Abstract
Liquid biopsy offers dynamic and noninvasive analysis of cellular biomarkers, thereby presenting enormous potential for early detection of cancer, cancer staging, prediction of relapse, real-time examination of therapeutic efficacy, perception of therapeutic targets, and understanding the resistance mechanisms. Nanotechnology has emerged as a novel tool to widen the application horizon of liquid biopsy. Several nanomaterials, nanodevices, nanostructures, and nanosensors have been explored for improved application of liquid biopsy for biomarker detection. The circulating tumor cells (CTCs), circulating tumor proteins (CTP), miRNA and extracellular vesicles (EVs) are some of the important biomarkers for detection by liquid biopsy in bodily fluids. Herein, we have discussed the state of the art and beyond in advances in nanotechnology and in increasing the specificity, sensitivity, and purity with which we detect liquid biopsy biomarkers. The opportunities and prospects of these advanced innovative nanomaterials and technologies in clinical applications are explored. Furthermore, various isolation and biosensing strategies for visualization and signal amplification using nanomaterials are summarized. The utilization of nanotechnology-based liquid biopsy may provide greater insights for improved treatment, diagnosis, and prognosis of cancer.
1. Introduction: Crystal Clear Nano and Liquid Biopsy
Cancer is one of the most devastating diseases and a notable cause of mortality across the globe, and the numbers of cases are increasing each year at an alarming pace. Until now, the majority of cancer patients have experienced poor prognosis, especially when diagnosed in an advanced stage with metastasis [1,2]. One of the chief reasons for rapidly increasing death is failure in diagnosing at an early stage. So, to resolve this issue, detection at earlier stages is highly desirable. In this context, liquid biopsy provides a unique platform for early detection of cancer without any invasive procedures or surgical intervention [3,4,5,6]. “Liquid biopsy” is a minimally invasive method for finding biomarkers of cancer in bodily fluids, most commonly blood but also urine, saliva, and cerebrospinal fluid. Although hematologic malignancies are also being investigated, liquid biopsy is applicable to a broad spectrum of cancers, including solid tumors like breast, lung, and colorectal cancer. Liquid biopsy deals with analysis or detection of genetic abnormalities (mutations, epigenetic modifications, etc.) and exploration of biomarkers such as circulating tumor cells (CTCs), cell-free nucleic acids (cfNAs), circulating tumor proteins (CTPs), miRNA, and extracellular vesicles (EVs) in bodily fluids, specifically the peripheral blood (Figure 1) [7]. In cases of metastasis, CTCs escape from the original site and enter the bloodstream. These cells move along with the circulation and subsequently arrive at a secondary site, forming a tumor colony. CTCs are found to be linked with tumor initiation and metastasis and have shown excellent prognostic and diagnostic potential [8,9]. Additionally, cell-free nucleic acids (cfNAs) are present in human blood, and it has been found that alterations in cfNAs can lead to cancer [10,11]. Small-noncoding RNAs, i.e., microRNAs (miRNAs), regulate the translation of protein-coding genes, and it has been found that those alterations or modifications in miRNA expression are associated with cancer progression. Furthermore, endosome-derived extracellular vesicles (EVs) play an important role in antigen presentation and in immune response. These EVs are generally secreted by all the mammalian cells and are abundantly present in bodily fluids including the blood. They are involved in various physiological processes; they function as communicating agents by transferring several biomolecules such as lipids, proteins, and nucleic acids (mRNA and microRNAs (miRNA)) from their origin to target cells; and they activate tumor progression and metastasis. They are thus considered an emerging liquid biopsy tool [12,13,14]. Liquid biopsy holds immense potential for early noninvasive diagnosis, prognosis, disease progression monitoring, assessment of treatment effectiveness, and detection of therapeutic targets for development of effective drug targeting [15]. Various techniques and methodologies have been utilized for the detection and analysis of these liquid biopsies, and the importance of liquid biopsies in the field of cancer has been demonstrated. Nanotechnology has made significant progress in further improving the potential of liquid biopsy. Advancements in nanomaterials can aid in the improvement of specificity, sensitivity, and selective detection of CTCs, cfNAs, TEPs, and EVs [7,16,17].
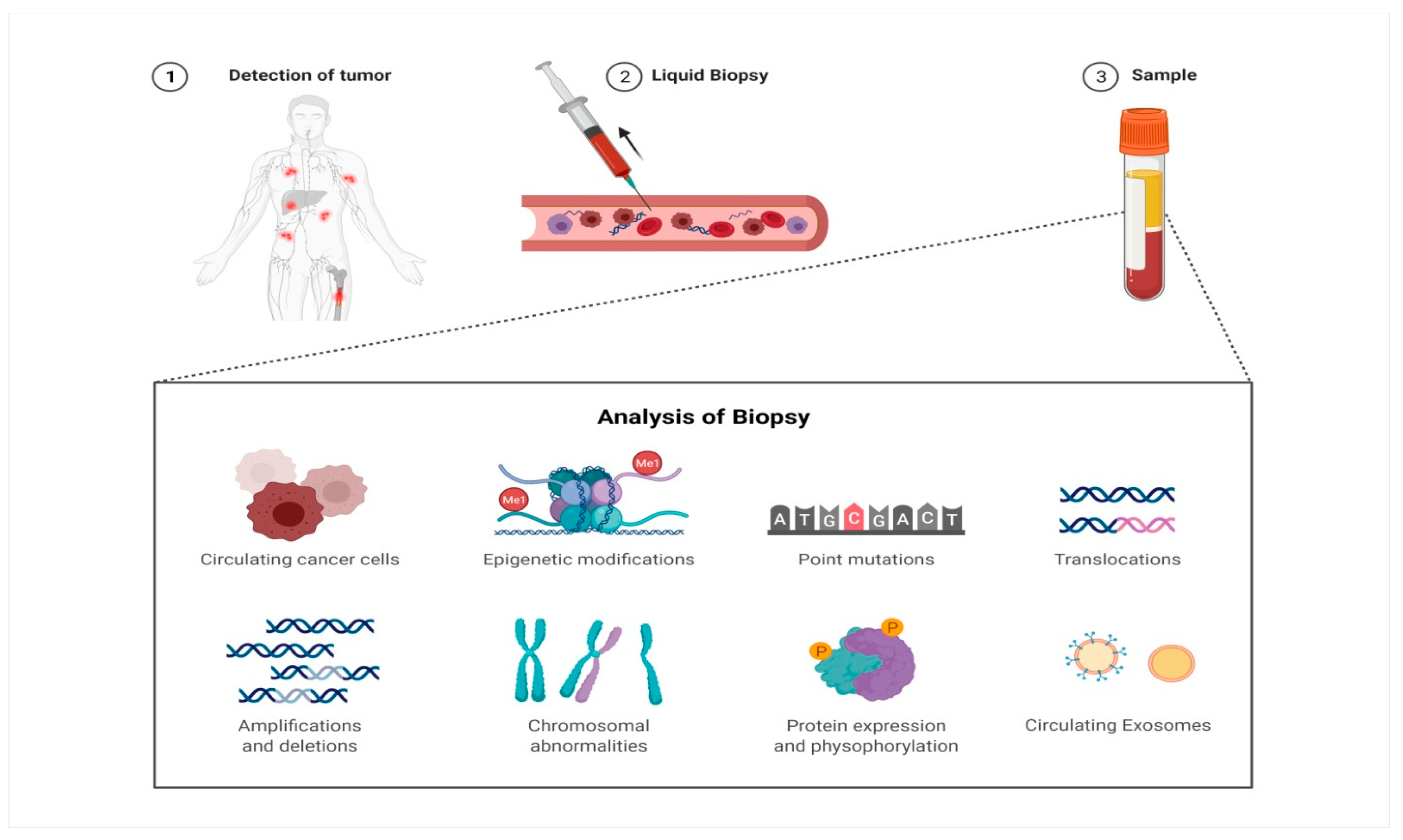
Figure 1.
The blood sample is collected from the cancer patient and is screened for the presence of circulating tumor cells (CTCs), alterations at the genetic level (mutations, epigenetic modifications), and changes in proteomic profile and circulating exosomes for detection of cancer and monitoring of given cancer therapy.
Nanotechnology—which is distinguished by the use of structures, devices, and materials whose size ranges from 1 nm to 1000 nm—possesses potential advantages in liquid biopsy like prominent sensitivity, simplicity, rapid detectability, analysis, lower cost, and the possibility for portability and personalized medicine [18,19,20,21]. The present review discusses the role of nanotechnology in liquid biopsy applications. This narrative review is based on works published between 2013 and 2024 that were retrieved from PubMed, Web of Science, and Google Scholar. We focused on English-language studies that examined tools based on nanotechnology for liquid biopsy applications in cancer. The search terms “nanotechnology,” “liquid biopsy,” “cancer diagnostics,” “extracellular vesicles,” “ctDNA,” “ctRNA,” “nanoparticles,” and “biosensors” were used for this review. Studies involving recent technology advancements, which showed preclinical or clinical relevance, or provided sensitivity/specificity measurements were preferred. Only when important to provide historical background and when referring to foundational publications were studies more than ten years old included.
2. Using Nanotechnology to Aid the Clinical Translation of Liquid Biopsy-Based Diagnostics
Nanotechnology offers a variety of advantages because of their nanodimensional structures which display enhanced optical, electrical, mechanical, magnetic and thermal features to generate empirical signals for diagnosis at an earlier stage and for concurrent monitoring during the treatment phase [22,23]. It provides highly sensitive and efficient detection of liquid biopsy biomarkers, as their concentration is very low in biological fluids. To overcome these limitations, nanotechnology has been used to increase the surface-area-to-volume ratio that in turn facilitates enhanced association of nanomaterials and biochemical analytes to increase the capturing of targeted cells in the blood associated with cancer [24]. Moreover, the extremely tiny dimensions of nanostructures promote the capturing and enrichment of nanoscale biomarkers like EVs (30–100 nm) and facilitate the amplification of localized signals from EVs [18]. Detection of CTCs, EVs, and cfNAs is very difficult in the bloodstream, as the bloodstream contains normal blood cells, cellular debris, and many other things that shield the signals originating from the CTCs and EVs, thereby impeding their capturing. In this regard, nanomaterials and nanostructures offer a unique platform for precisely detecting liquid biopsy biomarkers [18]. Additionally, cancers involve different cell populations, which are closely associated with cancer progression and drug resistance [25]. The detailed roles of CTCs and EVs are not fully known yet. However, in this context, nanotechnology-based approaches provide versatile and integrated platforms such as the lab-on-chip detection and further downstream molecular analysis of liquid biopsy biomarkers such as EVs and CTCs for future drug discovery, which hold enormous potential for clinical translation. The safety profile and biocompatibility of nanostructures not only enable the efficient capture of CTCs and EVs but also preserve the integrity of cellular biomarkers, which is crucial for in vitro and downstream molecular characterization [26]. Several nanomaterials are being developed and applied in clinical settings that strengthen the potential of nano-based approaches for improving the sensitivity of liquid biopsy for early detection, treatment response monitoring, disease prediction, and point-of-care testing [27,28,29]. Nanomaterials like nanorods, nanofibers, nanowires, and nanoparticles have been synthesized to increase the capacity to capture cancer-related abnormalities in fluids. By virtue of their nanoscale size, nanostructures exhibit amplified responses and increased surface area for interactions between materials and biomolecules, resulting in enhanced specificity and binding efficiency, as shown in Table 1.

Table 1.
Utility of nanotechnology for the isolation of CTCs and EVs.
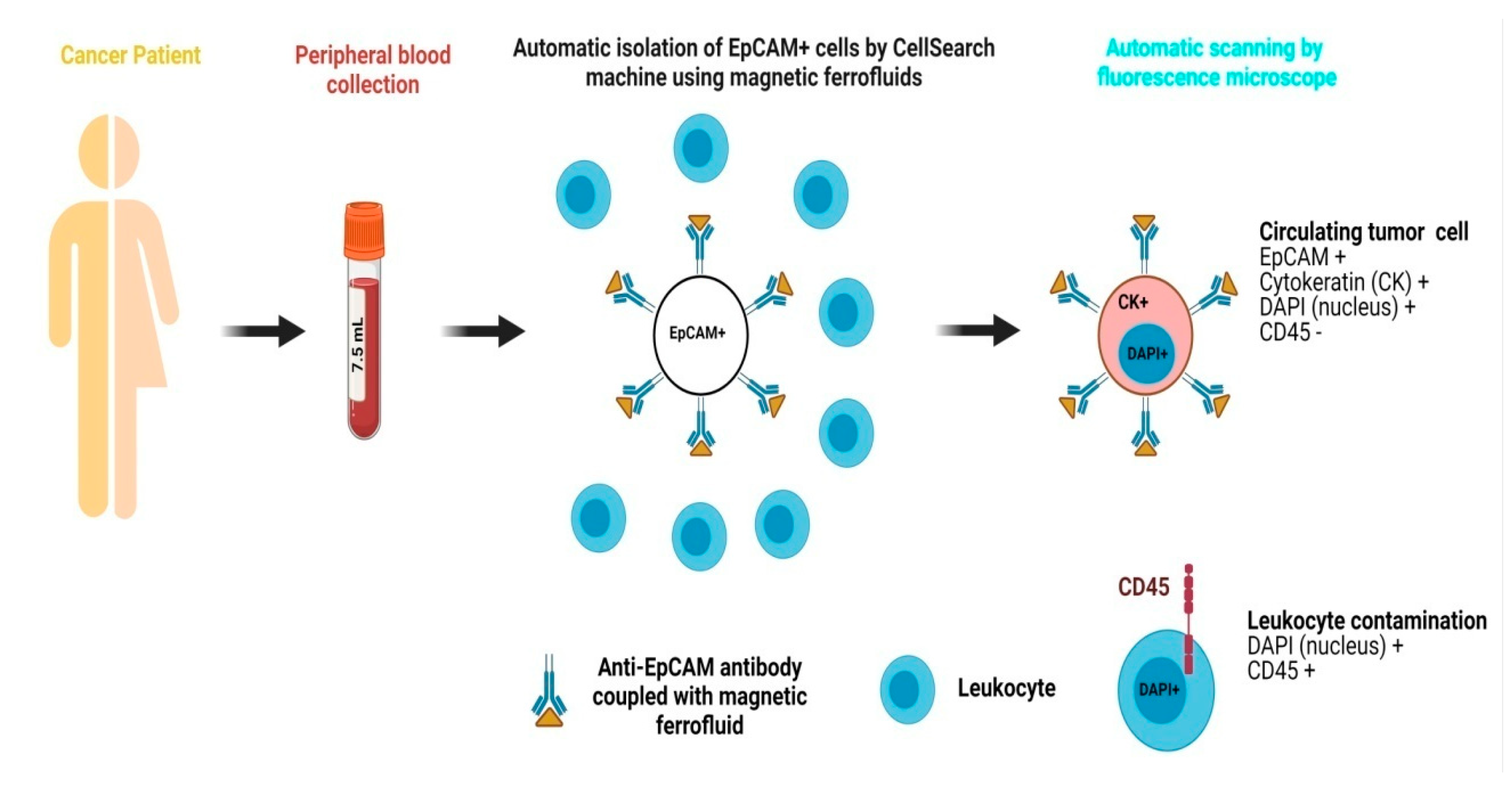
In recent times, various metallic and non-metallic nanostructures have been fabricated and demonstrated for enhanced CTC, EV, cfNA, etc. capture and detection. Magnetic nanoparticles (MNPs) are the classically used nanomaterials for detecting CTCs. Due to their high surface-to-volume ratio, rapid binding kinetics, and stability in solution, nanostructures offer high binding capacity, efficient target capture, and rapid enrichment, making them ideal for use in liquid biopsy to detect CTCs and related components [31,50,51]. The CellSearch system is clinically approved for the detection of circulating tumor cells (CTCs) in patients with breast and colorectal cancers. This system utilizes magnetic ferrofluids for the detection and capture of CTCs from blood samples. It is based on automated immunomagnetic enrichment of epithelial tumor cells that express the epithelial cell adhesion molecule (EpCAM). Specifically, magnetic ferrofluids conjugated with anti-EpCAM antibodies are added to 7.5 mL of peripheral blood collected from the patient, enabling selective isolation of EpCAM-positive CTCs [52,53]. Subsequently, the enriched sample is stained for nuclear staining by immunocytochemistry for cells expressing cytokeratin 8/18/19, whereas pan-leukocyte marker CD45 is utilized for eliminating contaminating leukocytes. Finally, based on consensus guidelines, a trained user using an automated fluorescence microscope captures and quantifies all the cells that meet the criteria for CTCs accordingly. The CellSearch system is highly specific and very sensitive, as it can capture even a single CTC in 7.5 mL of blood with minimal inter-reader variability [54,55,56]. A schematic of the CellSearch system is shown in Figure 2. Recently, Peng et al. and Bai et al. used the HER2 and EpCAM recognition peptide conjugated with MNPs to capture the CTCs expressing these two biomarkers on their surface. These conjugated MNPs demonstrated high capture efficiency and high sensitivity for CTC detection in breast, liver, and prostate cancers, achieving a capturing rate of over 90% for CTCs and 68% for HER2-expressing CTCs and significantly outperforming antibody-functionalized MNPs [30,31].
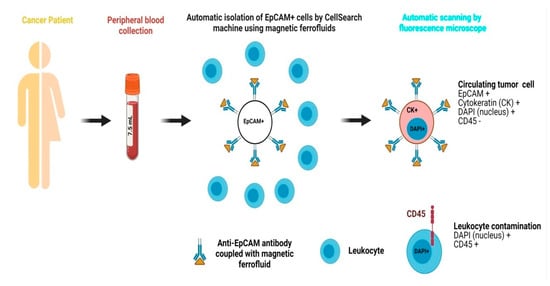
Figure 2.
The CellSearch method is intended for the detection of CTCs and is based on the automatized immunomagnetic enrichment of epithelial cells expressing EpCAM by addition of magnetic ferrofluids conjugated with anti-EpCAM antibodies. The whole blood sample (7.5 mL) is stained for positive markers, i.e., cytokeratins (8/18/19), with CD45 as a negative marker to exclude leukocyte antigen and a nuclear stain 4′,6-diamidino-2-phenylindole (DAPI). Finally, based on the data generated, an expert identifies and quantifies all the cells meeting the criteria for CTCs according to established guidelines using an automatic fluorescence microscope.
Gold nanoparticles are extensively employed for detection in biological systems and display great sensitivity and specificity at very low concentrations [57]. They have enhanced cell adhesion properties in their nano-form because of their increased surface interface. Furthermore, when they are functionalized with antibodies with different biological domains like peptides, antibodies, aptamers, etc., they show enhanced detection efficiency with an increased capture rate. Hyeun et al. tested the capture efficiency and sensitivity of functionalized graphene oxide (GO) nanosheets on a patterned gold surface for CTCs in breast, lung, and pancreatic cancer patients’ blood. Using the principle of electrostatic attraction, nanosheets of GO were adsorbed on the patterned gold surface followed by functionalization with biotinylated EpCAM antibodies. This fabricated material was examined to capture CTCs and was found to be successful for the detection of CTCs even at low concentrations of CTCs in patient-derived blood with promising sensitivity (3 ± 32.4% at 3–5 cells/mL blood) [36]. Furthermore, gold nanoparticle-based biosensors and microfluidic devices have been developed and have attracted interest in recent years [58]. The fabricated devices were able to detect the different biomolecular markers for different diseases from diverse array of samples that include serum, whole blood, saliva, and urine.
Recently, gold nanoparticle-based graphene quantum dots conjugated with the neuron-specific enolase (NSE) antibodies showed remarkable diagnosis of small cell lung cancer with as low as 0.09 pg/mL of NSE in blood [59]. Surface-enhanced Raman scattering (SERS) is widely used for the highly sensitive detection of multiplex biological components, offering molecular specificity, single-molecule sensitivity, and resistance to quenching. Leveraging these advantages, various metallic nanostructures with well-controlled sizes and shapes have been developed [60]. For direct detection of CTCs without a pre-purification step from the patient’s blood, Wu et al. designed gold-based SERS nanoparticles. They used the Raman reporter molecule 4-mercaptobenzoic acid (4-MBA) and modified the nanoparticles to attain strong SERS signals. For specific CTC detection, they conjugated the nanoparticles with folic acid (FA) and stabilized the nanoparticles with bovine serum albumin (BSA) to avoid any non-specified cell binding. The authors synthesized gold nanoparticles of various shapes and found that gold nanostars exhibited the highest sensitivity, achieving a detection limit of 1 cell/mL of blood, which is significantly lower than the previously reported limit of 5 cells/mL [61].
Similarly, graphene oxide (GO)-based materials were also assessed in various studies to examine their potential in improving liquid biopsy-based capturing of cancer biomarkers. In this regard, Zhang et al. proved the tremendous potential of GO to enhance sensitivity as well detection efficiency. They fabricated a nano-interfaced microfluidic EV device developed by coating a layer-by-layer channel and Y-shaped micro posts with antibody-conjugated GO-induced nanostructured polydopamine. This system captured the EVs with a lowest limit of 50/μL with a 4-log dynamic range. For clinical translational purposes, they used this device to differentiate ovarian cancer patients from healthy blood donor individuals, and they were successfully able to identify the cancer-bearing patients [47]. As a cell affinity substrate, NanoVelcro offers an exclusive method of capturing CTCs. In this system, compactly arranged and vertically oriented silicon nanopillars (SiNPs) are used and conjugated with specific antibodies, which results in enhanced capture of CTCs. SiNPs improve the interactions or binding kinetics between cells and substrates, which remarkably increases the capture efficiency in comparison with unstructured substrates [45,46,62]. Similarly, TiO2 nanofibers (TiNFs) were synthesized via electrospinning and subsequently functionalized with antibodies for CTC capture. The horizontally aligned electrospun TiNFs provided enhanced interaction and binding affinity with cells and cellular components, thereby improving detection sensitivity and capture efficiency. Using this approach, TiNFs substrates conjugated with anti-EpCAM were utilized to detect the CTCs in blood samples of three colorectal and seven gastric cancer patients. Of the three colorectal cancer patients, in two patients, 0–2 CTCs/0.5 mL of blood sample was detected, and in all seven gastric cancer patients, CTCs in the range of 3–19 were detected per 0.5 mL of blood sample. This highly efficient and sensitive approach indicated the potential of nanotechnology in improving liquid biopsy-based cancer diagnosis especially at early stages when only a few CTCs are roaming around in the patient’s blood [44].
Some in vitro studies have demonstrated that DNA catalytic reactions can detect nucleic acids with a high degree of sensitivity [63,64,65]. Inspired by those findings, Hu et al. fabricated a lipid–polymer hybrid nanoparticle (LPHN)-catalyzed hairpin DNA circuit (CHDC) biochip which offers very sensitive quantification and imaging of target RNAs in EVs. A specific CHDC containing two hairpin DNAs and a reporter for glypican-1 (GPC1) mRNA was encased in LPHNs and then fixed on a thin glass slide. These cationic charged LPHNs were capable to target negatively charged EVs by virtue of electrostatic interactions followed by formation of a used LPHN–EV nanocomplex, resulting in the mixing of CHDC with target RNA. CHDC is prompted by hybridizing with target RNA and rapidly generates amplified signals. The ultrahigh sensitivity of the fabricated LPHN–CHDC fusion complexes was evaluated by targeting GPC1 mRNA in EVs from pancreatic cancer. This design was capable of capturing EVs at concentrations as low as 105/mL, which is equivalent to approximately 60 EVs/μL. Furthermore, LPHN-CHDC also distinguished the early-to-late-stage PDAC patients from healthy donors and other patients with benign pancreatic disorders [66]. Moreover, apart from the discussed applications, there are several ongoing clinical trials for liquid biopsy which are tabulated in Table 2.

Table 2.
Ongoing clinical trials for application of liquid biopsy for cancer diagnosis.
3. Nanomaterials Utilized for Liquid Biopsy Applications
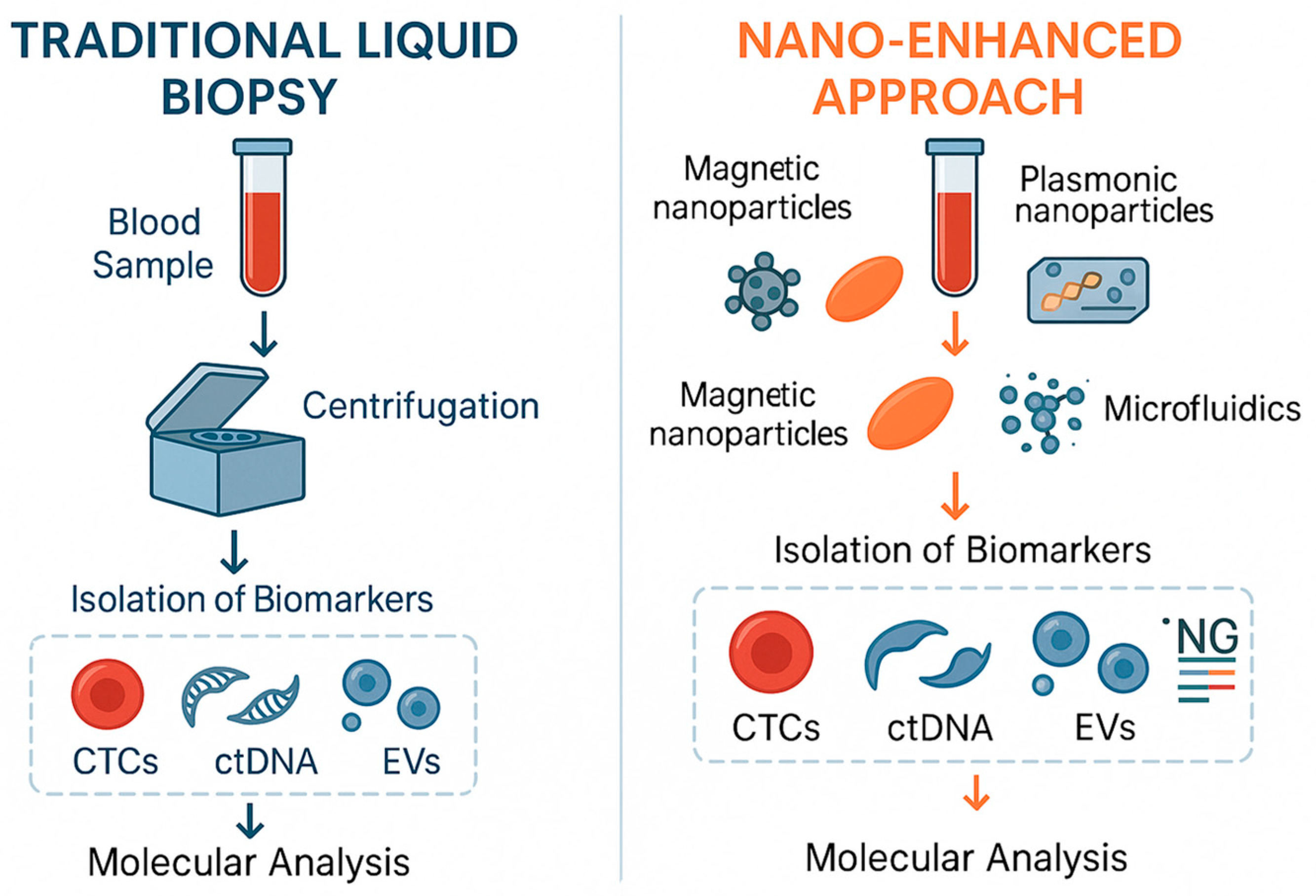
Nanotechnology-based techniques have developed rapidly in recent years and, compared to traditional analytical methods, offer advantages such as high sensitivity, simplicity, multiplex detection, automation, and miniaturization, supporting their potential use in clinical practice. Figure 3 compared the workflows of conventional and nano-based liquid biopsy. Nanoparticles such as gold, carbon-based nanomaterials like graphene oxide and quantum dots (QDs), as well as silver, silica, magnetic, and iron oxide nanocrystals hold tremendous potential due to their unique optical, electronic, structural, and magnetic properties, which aid in the sensitive detection of liquid biopsy biomarkers [67]. Some of the nanotechnological applications for detection of liquid biopsy are tabulated in Table 3.
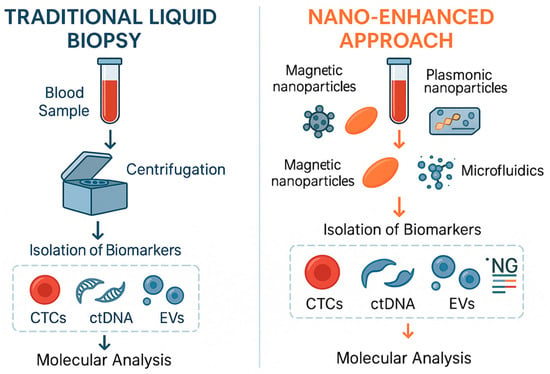
Figure 3.
Comparison between the workflows of conventional liquid biopsy and the nano-enhanced approach for liquid biopsy.

Table 3.
Nanotechnology in detection of liquid biopsy biomarkers.
3.1. Gold Nanostructures
Gold nanoparticles are commonly utilized for various applications of liquid biopsy. Their distinctive electrical and optical features, biocompatibility, ability for conjugation with various biomolecules, high stability, and large surface-area-to-volume ratio make them a unique platform for the development of versatile biosensors [99]. Additionally, they possess several analytical applications in sensing which include SPR, fluorescence resonance energy transfer (FRET), surface-enhanced Raman spectroscopy (SERS), lateral flow assays, and colorimetric and fluorescence methods [99,100].
3.1.1. MicroRNAs
Gold nanoparticles are utilized for detecting microRNA (miRNA), which are associated with the emergence of cancer. In one study, gold nanoparticles (AuNPs) were conjugated with DNA probes and allowed to hybridize with the miRNA target in the presence of Mg2+, resulting in the formation of a DNAzyme that digests the hybrids, generating fluorescent fragments. This process presented a limit of detection (LOD) of 50 fM [100]. Nossier et al. revealed a colorimetric method for detecting miRNA. This method was dependent on the stability of AuNPs stimulated by the hybridization of the miRNA target with short complementary DNA probes [101,102]. Moreover, lateral flow strips were employed to detect miRNA using AuNPs as reporters for visual detection, based on a sandwich-type hybridization assay on a nitrocellulose membrane [103]. Additionally, AuNPs coupled with rolling circle mechanism of replication for dual signal enhancement were utilized for simultaneous detection of two miRNAs [104]. AuNPs can also be conjugated with detection probes and many horseradish peroxide (HRP) molecules, which aid in enhancing the optical output by adding a chromogenic substrate [105]. Mohammad Niaei et al. developed a three-way junction RNA structure for detection of miRNA which contained a methylene blue-altered hairpin structure at one end (which works as a sensing moiety); the other ends were hybridized with DNA-barcoded AuNPs for amplifying the signals. Upon hybridization with target miRNA, the hairpin moiety was opened with successive hybridization onto a DNA-modified platinum/gold electrode, resulting in oxidation of methylene blue. This method required a very small amount of sample i.e., 4 μL, and showed an LOD of 135 Am, which is 230 times greater than RT-PCR [106]. Fredj et al. developed a voltammetric detection method for miRNA by hybridizing it with biotinylated beacons linked to gold nanoparticles, which were then captured via avidin–biotin interaction. This method can detect miRNA at concentrations as low as 4 fM [107].
AuNPs in association with magnetic beads and strand displacement amplification (SDA) were used for detecting miRNA. In this method, AuNPs coupled with magnetic beads were allowed to conjugate with the DNA probe complementary to the target miRNA sequence via a sandwich-like hybridization. The target miRNA was hybridized with its complementary probe via strand displacement and resulted in the release of AuNPs from the magnetic beads. The proportion of free AuNPs is related to the concentration of target miRNA and was observed through dark-field microscopy [108]. Moreover, SPR has been utilized to detect miRNA by hybridizing it with complementary DNA probes immobilized on the sensor chip. The resulting hybrids were captured with a biotinylated antibody that recognizes DNA-RNA complexes, and neutravidin-conjugated AuNPs were used for signal enhancement [109].
3.1.2. Circulating Tumor DNA
Gold nanorods and AuNPs conjugated with peptide nucleic acid (PNA) probes on a SPR sensor have been utilized to detect at least two nanograms of synthetic single-stranded ctDNA/mL within ten to fifteen minutes [110]. A colorimetric assay based on a three-way target CHA was employed for the detection of ctDNA. Following hairpin assembly, DNA fragments induced the aggregation of gold nanoparticles, causing a color change from red to blue [67]. One more colorimetric detection method was employed, where accumulation of unlabelled AuNPs took place and isolated ctDNA from blood samples bound to AuNPs resulted in a visible change in color [111]. Lin et al. utilized an SERS nanosensor for signal amplification in detecting ctDNA based on silica-coated Au nanorods. This method utilized metal-carbonyl SERS labels for enhancing the sensitivity of detection [67]. Additionally, to detect mutations in ctDNA, a lateral flow strip test was designed using AuNPs. In this method, the targeted DNA was allowed to amplify via allele-specific PCR with a specific oligonucleotide tail at one end. The resulting amplified DNA was placed on the strip and examined by AuNPs through hybridization with the oligonucleotide tail [112].
3.1.3. Circulating Tumor Cells
To detect CTCs, a dual amplification methodology was utilized by employing AuNPs and rolling circle amplification (RCA). Long ssDNA sequences were generated by RCA which were then captured by DNA-AuNPs conjugates, and the arrested AuNPs were analyzed using inductively coupled plasma mass spectrometry (ICP-MS) [113]. Moreover, the targeting of CTCs in blood samples was performed by employing aptamer-modified gold nanofilms (10–100 nm) utilizing pulsed-laser desorption/ionization mass spectrometry (LDI-MS). This method was able to identify at least ten cancer cells in a blood sample [114]. For visual detection of CTCs, an aptamer-AuNPs-based strip was utilized in which the positive signal at the strip was induced by other biotinylated aptamer conjugated with immobilized streptavidin at the test zone by interacting with CTC–aptamer–gold nanoparticle complexes. This method was able to detect at least 4 × 103 CTCs with the naked eye and as few as 800 CTCs using a portable reader, all within a few minutes [115].
3.2. Graphene Oxide (GO) Nanostructures
Two-dimensional graphene oxide (GO) is a great carbon-based nanomaterial with several advantages in bioimaging, biosensing, and drug delivery. Various spectrometric and electrochemical biosensors have been constructed using GO for biosensing. GO offers a good surface area which helps in providing a new platform for signal enhancement, possesses good electrical properties, and offers distinct functional groups for bioconjugation. GO provides excellent quenching ability and adsorption ability for ssDNA which can be utilized for various fluorescent applications [113].
3.2.1. MicroRNAs
GO can be employed for detecting miRNA by developing a GO-fluorescence-based assay which involves rolling circle amplification and ssDNA [116]. Treerattrakoon et al. proposed a multiplex miRNA-sensing method by utilizing various ssDNA, RCA, and GO, which was used as a fluorescence quencher [117]. Further, the GO surface was covalently coupled with the ssDNA, and fluorescently labelled DNA probes were used, containing two segments among which one is complementary to the immobilized DNA sequence and the other is complementary to the miRNA sequence. When DNA probes are in close proximity to GO, due to hybridization with single-stranded DNA sequences on its surface, fluorescence quenching occurs. However, upon addition of the miRNA target, the fluorescence is restored, with the recovered signal correlating to the amount of miRNA present [118]. Avila et al. investigated a new method for detecting miRNA by employing labeled GO/ssDNA complexes that are coated with gold nanowires that have the potential to capture cancer cells [119]. Ryoo et al. utilized the same mechanism by using a labelled peptide nucleic acid (PNA) probe and GO for detecting miRNA, and the fluorescence signal was recovered when the labelled PNA probe or ssDNA bound to the targeted miRNA [119,120]. Duplex molecular beacons are utilized for detecting miRNA colorimetrically where the target miRNAs are hybridized using strand displacement, causing the release of peroxidase-mimicking DNAzyme, which was collected on the surface of GO. The released DNAzyme is responsible for catalyzing a colorimetric reaction and provided an LOD of 12.9 nM [121]. The electrochemical sensing of miRNA included immobilization of DNA probe on an electrode. The target miRNA was allowed to hybridize with specified probes, and GO was accumulated on the hybrids, aggregating methylene blue dye which can reduce the pulse voltametric signal [122]. In this method, duplex-specific nucleases were used for the recycling of target and amplifying the signal. Similarly, an electrochemical sensing method for detecting miRNA was developed using the same approach by utilizing Prussian Blue nanoparticles on the surface of GO [123].
3.2.2. Circulating Tumor Cells
CTCs can be detected by employing an antibody-modified reduced GO, and the captured cells can be observed by immunostaining, thus detecting 2 CTCs/4 mL blood [124]. An aptasensor was developed by utilizing tetra-(4-aminophenyl) porphyrin-dependent reduced GO for the electrochemical identification of a minimum of 10 CTCs/mL [125]. Xiao et al. developed an aptasensor using hairpin aptamer probes, nicking endonuclease, linker DNA probes with dye labelling, and GO to detect as least 25 CTCs in a blood sample [126]. Furthermore, GO modified with AuNPs can be used for electrochemical detection of as few as 40 CTCs/mL by employing β-cyclodextrin–AuNPs/GO complexes or ferrocene–aptamer/Ru(bpy)32+ systems. These electrochemical aptasensors can be reused for six to seven cycles [127].
3.3. Quantum Dot (QD) Nanostructures
QDs offer numerous advantages for use in liquid biopsy. They possess excellent optical properties, such as broad excitation spectra, narrow emission peaks, high photostability, and strong fluorescence, all of which contribute to enhanced biosensing sensitivity compared to conventional fluorescent dyes. The use of QDs for the detection of miRNA generally employs lateral flow assays, electrochemiluminescence, and FRET. An electro-chemiluminescent biosensor was proposed for detecting miRNA using target recycling and a Zn2+-driven DNA rolling mechanism for amplifying the signals. The DNA nanomachine was developed by AuNPs conjugated with DNA probes which were hybridized and moved along the bound complementary DNA probes with Zn2+ recognition sites over the electrode surface. On the electrode surface, CdS@Mn QDs were used for electro-chemiluminescent signalling with signal quenching by ferrocene. The electro-chemiluminescent signal was restored for detecting miRNA [128]. Moreover, for the photoelectrochemical detection of miRNA, a DNA tetrahedron was developed as a nanocarrier for immobilizing CdTe QDs. This method utilized enzyme-assisted target cycling amplification and a QD–dye complex for detecting miRNA with an LOD of as low as 17 Am [129,130]. A dual-channel ratiometric nanoprobe was constructed for imaging and detecting miRNA by using molecular beacons with fluorescent dye and MoS2-QDs. FRET efficiency was low when the molecular beacon was coupled with QDs, but once miRNA hybridized with molecular beacons, the distance between dye and QDs increased, providing a strong FRET signal [131]. QDs are also used for detecting CTCs. Xie et al. developed a multifunctional nanobioprobe by coating the surface of QDs with alginate activated by Ca2+, followed by immobilization of biotinylated anti-EpCAM antibodies. This multifunctional nanoprobe has the potential for specific cell capture [132]. Similarly, Min et al. developed anti-EpCAM-modified QDs to capture CTCs and secondary anti-IgG-magnetic beads for linking the attached cells. They showed that the fluorescence intensity of QDs was equivalent to the captured number of cells. This method possesses high target efficiency and an easy quantification method for CTCs [133].
3.4. Copper Nanoparticles (CuNPs)
Metallic nanoclusters possess distinctive features which can be utilized in various applications of liquid biopsy. They are smaller in size and exhibit excellent conductivity, quantum effects, intrinsic fluorescence, and ferromagnetic and chirality features. Various copper nanostructures have peroxidase-like activity, and in the presence of hydrogen peroxide, they can oxidize the chromogenic substrate. Copper was used in the form of DNA-Cu nanoclusters by utilizing peroxidase-like catalytic activity to visually detect concentrations of miRNA [134]. Xu et al. designed fluorescence-based detection of miRNA by using duplex-specific nucleases (DSNs) that stimulate amplification of the target, thus leading to terminal deoxynucleotidyl transferase-mediated synthesis of polyT-CuNPs, which can carry out fluorescence-based detection of miRNA [135]. Additionally, the synthesis of copper nanoblocks linked with DNA probes was followed by hybridization, and finally, magnetic isolation resulted in fluorescent and electrochemical detection of miRNA in solution [136]. It has been shown that the target miRNA hybridized with a hairpin bound to an electrode started a hybridization chain reaction (HCR). The copper nanoclusters were generated on AT-rich dsDNA produced as a result of HCR and provided an electro-chemiluminescent signal for detecting miRNA [137].
3.5. Silver Nanoparticles (AgNPs)
Silver nanoparticles have been extensively employed for enhancing signals in SPR, SERS, and electrochemical detection because of their easy oxidation process and greater extinction coefficient value. Silver nanoclusters were employed for detection of miRNA by triggering catalytic hairpin assembly utilizing two hairpins. One of the hairpins consisted of silver nanoclusters, and the second hairpin bore G-rich DNA sequences. CHA took place when, upon hybridization of target miRNA with first hairpin—which was followed by close association among silver nanoclusters and G-rich regions, which increased the fluorescence of nanoclusters— as little as 0.3 nMmiRNA was detected [138]. Moreover, an easy method was developed for detecting miRNA in which silver nanoclusters were coupled with a DNA probe, which was allowed to hybridize with target miRNA. The negatively charged DNA probe interacted with positively charged gold nanoparticles, thereby quenching the fluorescence of silver nanoclusters. This was followed by hydrolysis of the RNA-DNA hybrid by duplex-specific nucleases, which removed the gold nanoparticles, leading to fluorescence recovery and enabling miRNA detection [139]. Similarly, silver nanorods were used for ctDNA detection via an SERS system combined with target-triggered enzyme-free recycling and a signal enhancement step involving various reporters, enabling detection of ctDNA at 40.4 aM [140]. For the detection of CTCs, silver nanoprisms in association with magnetic iron oxide nanoparticles were used by employing magnetic enrichment and SERS detection system [141].
3.6. Silica Nanoparticles (SiNPs)
Silica nanoparticles (SiNPs) are widely used in various analytical detection techniques due to their ability to encapsulate multiple molecules within a single nanoparticle. They can also encapsulate dye or reporter molecules, enhancing the detection process. SiNPs have been employed for miRNA detection by coupling them with fluorescent dye-labeled DNA probes. The fluorescence of the dye is initially quenched by the SiNPs and is restored upon hybridization with the target miRNA [142]. Similarly, a DNA–SiNPs complex was used for miRNA detection. A fluorophore is attached to the immobilized DNA probe, while a quencher molecule is attached to a complementary probe. When the quencher-carrying probe hybridizes with the immobilized DNA probe, fluorescence is quenched. This fluorescence is restored when miRNA displaces the quencher probe upon hybridization [143].
3.7. Iron and Magnetic Nanostructures
Iron and magnetic nanoparticles have been extensively used in biosensing and detection due to their ability to enhance signals, thereby improving the detectability of biomarkers. Additionally, their peroxidase-like activity enables colorimetric detection. Au-coated paramagnetic nanoparticles have been utilized for electrocatalytic determination of miRNA through SERS, employing conjugated DNA probes [144,145]. Additionally, magnetic Fe@SiO2 nanoparticles were attached to DNA probes, while gold nanoparticles were conjugated with complementary DNA probes for detecting ctDNA. The silica coating prevented oxidation of the magnetic nanoparticles and enhanced their biocompatibility and solubility. When both nanoparticles come into close proximity through hybridization, magnetic isolation is performed, followed by measuring gold concentration via inductively coupled plasma mass spectrometry (ICP-MS), enabling detection of ctDNA at levels as low as 0.1 pg/mL [146]. Magnetic Fe3O4 nanoparticles conjugated with silicon nanoparticles linked to anti-MUC1 aptamers have been utilized to detect cancer cells at concentrations as low as 100 cells/mL. Similarly, Fe3O4 nanostructures coupled with cross-linked PEG have been employed for detecting CTCs [147].
4. Limitation
While nanotechnology holds great promise for advancing liquid biopsy diagnostics, several limitations must be carefully addressed to ensure successful clinical translation. One major concern is cytotoxicity, particularly with inorganic nanomaterials such as quantum dots and silver nanoparticles, which can induce oxidative stress or release toxic ions. Additionally, many nanoparticles exhibit poor biodegradability and may accumulate in organs such as the liver and spleen, raising safety concerns for long-term use. In diagnostic assays, off-target effects and non-specific binding can reduce sensitivity and specificity, potentially leading to false-positive or false-negative results, especially in complex biological fluids. Moreover, challenges in scalability and reproducibility continue to hinder regulatory approval, as clinical-grade nanoparticles must meet stringent quality standards under Good Manufacturing Practices (GMPs). To date, only a limited number of nano-enabled liquid biopsy platforms have received FDA or EMA approval, largely due to unclear regulatory pathways and the need for extensive clinical validation. Additionally, the high cost and complexity of some nanomaterial-based systems may limit their widespread adoption, particularly in low-resource settings. Addressing these challenges through the development of biocompatible materials, standardized manufacturing protocols, and collaborative translational research will be critical to realizing the full potential of nanotechnology in cancer diagnostics.
5. Conclusions
The emerging field of liquid biopsy has opened up new avenues for cancer diagnosis, with significant clinical applications in personalized oncology. Advances in nanotechnology have greatly enhanced the detection, characterization, and sensing of liquid biopsy biomarkers such as circulating tumor cells (CTCs), cell-free nucleic acids (cfNAs), circulating tumor proteins (CTPs), microRNAs (miRNAs), and extracellular vesicles (EVs). Nanomaterials provide excellent platforms for advanced cancer diagnostics, treatment evaluation, and monitoring of disease progression, offering high sensitivity, selectivity, and rapid analysis. Nanotechnology has evolved substantially from the use of individual nanostructures or nanomaterials to the development of multifunctional, integrated nano-based devices and platforms. Future directions in this field include the development of point-of-care nano-enabled liquid biopsy devices, integration with artificial intelligence for data interpretation, and the design of personalized nanosensors for real-time monitoring. Emerging research is also exploring biodegradable and stimulus-responsive nanomaterials that can adapt to dynamic tumor environments, thereby improving diagnostic precision and safety. However, the routine application of nanotechnology-based liquid biopsy in clinical settings remains a work in progress. Ethical considerations—such as data privacy, equitable access to advanced diagnostics, and the long-term biocompatibility of nanomaterials—must be addressed to ensure responsible clinical translation. In parallel, regulatory frameworks must evolve to guide the safe and effective implementation of these technologies. Finally, more extensive research and collaborative efforts are urgently needed to promote translational research and facilitate the clinical adoption of nanotechnology-enabled liquid biopsy tools.
Author Contributions
Conceptualization, P.A. and A.K. Writing—original draft preparation, P.A. and S.A. Writing—review and editing, V.S., A.K.B., K.K.B., and A.K. Supervision, A.K. Project administration, A.K. All authors have read and agreed to the published version of the manuscript.
Funding
The authors did not receive any funding for this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Real-time liquid biopsy in cancer patients: Fact or fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef]
- Alix-Panabieres, C. The future of liquid biopsy. Nature 2020, 579, S9. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Li, W.; Wang, H.; Zhao, Z.; Gao, H.; Liu, C.; Zhu, L.; Wang, C.; Yang, Y. Emerging nanotechnologies for liquid biopsy: The detection of circulating tumor cells and extracellular vesicles. Adv. Mater. 2019, 31, 1805344. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Sorenson, G.; Pribish, D.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single—Copy genes in human blood. Cancer Epidemiol. Biomark. Prev. 1994, 3, 67–71. [Google Scholar]
- Nawroz, H.; Koch, W.; Anker, P.; Stroun, M.; Sidransky, D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat. Med. 1996, 2, 1035–1037. [Google Scholar] [CrossRef]
- Chaput, N.; Théry, C. Exosomes: Immune properties and potential clinical implementations. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 419–440. [Google Scholar]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Andaloussi, S.E.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Chen, X.; Wang, Y.; Li, Z.; Du, S.; Wang, L.; Chen, S. Nanotechnology-based strategies for early cancer diagnosis using circulating tumor cells as a liquid biopsy. Nanotheranostics 2018, 2, 21. [Google Scholar] [CrossRef]
- Lin, D.; Wu, Q.; Qiu, S.; Chen, G.; Feng, S.; Chen, R.; Zeng, H. Label-free liquid biopsy based on blood circulating DNA detection using SERS-based nanotechnology for nasopharyngeal cancer screening. Nanomed. Nanotechnol. Biol. Med. 2019, 22, 102100. [Google Scholar] [CrossRef]
- Wong, I.Y.; Bhatia, S.N.; Toner, M. Nanotechnology: Emerging tools for biology and medicine. Genes Dev. 2013, 27, 2397–2408. [Google Scholar] [CrossRef]
- Whitesides, G.M. The’right’size in nanobiotechnology. Nat. Biotechnol. 2003, 21, 1161–1165. [Google Scholar] [CrossRef]
- Khurana, A.; Allawadhi, P.; Khurana, I.; Allwadhi, S.; Weiskirchen, R.; Banothu, A.K.; Chhabra, D.; Joshi, K.; Bharani, K.K. Role of nanotechnology behind the success of mRNA vaccines for COVID-19. Nano Today 2021, 38, 101142. [Google Scholar] [CrossRef]
- Allawadhi, P.; Khurana, A.; Allwadhi, S.; Joshi, K.; Packirisamy, G.; Bharani, K.K. Nanoceria as a possible agent for the management of COVID-19. Nano Today 2020, 35, 100982. [Google Scholar] [CrossRef]
- Gogotsi, Y. Nanomaterials Handbook; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Ozin, G.A.; Arsenault, A. Nanochemistry: A Chemical Approach to Nanomaterials; Royal Society of Chemistry: London, UK, 2015. [Google Scholar]
- Ravishankar Rai, V. Nanoparticles and their potential application as antimicrobials. In Science Against Microbial Pathogens: Communicating Current Research and Technological Advances; FORMATEX: Badajoz, Spain, 2011. [Google Scholar]
- Lohr, J.G.; Adalsteinsson, V.A.; Cibulskis, K.; Choudhury, A.D.; Rosenberg, M.; Cruz-Gordillo, P.; Francis, J.M.; Zhang, C.-Z.; Shalek, A.K.; Satija, R. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014, 32, 479. [Google Scholar] [CrossRef]
- Qian, W.; Zhang, Y.; Chen, W. Capturing cancer: Emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small 2015, 11, 3850–3872. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef]
- Sestini, S.; Boeri, M.; Marchiano, A.; Pelosi, G.; Galeone, C.; Verri, C.; Suatoni, P.; Sverzellati, N.; La Vecchia, C.; Sozzi, G. Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget 2015, 6, 32868. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- Bai, L.; Du, Y.; Peng, J.; Liu, Y.; Wang, Y.; Yang, Y.; Wang, C. Peptide-based isolation of circulating tumor cells by magnetic nanoparticles. J. Mater. Chem. B 2014, 2, 4080–4088. [Google Scholar] [CrossRef]
- Peng, J.; Zhao, Q.; Zheng, W.; Li, W.; Li, P.; Zhu, L.; Liu, X.; Shao, B.; Li, H.; Wang, C. Peptide-functionalized nanomaterials for the efficient isolation of HER2-positive circulating tumor cells. ACS Appl. Mater. Interfaces 2017, 9, 18423–18428. [Google Scholar] [CrossRef]
- Xiong, K.; Wei, W.; Jin, Y.; Wang, S.; Zhao, D.; Wang, S.; Gao, X.; Qiao, C.; Yue, H.; Ma, G. Biomimetic Immuno-Magnetosomes for High-Performance Enrichment of Circulating Tumor Cells. Adv. Mater. 2016, 28, 7929–7935. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. Clin. Oncol 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Haun, J.B.; Devaraj, N.K.; Marinelli, B.S.; Lee, H.; Weissleder, R. Probing intracellular biomarkers and mediators of cell activation using nanosensors and bioorthogonal chemistry. ACS Nano 2011, 5, 3204–3213. [Google Scholar] [CrossRef]
- Haun, J.B.; Devaraj, N.K.; Hilderbrand, S.A.; Lee, H.; Weissleder, R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat. Nanotechnol. 2010, 5, 660–665. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [Google Scholar] [CrossRef]
- Pang, C.; Koo, J.H.; Nguyen, A.; Caves, J.M.; Kim, M.G.; Chortos, A.; Kim, K.; Wang, P.J.; Tok, J.B.H.; Bao, Z. Highly skin-conformal microhairy sensor for pulse signal amplification. Adv. Mater. 2015, 27, 634–640. [Google Scholar] [CrossRef]
- Wang, S.; Liu, K.; Liu, J.; Yu, Z.T.F.; Xu, X.; Zhao, L.; Lee, T.; Lee, E.K.; Reiss, J.; Lee, Y.K. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem. 2011, 123, 3140–3144. [Google Scholar] [CrossRef]
- Shen, Q.; Xu, L.; Zhao, L.; Wu, D.; Fan, Y.; Zhou, Y.; OuYang, W.H.; Xu, X.; Zhang, Z.; Song, M. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv. Mater. 2013, 25, 2368–2373. [Google Scholar] [CrossRef]
- Chung, J.; Yokoyama, M.; Yamato, M.; Aoyagi, T.; Sakurai, Y.; Okano, T. Thermo-responsive drug delivery from polymeric micelles constructed using block copolymers of poly (N-isopropylacrylamide) and poly (butylmethacrylate). J. Control Release 1999, 62, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, Y.; Sun, K.; Fan, J.; Zhang, P.; Meng, J.; Wang, S.; Jiang, L. Dual-responsive surfaces modified with phenylboronic acid-containing polymer brush to reversibly capture and release cancer cells. J. Am. Chem. Soc. 2013, 135, 7603–7609. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Li, C.; Lu, S.; Zhou, W.; Tang, F.; Xie, X.S.; Huang, Y. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proc. Natl. Acad. Sci. USA 2015, 112, 11923–11928. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, L.; Wan, S.; Cansiz, S.; Cui, C.; Liu, Y.; Cai, R.; Hong, C.; Teng, I.-T.; Shi, M. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano 2017, 11, 3943–3949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Deng, Y.; Tai, Q.; Cheng, B.; Zhao, L.; Shen, Q.; He, R.; Hong, L.; Liu, W.; Guo, S. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv. Mater. 2012, 24, 2756–2760. [Google Scholar] [CrossRef]
- Jan, Y.J.; Chen, J.-F.; Zhu, Y.; Lu, Y.-T.; Chen, S.H.; Chung, H.; Smalley, M.; Huang, Y.-W.; Dong, J.; Chen, L.-C. NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, H.; Jiao, J.; Chen, K.J.; Owens, G.E.; Kamei, K.i.; Sun, J.; Sherman, D.J.; Behrenbruch, C.P.; Wu, H. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew. Chem. Int. Ed. 2009, 48, 8970–8973. [Google Scholar] [CrossRef]
- Zhang, P.; He, M.; Zeng, Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip 2016, 16, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Zhang, J.; Sluyter, R.; Zhao, Q.; Yan, S.; Alici, G.; Li, W. Continuous plasma extraction under viscoelastic fluid in a straight channel with asymmetrical expansion–contraction cavity arrays. Lab Chip 2016, 16, 3919–3928. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.-C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V. The exosome total isolation chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef]
- Galanzha, E.I.; Shashkov, E.V.; Kelly, T.; Kim, J.-W.; Yang, L.; Zharov, V.P. In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells. Nat. Nanotechnol. 2009, 4, 855–860. [Google Scholar] [CrossRef]
- Bamrungsap, S.; Chen, T.; Shukoor, M.I.; Chen, Z.; Sefah, K.; Chen, Y.; Tan, W. Pattern recognition of cancer cells using aptamer-conjugated magnetic nanoparticles. ACS Nano 2012, 6, 3974–3981. [Google Scholar] [CrossRef]
- Jiang, Y.; Palma, J.F.; Agus, D.B.; Wang, Y.; Gross, M.E. Detection of androgen receptor mutations in circulating tumor cells in castration-resistant prostate cancer. Clin. Chem. 2010, 56, 1492–1495. [Google Scholar] [CrossRef]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef]
- Kraan, J.; Sleijfer, S.; Strijbos, M.H.; Ignatiadis, M.; Peeters, D.; Pierga, J.Y.; Farace, F.; Riethdorf, S.; Fehm, T.; Zorzino, L. External quality assurance of circulating tumor cell enumeration using the CellSearch® system: A feasibility study. Cytom. Part B Clin. Cytom. 2011, 80, 112–118. [Google Scholar] [CrossRef]
- Welinder, C.; Jansson, B.; Lindell, G.; Wenner, J. Cytokeratin 20 improves the detection of circulating tumor cells in patients with colorectal cancer. Cancer Lett. 2015, 358, 43–46. [Google Scholar] [CrossRef]
- Gradilone, A.; Iacovelli, R.; Cortesi, E.; Raimondi, C.; Gianni, W.; Nicolazzo, C.; Petracca, A.; Palazzo, A.; Longo, F.; Frati, L. Circulating tumor cells and “suspicious objects” evaluated through CellSearch® in metastatic renal cell carcinoma. Anticancer Res. 2011, 31, 4219–4221. [Google Scholar]
- Neely, A.; Perry, C.; Varisli, B.; Singh, A.K.; Arbneshi, T.; Senapati, D.; Kalluri, J.R.; Ray, P.C. Ultrasensitive and highly selective detection of Alzheimer’s disease biomarker using two-photon Rayleigh scattering properties of gold nanoparticle. ACS Nano 2009, 3, 2834–2840. [Google Scholar] [CrossRef]
- Khurana, I.; Allawadhi, P.; Neeradi, D.; Banothu, A.K.; Thalugula, S.; Naik, R.R.; Packirisamy, G.; Bharani, K.K.; Khurana, A. Introduction to nanoengineering and nanotechnology for biomedical applications. In Emerging Nanotechnologies for Medical Applications; Elsevier: Amsterdam, The Netherlands, 2023; pp. 1–34. [Google Scholar]
- Kalkal, A.; Pradhan, R.; Kadian, S.; Manik, G.; Packirisamy, G. Biofunctionalized Graphene Quantum Dots Based Fluorescent Biosensor toward Efficient Detection of Small Cell Lung Cancer. ACS Appl. Bio Mater. 2020, 3, 4922–4932. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Su, S.; Guo, Y.; Jiang, X.; Zhong, Y.; Su, Y.; Fan, C.; Lee, S.-T.; He, Y. A Molecular Beacon-Based Signal-Off Surface-Enhanced Raman Scattering Strategy for Highly Sensitive, Reproducible, and Multiplexed DNA Detection. Small 2013, 9, 2493–2499. [Google Scholar] [CrossRef]
- Wu, X.; Xia, Y.; Huang, Y.; Li, J.; Ruan, H.; Chen, T.; Luo, L.; Shen, Z.; Wu, A. Improved SERS-Active Nanoparticles with Various Shapes for CTC Detection without Enrichment Process with Supersensitivity and High Specificity. ACS Appl. Mater. Interfaces 2016, 8, 19928–19938. [Google Scholar] [CrossRef]
- Hou, S.; Zhao, L.; Shen, Q.; Yu, J.; Ng, C.; Kong, X.; Wu, D.; Song, M.; Shi, X.; Xu, X.; et al. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew. Chem. Int. Ed. Engl. 2013, 52, 3379–3383. [Google Scholar] [CrossRef]
- Silverman, S.K. Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Turberfield, A.J.; Yurke, B.; Winfree, E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 2007, 318, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Choi, H.M.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef]
- Hu, J.; Sheng, Y.; Kwak, K.J.; Shi, J.; Yu, B.; Lee, L.J. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat. Commun. 2017, 8, 1683. [Google Scholar] [CrossRef]
- Kalogianni, D.P. Nanotechnology in emerging liquid biopsy applications. Nano Converg. 2021, 8, 13. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, L.; Huang, Y.; Yang, Y.; He, Y.; Lu, C.; Yang, H. DNA-mediated reversible capture and release of circulating tumor cells with a multivalent dual-specific aptamer coating network. Chem. Commun. 2019, 55, 5387–5390. [Google Scholar] [CrossRef]
- Hu, X.; Wei, C.-W.; Xia, J.; Pelivanov, I.; O’Donnell, M.; Gao, X. Trapping and photoacoustic detection of CTCs at the single cell per milliliter level with magneto-optical coupled nanoparticles. Small 2013, 9, 2046. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, F.; Dan, W.; Fu, Y.; Liu, S. Construction of carbon nanotube based nanoarchitectures for selective impedimetric detection of cancer cells in whole blood. Analyst 2014, 139, 5086–5092. [Google Scholar] [CrossRef]
- Hou, S.; Zhao, H.; Zhao, L.; Shen, Q.; Wei, K.S.; Suh, D.Y.; Nakao, A.; Garcia, M.A.; Song, M.; Lee, T.; et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 2013, 25, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-y.; Hoshino, K.; Chen, P.; Wu, C.-h.; Lane, N.; Huebschman, M.; Liu, H.; Sokolov, K.; Uhr, J.W.; Frenkel, E.P. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed. Microdevices 2013, 15, 673–681. [Google Scholar] [CrossRef]
- Hoshino, K.; Huang, Y.-Y.; Lane, N.; Huebschman, M.; Uhr, J.W.; Frenkel, E.P.; Zhang, X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 2011, 11, 3449–3457. [Google Scholar] [CrossRef]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.-i.; Morgan, B.; Trautwein, J. Inertial focusing for tumor antigen–dependent and–independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, ra147–ra179. [Google Scholar] [CrossRef] [PubMed]
- Poudineh, M.; Aldridge, P.M.; Ahmed, S.; Green, B.J.; Kermanshah, L.; Nguyen, V.; Tu, C.; Mohamadi, R.M.; Nam, R.K.; Hansen, A. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 2017, 12, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.-Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef]
- Oliveira-Rodríguez, M.; López-Cobo, S.; Reyburn, H.T.; Costa-García, A.; López-Martín, S.; Yáñez-Mó, M.; Cernuda-Morollón, E.; Paschen, A.; Valés-Gómez, M.; Blanco-López, M.C. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J. Extracell. Vesicles 2016, 5, 31803. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, Y.; Lu, Y.; Xing, W. Isolation and visible detection of tumor-derived exosomes from plasma. Anal. Chem. 2018, 90, 14207–14215. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Liu, J.-W.; Adkins, G.B.; Shen, W.; Trinh, M.P.; Duan, L.-Y.; Jiang, J.-H.; Zhong, W. Enhancement of the intrinsic peroxidase-like activity of graphitic carbon nitride nanosheets by ssDNAs and its application for detection of exosomes. Anal. Chem. 2017, 89, 12327–12333. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, J.; Wu, Y.; Xing, S.; Lai, Y.; Zhang, G. Target-induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens. Bioelectron. 2018, 102, 204–210. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, D.; Yue, S.; Lu, Y.; Yang, C.; Fang, J.; Xu, Z. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019, 4, 3210–3218. [Google Scholar] [CrossRef]
- He, F.; Wang, J.; Yin, B.-C.; Ye, B.-C. Quantification of exosome based on a copper-mediated signal amplification strategy. Anal. Chem. 2018, 90, 8072–8079. [Google Scholar] [CrossRef]
- Gao, M.-L.; Yin, B.-C.; Ye, B.-C. Construction of a DNA-AuNP-based satellite network for exosome analysis. Analyst 2019, 144, 5996–6003. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.; Zong, J.; Chen, C.; Jiang, X.; Zhang, Y.; Wang, Z.; Cui, Y. Single molecule localization imaging of exosomes using blinking silicon quantum dots. Nanotechnology 2018, 29, 065705. [Google Scholar] [CrossRef]
- Tayebi, M.; Yaraki, M.T.; Yang, H.Y.; Ai, Y. A MoS 2–MWCNT based fluorometric nanosensor for exosome detection and quantification. Nanoscale Adv. 2019, 1, 2866–2872. [Google Scholar] [CrossRef]
- Jin, D.; Yang, F.; Zhang, Y.; Liu, L.; Zhou, Y.; Wang, F.; Zhang, G.-J. ExoAPP: Exosome-oriented, aptamer nanoprobe-enabled surface proteins profiling and detection. Anal. Chem. 2018, 90, 14402–14411. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, F.; Zhang, H.; Zhang, Y.; Liu, M.; Liu, Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal. Chem. 2018, 90, 12737–12744. [Google Scholar] [CrossRef] [PubMed]
- Boriachek, K.; Masud, M.K.; Palma, C.; Phan, H.-P.; Yamauchi, Y.; Hossain, M.S.A.; Nguyen, N.-T.; Salomon, C.; Shiddiky, M.J. Avoiding pre-isolation step in exosome analysis: Direct isolation and sensitive detection of exosomes using gold-loaded nanoporous ferric oxide nanozymes. Anal. Chem. 2019, 91, 3827–3834. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.-T.; Shiddiky, M.J. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst 2017, 142, 2211–2219. [Google Scholar] [CrossRef]
- Zhou, Y.G.; Mohamadi, R.M.; Poudineh, M.; Kermanshah, L.; Ahmed, S.; Safaei, T.S.; Stojcic, J.; Nam, R.K.; Sargent, E.H.; Kelley, S.O. Interrogating circulating microsomes and exosomes using metal nanoparticles. Small 2016, 12, 727–732. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.; Zhang, Q.; Wang, F.; Liu, Y. Ti3C2 MXenes nanosheets catalyzed highly efficient electrogenerated chemiluminescence biosensor for the detection of exosomes. Biosens. Bioelectron. 2019, 124, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.; Wang, L.; Chen, C.; Lu, J.; Zhu, D.; Zhang, Y.; Wang, Z.; Cui, Y. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal. Methods 2016, 8, 5001–5008. [Google Scholar] [CrossRef]
- Jeong, S.; Park, J.; Pathania, D.; Castro, C.M.; Weissleder, R.; Lee, H. Integrated magneto–electrochemical sensor for exosome analysis. ACS Nano 2016, 10, 1802–1809. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, X.; Liu, B.; Liu, J. Molecular imprinting on inorganic nanozymes for hundred-fold enzyme specificity. J. Am. Chem. Soc. 2017, 139, 5412–5419. [Google Scholar] [CrossRef]
- Chen, X.; Lan, J.; Liu, Y.; Li, L.; Yan, L.; Xia, Y.; Wu, F.; Li, C.; Li, S.; Chen, J. A paper-supported aptasensor based on upconversion luminescence resonance energy transfer for the accessible determination of exosomes. Biosens. Bioelectron. 2018, 102, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Yang, K.S.; Im, H.; Hong, S.; Pergolini, I.; Del Castillo, A.F.; Wang, R.; Clardy, S.; Huang, C.-H.; Pille, C.; Ferrone, S. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med. 2017, 9, eaal3226. [Google Scholar] [CrossRef]
- Liang, K.; Liu, F.; Fan, J.; Sun, D.; Liu, C.; Lyon, C.J.; Bernard, D.W.; Li, Y.; Yokoi, K.; Katz, M.H. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 2017, 1, 21. [Google Scholar] [CrossRef]
- Coutinho, C.; Somoza, Á. MicroRNA sensors based on gold nanoparticles. Anal. Bioanal. Chem. 2019, 411, 1807–1824. [Google Scholar] [CrossRef]
- He, W.; Li, S.; Wang, L.; Zhu, L.; Zhang, Y.; Luo, Y.; Huang, K.; Xu, W. AuNPs-DNAzyme molecular motor biosensor mediated by neighborhood click chemistry reactions for the ultrasensitive detection of microRNA-155. Sens. Actuators B Chem. 2019, 290, 503–511. [Google Scholar] [CrossRef]
- Borghei, Y.-S.; Hosseini, M. A New eye Dual-readout Method for MiRNA Detection based on Dissolution of Gold nanoparticles via LspR by Cdte QDs photoinduction. Sci. Rep. 2019, 9, 5453. [Google Scholar] [CrossRef] [PubMed]
- Nossier, A.I.; Abdelzaher, H.; Matboli, M.; Eissa, S. Dual approach for the colorimetric determination of unamplified microRNAs by using citrate capped gold nanoparticles. Microchim. Acta 2018, 185, 236. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wang, X.; Chen, W. The formulation of a refined hybrid enhanced assumed strain solid shell element and its application to model smart structures containing distributed piezoelectric sensors/actuators. Smart Mater. Struct. 2004, 13, N43. [Google Scholar] [CrossRef]
- Yao, M.; Lv, X.; Deng, Y.; Rasheed, M. Specific and simultaneous detection of micro RNA 21 and let-7a by rolling circle amplification combined with lateral flow strip. Anal. Chim. Acta 2019, 1055, 115–125. [Google Scholar] [CrossRef]
- Gao, X.; Xu, L.-P.; Wu, T.; Wen, Y.; Ma, X.; Zhang, X. An enzyme-amplified lateral flow strip biosensor for visual detection of microRNA-224. Talanta 2016, 146, 648–654. [Google Scholar] [CrossRef]
- Mohammadniaei, M.; Go, A.; Chavan, S.G.; Koyappayil, A.; Kim, S.-E.; Yoo, H.J.; Min, J.; Lee, M.-H. Relay-race RNA/barcode gold nanoflower hybrid for wide and sensitive detection of microRNA in total patient serum. Biosens. Bioelectron. 2019, 141, 111468. [Google Scholar] [CrossRef] [PubMed]
- Fredj, Z.; Azzouzi, S.; Turner, A.P.; Ali, M.B.; Mak, W.C. Neutravidin biosensor for direct capture of dual-functional biotin-molecular beacon-AuNP probe for sensitive voltammetric detection of microRNA. Sens. Actuators B Chem. 2017, 248, 77–84. [Google Scholar] [CrossRef]
- Li, T.; Wu, X.; Tao, G.; Yin, H.; Zhang, J.; Liu, F.; Li, N. A simple and non-amplification platform for femtomolar DNA and microRNA detection by combining automatic gold nanoparticle enumeration with target-induced strand-displacement. Biosens. Bioelectron. 2018, 105, 137–142. [Google Scholar] [CrossRef]
- Sguassero, A.; Artiga, Á.; Morasso, C.; Jimenez, R.R.; Rapún, R.M.; Mancuso, R.; Agostini, S.; Hernis, A.; Abols, A.; Linē, A. A simple and universal enzyme-free approach for the detection of multiple microRNAs using a single nanostructured enhancer of surface plasmon resonance imaging. Anal. Bioanal. Chem. 2019, 411, 1873–1885. [Google Scholar] [CrossRef] [PubMed]
- Tadimety, A.; Zhang, Y.; Kready, K.M.; Palinski, T.J.; Tsongalis, G.J.; Zhang, J.X. Design of peptide nucleic acid probes on plasmonic gold nanorods for detection of circulating tumor DNA point mutations. Biosens. Bioelectron. 2019, 130, 236–244. [Google Scholar] [CrossRef]
- Khanna, S.; Padhan, P.; Das, S.; Jaiswal, K.S.; Tripathy, A.; Smita, S.; Tripathy, S.K.; Raghav, S.K.; Gupta, B. A Simple Colorimetric Method for Naked-Eye Detection of Circulating Cell-Free DNA Using Unlabelled Gold Nanoparticles. ChemistrySelect 2018, 3, 11541–11551. [Google Scholar] [CrossRef]
- Wang, C.; Chen, X.; Wu, Y.; Li, H.; Wang, Y.; Pan, X.; Tang, T.; Liu, Z.; Li, X. Lateral flow strip for visual detection of K-ras mutations based on allele-specific PCR. Biotechnol. Lett. 2016, 38, 1709–1714. [Google Scholar] [CrossRef]
- Li, X.; Chen, B.; He, M.; Hu, B. Immunodetection and counting of circulating tumor cells (HepG2) by combining gold nanoparticle labeling, rolling circle amplification and ICP-MS detection of gold. Microchim. Acta 2019, 186, 344. [Google Scholar] [CrossRef]
- Chiu, W.-J.; Ling, T.-K.; Chiang, H.-P.; Lin, H.-J.; Huang, C.-C. Monitoring cluster ions derived from aptamer-modified gold nanofilms under laser desorption/ionization for the detection of circulating tumor cells. ACS Appl. Mater. Interfaces 2015, 7, 8622–8630. [Google Scholar] [CrossRef]
- Liu, G.; Mao, X.; Phillips, J.A.; Xu, H.; Tan, W.; Zeng, L. Aptamer− nanoparticle strip biosensor for sensitive detection of cancer cells. Anal. Chem. 2009, 81, 10013–10018. [Google Scholar] [CrossRef]
- Khoothiam, K.; Treerattrakoon, K.; Iempridee, T.; Luksirikul, P.; Dharakul, T.; Japrung, D. Ultrasensitive detection of lung cancer-associated miRNAs by multiple primer-mediated rolling circle amplification coupled with a graphene oxide fluorescence-based (MPRCA-GO) sensor. Analyst 2019, 144, 4180–4187. [Google Scholar] [CrossRef]
- Treerattrakoon, K.; Jiemsakul, T.; Tansarawiput, C.; Pinpradup, P.; Iempridee, T.; Luksirikul, P.; Khoothiam, K.; Dharakul, T.; Japrung, D. Rolling circle amplification and graphene-based sensor-on-a-chip for sensitive detection of serum circulating miRNAs. Anal. Biochem. 2019, 577, 89–97. [Google Scholar] [CrossRef]
- Ou, X.; Zhan, S.; Sun, C.; Cheng, Y.; Wang, X.; Liu, B.; Zhai, T.; Lou, X.; Xia, F. Simultaneous detection of telomerase and miRNA with graphene oxide-based fluorescent aptasensor in living cells and tissue samples. Biosens. Bioelectron. 2019, 124, 199–204. [Google Scholar] [CrossRef]
- Esteban-Fernaández de AÁvila, B.; Martín, A.; Soto, F.; Lopez-Ramirez, M.A.; Campuzano, S.; Vasquez-Machado, G.M.; Gao, W.; Zhang, L.; Wang, J. Single cell real-time miRNAs sensing based on nanomotors. ACS Nano 2015, 9, 6756–6764. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.-R.; Lee, J.; Yeo, J.; Na, H.-K.; Kim, Y.-K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 2013, 7, 5882–5891. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.-k.; Lee, S.; Yoon, S.; Kim, W.-k. Graphene oxide-based NET strategy for enhanced colorimetric sensing of miRNA. Sens. Actuators B Chem. 2019, 282, 861–867. [Google Scholar] [CrossRef]
- Xiao, Q.; Li, J.; Jin, X.; Liu, Y.; Huang, S. Ultrasensitive electrochemical microRNA-21 biosensor coupling with carboxylate-reduced graphene oxide-based signal-enhancing and duplex-specific nuclease-assisted target recycling. Sens. Actuators B Chem. 2019, 297, 126740. [Google Scholar] [CrossRef]
- Wang, F.; Chu, Y.; Ai, Y.; Chen, L.; Gao, F. Graphene oxide with in-situ grown Prussian Blue as an electrochemical probe for microRNA-122. Microchim. Acta 2019, 186, 116. [Google Scholar] [CrossRef]
- Song, Y.; Li, L.; Yang, Z.; Zhao, G.; Zhang, X.; Wang, L.; Zheng, L.; Zhuo, F.; Yin, H.; Ge, X. Target of rapamycin (TOR) regulates the expression of lncRNAs in response to abiotic stresses in cotton. Front. Genet. 2019, 9, 690. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, Z.; Gu, Y.; Li, Y.; Jia, Y. Clinical available circulating tumor cell assay based on tetra (4-aminophenyl) porphyrin mediated reduced graphene oxide field effect transistor. Electrochim. Acta 2019, 313, 415–422. [Google Scholar] [CrossRef]
- Xiao, K.; Liu, J.; Chen, H.; Zhang, S.; Kong, J. A label-free and high-efficient GO-based aptasensor for cancer cells based on cyclic enzymatic signal amplification. Biosens. Bioelectron. 2017, 91, 76–81. [Google Scholar] [CrossRef]
- Kun, Q.; Lin, Y.; Peng, H.; Cheng, L.; Cui, H.; Hong, N.; Xiong, J.; Fan, H. A “signal-on” switch electrochemiluminescence biosensor for the detection of tumor cells. J. Electroanal. Chem. 2018, 808, 101–106. [Google Scholar] [CrossRef]
- Xu, Z.; Chang, Y.; Chai, Y.; Wang, H.; Yuan, R. Ultrasensitive electrochemiluminescence biosensor for speedy detection of microrna based on a DNA rolling machine and target recycling. Anal. Chem. 2019, 91, 4883–4888. [Google Scholar] [CrossRef]
- Zhu, H.-Y.; Ding, S.-N. Dual-signal-amplified electrochemiluminescence biosensor for microRNA detection by coupling cyclic enzyme with CdTe QDs aggregate as luminophor. Biosens. Bioelectron. 2019, 134, 109–116. [Google Scholar] [CrossRef]
- Hao, N.; Lu, J.; Chi, M.; Xiong, M.; Zhang, Y.; Hua, R.; Wang, K. A universal photoelectrochemical biosensor for dual microRNA detection based on two CdTe nanocomposites. J. Mater. Chem. B 2019, 7, 1133–1141. [Google Scholar] [CrossRef]
- Yu, X.; Hu, L.; Zhang, F.; Wang, M.; Xia, Z.; Wei, W. MoS2 quantum dots modified with a labeled molecular beacon as a ratiometric fluorescent gene probe for FRET based detection and imaging of microRNA. Microchim. Acta 2018, 185, 239. [Google Scholar] [CrossRef]
- Xie, M.; Lu, N.-N.; Cheng, S.-B.; Wang, X.-Y.; Wang, M.; Guo, S.; Wen, C.-Y.; Hu, J.; Pang, D.-W.; Huang, W.-H. Engineered decomposable multifunctional nanobioprobes for capture and release of rare cancer cells. Anal. Chem. 2014, 86, 4618–4626. [Google Scholar] [CrossRef]
- Min, H.; Jo, S.M.; Kim, H.S. Efficient capture and simple quantification of circulating tumor cells using quantum dots and magnetic beads. Small 2015, 11, 2536–2542. [Google Scholar] [CrossRef]
- Borghei, Y.-S.; Hosseini, M.; Ganjali, M.R. Visual detection of miRNA using peroxidase-like catalytic activity of DNA-CuNCs and methylene blue as indicator. Clin. Chim. Acta 2018, 483, 119–125. [Google Scholar] [CrossRef]
- Xu, F.; Luo, L.; Shi, H.; He, X.; Lei, Y.; Tang, J.; He, D.; Qiao, Z.; Wang, K. Label-free and sensitive microRNA detection based on a target recycling amplification-integrated superlong poly (thymine)-hosted copper nanoparticle strategy. Anal. Chim. Acta 2018, 1010, 54–61. [Google Scholar] [CrossRef]
- Koo, K.M.; Carrascosa, L.G.; Trau, M. DNA-directed assembly of copper nanoblocks with inbuilt fluorescent and electrochemical properties: Application in simultaneous amplification-free analysis of multiple RNA species. Nano Res. 2018, 11, 940–952. [Google Scholar] [CrossRef]
- Liao, H.; Zhou, Y.; Chai, Y.; Yuan, R. An ultrasensitive electrochemiluminescence biosensor for detection of MicroRNA by in-situ electrochemically generated copper nanoclusters as luminophore and TiO2 as coreaction accelerator. Biosens. Bioelectron. 2018, 114, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, S.; Park, K.S.; Park, H.G. Enzyme-free and label-free miRNA detection based on target-triggered catalytic hairpin assembly and fluorescence enhancement of DNA-silver nanoclusters. Sens. Actuators B Chem. 2018, 260, 140–145. [Google Scholar] [CrossRef]
- Miao, X.; Cheng, Z.; Ma, H.; Li, Z.; Xue, N.; Wang, P. Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters. Anal. Chem. 2018, 90, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; Jiang, X.; Dong, C.; Song, C.; Han, C.; Wang, L. Ultrasensitive SERS detection of nucleic acids via simultaneous amplification of target-triggered enzyme-free recycling and multiple-reporter. Biosens. Bioelectron. 2019, 141, 111402. [Google Scholar] [CrossRef]
- Ruan, H.; Wu, X.; Yang, C.; Li, Z.; Xia, Y.; Xue, T.; Shen, Z.; Wu, A. A supersensitive CTC analysis system based on triangular silver nanoprisms and SPION with function of capture, enrichment, detection, and release. ACS Biomater. Sci. Eng. 2018, 4, 1073–1082. [Google Scholar] [CrossRef]
- Zhang, Y.; Ning, X.; Mao, G.; Ji, X.; He, Z. Fluorescence turn-on detection of target sequence DNA based on silicon nanodot-mediated quenching. Anal. Bioanal. Chem. 2018, 410, 3209–3216. [Google Scholar] [CrossRef]
- Su, X.; Kuang, L.; Battle, C.; Shaner, T.; Mitchell, B.S.; Fink, M.J.; Jayawickramarajah, J. Mild two-step method to construct DNA-conjugated silicon nanoparticles: Scaffolds for the detection of microRNA-21. Bioconjugate Chem. 2014, 25, 1739–1743. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, C.; Wu, S.; Shen, Y.; Zhou, C.; Neng, J.; Yi, Y.; Jin, Y.; Zhu, Y. Magnetic-capture-based SERS detection of multiple serum microRNA biomarkers for cancer diagnosis. Anal. Methods 2019, 11, 783–793. [Google Scholar] [CrossRef]
- Liu, F.; Li, P.; Chen, M.; Luo, Y.; Prabhakar, M.; Zheng, H.; He, Y.; Qi, Q.; Long, H.; Zhang, Y. Fructooligosaccharide (FOS) and galactooligosaccharide (GOS) increase Bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci. Rep. 2017, 7, 11798. [Google Scholar] [CrossRef]
- Hu, P.; Zhang, S.; Wu, T.; Ni, D.; Fan, W.; Zhu, Y.; Qian, R.; Shi, J. Fe–Au Nanoparticle-Coupling for Ultrasensitive Detections of Circulating Tumor DNA. Adv. Mater. 2018, 30, 1801690. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, S.; Li, C.; Wang, Y.; Yan, C. Dual-target recognition sandwich assay based on core-shell magnetic mesoporous silica nanoparticles for sensitive detection of breast cancer cells. Talanta 2018, 182, 306–313. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).