biology Methods
A real-time PCR method based on the detection of genes specific for Salmonella spp. was used to confirm presumptive identification. DNA for real-time PCR was extracted from bacterial cells using a commercial Kylt® DNA Extraction-Mix II (Anicon, Emstek, Germany). For detection of Salmonella spp., a commercial Kylt® Salmonella spp. (Anicon, Emstek, Germany) kit was used, and, for simultaneous detection of Salmonella Enteritidis and Salmonella Typhimurium, a commercial Spp-Se-St PCR (BioChek, Reeuwijk, The Netherland) kits was used. Both real-time PCR tests to detect Salmonella were performed according to the manufacturer’s instructions using an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo, Waltham, MA, USA).
2.3. Antibiotic Resistance Test
Antimicrobial susceptibility was assessed by determining the MIC values using a VITEK® 2 System and an AST-GN96 card for Gram-negative bacteria (BioMérieux).
To analyze MIC patterns and detect phenotypes of Citrobacter braakii, an Advanced Expert System (AES, BioMérieux, Craponne, France) and VITEK® 2 GN 96 cards (BioMérieux, Craponne, France) were used. The MICs were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) and FDA breakpoints (CLSI M100-ED28, 2018).
2.4. Statistical Assessment
Statistical testing was performed with a Statistica 13.1 software package (StatSoft, Kraków, Poland). Descriptive statistics were computed to determine the proportions of isolates resistant to different antimicrobial agents. The chi-square test was adopted for the determination of the statistical significance of differences between the proportions.
3. Results
The accuracy of identification obtained with the first biochemical test, API 20E (BioMérieux, Craponne, France), for some of the isolates of Salmonella like colonies on selective agars (Figure 1) was unsatisfactory, e.g., Salmonella spp. 71.9% and Citrobacter freundii 25% [3] Sero-diagnosis was difficult because it showed autoagglutinations or the test with the group sera was positive (especially DO), which could cause presumptive Salmonella diagnosis. Out of 180 meat samples, 23.33% indicated Salmonella spp. or presumptive strains of Salmonella. After obtaining the biochemical pattern from all 42 strains, 35 were confirmed as belonging to Salmonella enterica spp. enterica, 6 proved to be Citrobacter braakii, and 1 Proteus mirabilis. The results of the occurrence of Salmonella spp., C. braakii, and Proteus mirabilis in the meat samples tested are presented in Table 1.
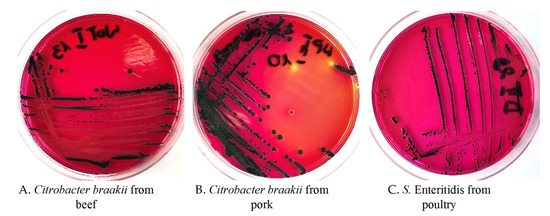
Figure 1.
Citrobacter braakii from a meat sample of (A) beef and (B) pork, and Salmonella Enteritidis from a meat sample of (C) poultry, on XLD media.

Table 1.
Incidence of Salmonella spp., Citrobacter braakii, and Proteus mirabilis in meat samples.
The accuracy of identification obtained with the first biochemical test, API 20E (BioMérieux, Craponne, France), for some of the isolates of Salmonella like colonies on selective agars (Figure 1) was unsatisfactory, e.g., Salmonella spp. 71.9% and Citrobacter freundii 25% [3] Sero-diagnosis was difficult because it showed autoagglutinations or the test with the group sera was positive (especially DO), which could cause presumptive Salmonella diagnosis. Out of 180 meat samples, 23.33% indicated Salmonella spp. or presumptive strains of Salmonella. After obtaining the biochemical pattern from all 42 strains, 35 were confirmed as belonging to Salmonella enterica spp. enterica, 6 proved to be Citrobacter braakii, and 1 Proteus mirabilis. The results of the occurrence of Salmonella spp., C. braakii, and Proteus mirabilis in the meat samples tested are presented in Table 1.
General results based on colony morphology on selective agars, biochemical properties, and the PCR technique are presented in Table 2.

Table 2.
Identification of Salmonella spp. isolates obtained from meat samples based on colony morphology on selective agars, biochemical properties, and the PCR technique.
No Salmonella spp. were isolated from beef meat samples but three Citrobacter braakii were isolated. From pork meat samples, 1.67% (n = 1) was Salmonella spp. positive, Citrobacter braakii were isolated from three samples, and Proteus mirabilis from one. In poultry, 56.67% of samples (n = 35) isolated were Salmonella spp. Amongst isolated species, Salmonella enterica subsp. enterica was detected. The most common serovars were S. Enteritidis (55.88%, n = 19), S. Derby (14.71%, n = 5), and S. Newport (14.71%, n = 5); nd less frequently isolated were S. Infantis (5.88%, n = 2), S. Kentucky (5.88%, n = 2), and S. Mbandaka (2.94%, n = 1).
The colonies formed by Citrobacter braakii and Salmonella Enteritidis on XLD selective media look almost identical (Figure 1).
The antibiotic resistance studies showed that all strains of Citrobacter braakii were susceptible to ampicillin, cephalosporins (III generation cefoperazone, ceftiofur, and IV generation cefquinome), aminoglycosides (gentamicin, neomycin), enrofloxacin, and trimethoprim/sulfamethoxazole, while resistance to other antibiotics was variable. For individual strains of Citrobacter braakii isolated from pork and beef samples, Table 3 presents several multi-drug resistance patterns.

Table 3.
Antimicrobial resistance common to the variously identified Citrobacter braakii isolated from samples of pork and beef.
4. Discussion
The phenotypic heterogeneity amongst bacteria is well-known, especially amongst closely related microorganisms. However, in the case of pathogens such as Salmonella, the misidentification may lead to a serious public health threat. In 2000, Manafi noticed that conventional media for the detection of Salmonella have a very poor specificity, creating an abundance of false positives such as Citrobacter or Proteus among the rare real positive Salmonella [4]. Twenty years later, despite many modifications and the development of further enriched media for the detection of Salmonella, there are still problems with the rapid identification of Salmonella spp. in meat.
There is no information in the world literature about difficulties in interpreting the results obtained when the result is “presumably Salmonella”. However, it should be remembered that the experience of a laboratory technician and their proficiency in conducting research are crucial for the correctness of the conducted research. At the same time, the necessity to introduce molecular diagnostics for the final confirmation of samples with the result “presumably Salmonella” should be obligatory, not only to protect public health, but also in connection with the handling of meat on the EU market. Mistakenly qualifying Citrobacter braakii as Salmonella spp. may lead to the withdrawal from the market of meat or meat products and may cause risk of economic loss for farmers and producers. The requirements of the PN-EN ISO 6579-1: 2017-04 standard indicate that the result should specify the serotype of Salmonella spp. with the provisions of the Commission Regulation (EC) No/ 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs [2,5]. However, in the Rapid Alert System for Food and Feed (RASFF) system, sometimes an alert notification occurs with a risk description of “serious” resulting in withdrawal from the customer because apathogenic microorganisms identified as “no definition–Salmonella” in beef was detected [6].
Reliable identification of Salmonella is crucial for ensuring public health security. Culture-based methods, biochemical identification, and serotyping are traditionally used and recommended by relevant standards. However, considerable variability is observed in the biochemical properties of some Salmonella, Proteus, and Citrobacter isolates. The consequence of this is that some “presumptive Salmonella” isolates cannot be identified by subsequent serotyping. Previous studies on the reliability of the API 20E test showed both good and inaccurate results. The presumptive Salmonella identification was confirmed by API 20E, but 24% of the isolates were recognized as Citrobacter spp. and 16.4% as Proetus spp. [7]. The results presented in this study are in accordance with the above observations. We found that 16.7% of 42 isolates recognized as presumptive Salmonella were confirmed as Citrobacter braakii.
Recently, many alternative non-culture methods have been described for Salmonella detection. These methods are based on DNA analysis or immunological reactions. A significant disadvantage of traditional methods is the time required for all culture procedures, which may take up to a few days for a presumptive identification. The available literature data indicate that the evident superiority of DNA-based methods is their high specificity and that results can be obtained after 24 h [8,9].
Bacteria belonging to the Citrobacter genus are closely related to Salmonella; thus, some similarities in cell surface antigens and biochemical properties occur between them. Pilar et al. proved that approximately one-third of Citrobacter and Salmonella genes are composed of core genes, confirming their close relationships [10]. This surprisingly high genotypic similarity can be explained by their common evolutionary history and genetic exchange [11]. All the properties of Citrobacter spp. listed above may result in false identification of these bacteria as Salmonella. It is also important to mention that it takes another day or two to confirm questionable identifications and, therefore, it takes longer to obtain the results and increases the costs of analysis.
Our results suggest that DNA-based methods should be used in cases of questionable results of Salmonella identification based on phenotypic properties.
On the other hand, further research on C. braakii is desirable in order to determine the current potential pathogenicity for humans, especially in view of the developing antibiotic resistance of C. braakii, which may contribute to the spread of resistance genes in the environment.
Author Contributions
Conceptualization, J.P.-C. and M.K.-Ś.; methodology, J.P.-C., K.W. and M.K.-Ś.; validation, J.P.-C., K.W., M.K.-Ś. and J.B.; formal analysis, J.P.-C., K.W., M.K.-Ś. and T.N.; investigation, J.P.-C. and K.A.; resources, J.P.-C.; data curation, J.P.-C., K.W. and M.K.-Ś.; writing—original draft preparation, J.P.-C., K.W., M.K.-Ś., T.N. and J.B.; writing—review and editing, J.P.-C., K.W., M.K.-Ś., K.A., P.K. and A.K.; visualization, J.P.-C. and M.K.-Ś.; supervision, J.P.-C. and K.A.; project administration, J.P.-C.; funding acquisition, K.A., P.K. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the image of the meat processing plants.
Acknowledgments
Special thanks to Jolanta Przybylska for help with the laboratory work.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- PN-EN ISO 6579-1:2017-04. Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.; ISO 6579-1:2017; ISO: Geneva, Switzerland, 2017. [Google Scholar]
- Bogdan, J.; Kizerwetter-Świda, M.; Wódz, K.; Rosiak, E.; Zarzyńska, J.; Anusz, K.; Pławińska-Czarnak, J. Occurrence of Salmonella spp. in raw meat-diagnostic difficulties. In Proceedings of the XX Conference on “Molecular Biology in Diagnostics of Infectious Diseases and Biotechnology”, Warsaw University of Life Science-SGGW, Warszawa, Poland, 23 November 2019; pp. 43–44, ISBN 978-83-7583-894-7. [Google Scholar]
- Manafi, M. New developments in chromogenic and fluorogenic culture media. Int. J. Food Microbiol. 2000, 60, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation (EC). No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs (Text with EEA Relevance: OJ L 338 22.12.2005, p. 1) 02005R2073—EN—08.03.2020—009.001 (2020). Available online: https://www.fsai.ie/uploadedFiles/Reg2073_2005(1).pdf (accessed on 8 March 2020).
- Rassf Notification. 2020.3317 Salmonella in Beef Shreds 2020. Available online: https://webgate.ec.europa.eu/rasff-window/screen/notification/438564 (accessed on 18 August 2020).
- Turki, Y.; Mehri, I.; Khessairi, A.; Agrebi, K.; Hassen, A.; Ouzari, H. Identification of three related genera, Salmonella, Citrobacter and Proteus using API 20E, 16S-23S rDNA intergenic transcribed spacer fingerprinting and 16S rDNA sequencing. AJMR 2013, 7, 3874–3884. [Google Scholar] [CrossRef]
- Bell, R.L.; Jarvis, K.G.; Ottesen, A.R.; McFarland, M.A.; Brown, E.W. Recent and emerging innovations in Salmonella detection: A food and environmental perspective. Microb. Biotechnol. 2016, 9, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Ricke, S.C.; Kim, S.A.; Shi, Z.; Park, S.H. Molecular-based identification and detection of Salmonella in food production systems: Current perspectives. J. Appl. Microbiol. 2018, 125, 313–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilar, A.V.C.; Petronella, N.; Dussault, F.M.; Verster, A.J.; Bekal, S.; Levesque, R.C.; Goodridge, L.; Tamber, S. Similar yet different: Phylogenomic analysis to delineate Salmonella and Citrobacter species boundaries. BMC Genom. 2020, 21, 377. [Google Scholar] [CrossRef] [PubMed]
- Retchless, A.C.; Lawrence, J.G. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11453–11458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).