Abstract
Glutamine is an essential biomolecule that plays a pivotal role in many diseases, such as cancer, where it can serve as fuel for rapid proliferation. Treatments for these diseases can be monitored and optimized through the detection of glutamine, though standard glutamine detection procedures are costly and require complex instrumentation. Cell-free protein synthesis (CFPS) has recently enabled a paper-based, colorimetric glutamine sensor that carries the potential to increase test accessibility while dramatically reducing consumer cost to enable at-home, rapid treatment monitoring. Test sensitivity remained limited by residual assay background, thus motivating this work where CFPS reactions traditionally formulated with glutamate salts were compared to systems using alternative salts, including aspartate, acetate, citrate, and sulfate, to reduce the background generation of glutamine. This led to the discovery of a novel aspartate-based CFPS system that boasts a high signal strength and indetectable background noise over 225 min. Acetate-, citrate-, and sulfate-based systems also yielded zero background glutamine detection but at a lower signal response compared to the aspartate-based system. These findings mark crucial advancements in producing a cost-effective, simple glutamine monitor while simultaneously showcasing the adaptability of CFPS’s open reaction environment for solving complex challenges in next-generation biosensor development.
1. Introduction
Healthcare systems worldwide are increasingly reliant on biomarker analysis for disease diagnosis, treatment monitoring, and therapeutic decision making [1]. Such monitoring and diagnostics can significantly impact disease management and survival rates for metabolic diseases like cancer [2,3]. Metabolic biomarkers are particularly valuable as they reflect functional changes in cellular processes that often precede clinical manifestations of disease [4,5]. Among these, amino acid metabolism has gained significant attention in oncology with glutamine emerging as a critical metabolite in cancer pathophysiology [5,6,7,8]. Unlike normal cells that primarily use glucose through oxidative phosphorylation, many cancer cells exhibit “glutamine addiction”, redirecting this amino acid to fuel rapid proliferation, support redox homeostasis, and enable metabolic adaptations under hypoxic conditions [9,10,11]. This metabolic reprogramming presents both diagnostic opportunities and therapeutic vulnerabilities that remain underexploited in clinical settings.
Glutamine serves as a primary nitrogen donor for biosynthesis and a carbon replenisher for the tricarboxylic acid (TCA) cycle in rapidly dividing cancer cells [12,13]. Multiple studies have demonstrated that glutamine metabolism is upregulated in various cancer types [14,15,16]. This metabolic dependency has sparked the development of specialized cancer therapies that target glutamine utilization [17,18]. Glutaminase inhibitors represent one therapeutic approach that has shown promise in clinical trials [19] with demonstrated efficacy in multiple cancer types that exhibit particular dependence on glutamine metabolism [20].
However, glutamine metabolism in healthy cells must remain adequately balanced to prevent dangerous side effects. Clinical trials targeting glutamine metabolism have yielded mixed results, indicating that personalized treatment approaches with regular monitoring of glutamine levels could optimize the balance between treatment efficacy and adverse effects [21]. The frequent testing of patient glutamine levels would enable appropriate treatment dosage adjustments to maintain this critical balance.
Developing clinically relevant glutamine detection methods presents significant analytical challenges. Current techniques rely on separation technologies such as ion-exchange chromatography coupled with spectrophotometric detection or liquid chromatography-mass spectrometry (LC-MS) [22,23]. While these methods offer excellent sensitivity and specificity, their implementation requires sophisticated instrumentation, specialized operator training, and complex sample preparation protocols that limit accessibility in resource-constrained environments.
Recent innovations have attempted to address these limitations through various approaches, including optical sensors (split fluorescent proteins [24], environmentally sensitive fluorophores [25], and fluorescent protein-based sensors for in vivo monitoring [26]), nanomaterial-based methods (zinc oxide nanorod-based sensors [27] and enzyme-nanomaterial conjugates [28]), and surface plasmon resonance platforms [29,30]. Other novel approaches include electrochemical methods utilizing modified binding proteins [31] and biomolecular interactions such as TORC1-activating glutamine sensors [32]. Despite these advances, many of these sensing platforms remain unsuitable for widespread patient use due to the equipment requirements, expertise needed, and lengthy analysis times. There remains a critical need for truly accessible diagnostic platforms that combine simplicity, speed, and affordability for point-of-care implementation.
A particularly promising approach to address these limitations leverages cell-free protein synthesis (CFPS) technology, which has gained recognition for its versatility in biosensor development [33,34]. By eliminating the constraints of working with intact cells, CFPS systems provide an open reaction environment where sensor components and analytes can interact directly [35,36]. A major advantage of CFPS-based biosensors is their compatibility with lyophilization (freeze drying), which enables the development of shelf-stable diagnostic formats for point-of-care and at-home applications. Lyophilized CFPS reactions can remain viable for extended periods without refrigeration, simplifying distribution and storage logistics, particularly in resource-limited settings [37,38,39,40,41]. When rehydrated with a patient sample, these stabilized reactions rapidly produce detectable signals, allowing for the real-time monitoring of biomarker levels. In the context of glutamine detection for cancer treatment monitoring, this technology could enable patients to perform frequent, affordable testing at home without specialized equipment or technical expertise. Such capabilities would allow for timely adjustments to treatment regimens based on individual metabolic responses, potentially improving therapeutic outcomes while reducing hospital visits.
In our previous work, we developed a paper-based, colorimetric, cell-free glutamine biosensor requiring only the addition of a liquid sample to generate a visually interpretable result, which is similar to common consumer diagnostics like pregnancy tests [42,43,44]. This approach used the dependence of protein synthesis on amino acid availability to formulate reactions lacking glutamine while supplying all other amino acids in excess. When mixed with a sample, the limiting reactant in protein translation became the glutamine present in the sample. However, we encountered a significant challenge: reporter protein production occurred even in the absence of exogenous glutamine, creating a substantial background signal that limited assay sensitivity and dynamic range.
We partially addressed this issue through the genetic engineering of E. coli BL21-Star™ DE3 bacteria to create a knockout strain (ΔlacZ ΔglnA) lacking β-galactosidase and glutamine synthetase, thereby preventing endogenous β-gal production and reducing background glutamine synthesis [42]. Additionally, we incorporated L-methionine sulfoximine (MSO), a competitive inhibitor of glutamine synthetase, and protease inhibitors to further reduce background glutamine generation. These combined approaches significantly improved the signal-to-background ratio, enabling the development of a paper-based colorimetric glutamine test with enhanced signal resolution.
Despite prior improvements, residual background signal persisted, suggesting additional glutamine-generating pathways remained active in the cell extract. This work reports alternatively engineered formulations of CFPS to eliminate the background signal to enhance sensor specificity. The enzyme glutaminase, which catalyzes the conversion between glutamine and glutamate, represents a likely contributor to this background. Traditional cell-free protein synthesis systems rely heavily on glutamate-based energy regeneration solutions, typically containing high concentrations of glutamate (~200 mM) to facilitate ATP regeneration via the conversion of phosphoenolpyruvate to pyruvate. This abundance of glutamate in standard CFPS formulations potentially drives the glutaminase reaction toward glutamine production, contributing to the persistent background signal observed in our previous sensor. This background limits the sensitivity and dynamic range of glutamine biosensors, constraining their utility for monitoring cancer treatment responses where precise measurements across physiologically relevant concentrations are essential.
Cell-free systems have historically been formulated with various energy sources and buffer compositions with glutamate being predominant due to its effectiveness in supporting high-yield protein production. Laohakunakorn [45] highlighted the potential of cell-free systems as platforms for rational biodesign, emphasizing their flexibility and adaptability for specific applications. Aw and Polizzi [46] demonstrated the utility of biosensor-assisted engineering to optimize cell-free protein synthesis, showing how alternative formulations can enhance specific performance characteristics.
To our knowledge, a zero-background, low-cost, portable glutamine biosensor has yet to be reported. Thus, a critical goal of this work is to eliminate the background signal by finding alternatives to glutamate-based systems that overcome metabolic conversion. Aspartate represents a particularly promising candidate for replacing glutamate in CFPS formulations. Unlike glutamate, aspartate is not directly connected to glutamine biosynthesis through major metabolic enzymes like glutaminase, potentially offering a pathway to reduce background signal while maintaining protein synthesis capacity. Aspartate participates in the malate–aspartate shuttle and can contribute to energy metabolism through conversion to oxaloacetate [47]. Recent evidence suggests that supplementing culture media with aspartate can significantly enhance recombinant protein production with one study demonstrating a nearly tenfold increase in enzyme yields when combined with arginine in defined medium formulations [48]. The metabolic relationship between aspartate and glutamate has been explored in various contexts with studies demonstrating that L-aspartate serves as a high-quality nitrogen source in E. coli [49] and that aspartate aminotransferase plays a critical role in cellular metabolic networks [50].
In this work, we systematically compared the traditional glutamate-based system to alternative salt sources to identify formulations that provide zero background while maintaining sufficient protein yield for sensing applications. Building on our investigation of aspartate-based systems, we expanded our approach to include several other alternatives, which were each selected for specific biochemical properties. Acetate-based cell-free systems were included despite their historically lower translation efficiency [51] as they represent a metabolically distant alternative to glutamate that could potentially eliminate background glutamine formation completely. In addition to aspartate-based and acetate-based systems, our investigation expanded to include citrate and sulfate systems, which were each selected for specific biochemical properties. Citrate was included due to its role as both a metabolic intermediate and a metal chelator, which can help regulate the availability of divalent cations essential for enzymatic function [52]. Sulfate-based formulations were incorporated as they provide distinct ionic properties that can influence protein–protein interactions and maintain an optimal redox environment for enzymatic activity [53]. The four alternative salt systems investigated in this study (aspartate, acetate, citrate, and sulfate) offer potential advantages for specific applications by altering the metabolic landscape of the reaction environment [54]. Our approach aims to disrupt the metabolic pathways leading to glutamine formation while maintaining the protein synthesis capacity necessary for biosensor functionality. These cell-free biosensor systems are inherently cost-effective, with low production costs compared to conventional diagnostic platforms, making them suitable for frequent, affordable testing by patients. This advancement represents an important step toward enabling personalized cancer treatment regimens that optimize therapeutic efficacy while minimizing adverse effects.
2. Materials and Methods
2.1. Cell-Free Protein Synthesis
Cell extract for protein synthesis reactions was prepared using E. coli BL21-Star™ DE3 (Invitrogen, Carlsbad, CA, USA) cells with Δlac and ΔglnA frameshift mutations [42]. The extract was prepared following previously reported protocols [55]. Cells were grown in 2× YT media supplemented with 1 g/L glutamine to sustain cell growth given the ΔglnA mutation. The 1 L cultures were induced with 1 mM IPTG at an OD600 of ~0.5 and harvested at an OD600 of ~3.
Cells were washed three times by resuspension in 10 mL of Buffer S30 per gram of wet cells followed by centrifugation at 6000 relative centrifugal force (RCF) units for 15 min. The supernatant was discarded each time. Buffer S30 was made as previously reported [43], except in this work, we used various potassium-based salts in place of potassium glutamate. For the glutamate-based system, 60 mM potassium glutamate was used as the control. For both the citrate- and acetate-based systems, 60 mM potassium acetate was used. For the sulfate-based system, 60 mM potassium sulfate was used. For the aspartate-based system, 60 mM potassium hydroxide and 60 mM aspartic acid were used. After washing, the cells were resuspended in 1 mL of Buffer S30 per gram of wet cells. Cell lysis was performed using sonication following previously reported protocols [42,55]. Cells were lysed with a Vibra-cell VCX 400 probe sonicator with a CV 26 probe (tip diameter of 3 mm; Sonics and Materials, Newtown, CT, USA) at a tip frequency of 20 kHz at 15% amplitude. Samples were kept in an ice bath to prevent overheating. Sonication was performed in 2 min intervals followed by a 3–6 min rest period for a total of 12 cycles. All lysates were subsequently centrifuged at 12,000 RCF for 30 min. The extracts were then incubated in a runoff reaction for 30 min at 37 °C and 280 RPM. Extracts were treated with 4× MSO at a final concentration of 320 mM in the cell extract and 4× protease inhibitor (EDTA-free PierceTM Protease Inhibitor Tablets, ThermoScientific (Waltham, MA, USA)).
PANOxSP was prepared as previously reported [43,44], except in this work, we replaced glutamate-based salts with other salts to eliminate glutamate from the system. For the glutamate-based system, ammonium glutamate and potassium glutamate were used at final concentrations of 10 and 175 mM, respectively, in the cell-free reactions. For the acetate-based system, ammonium acetate and potassium acetate were used at final concentrations of 10 and 175 mM, respectively. For the citrate-based system, ammonium acetate, potassium hydroxide, and citric acid were used at final concentrations of 10, 300, and 100 mM, respectively. For the sulfate-based system, ammonium sulfate and potassium sulfate were used at final concentrations of 10 and 100 mM, respectively. For the aspartate-based system, ammonium hydroxide, potassium hydroxide, and aspartic acid were used at final concentrations of 10, 175, and 185 mM, respectively. The citrate and aspartate PANOxSP mixtures required titration with potassium hydroxide to reach a pH of ~7 followed by the addition of 94 mM HEPES free acid to maintain a stable pH during protein synthesis.
Under normal conditions, 2 mM of each of the 20 canonical amino acids is added to the PANOxSP preparation; however, in this work, glutamine, glutamate, asparagine, and aspartate were excluded. Glutamate, asparagine, and aspartate were added separately at 2 mM concentrations in each of the cell-free reactions, whereas glutamine was added only where expressly noted. DNA plasmids encoding green fluorescent protein (GFP) or β-galactosidase at a concentration of 12 nM were used in the cell-free protein synthesis reactions where expressly noted. For β-galactosidase-based reactions, the substrate chlorophenol red-β-D-galactopyranoside (CRPG) (Gold Bio, St. Louis, MO, USA), which changes color from yellow to purple in the presence of β-galactosidase, was used at a concentration of 0.6 mg/mL. In place of magnesium glutamate, reactions used magnesium sulfate where expressly noted. Additionally, cell extract and PANOxSP were used at 25% (v/v) amounts with the balance being water. For GFP reactions, 30 μL liquid volumes in 2 mL microcentrifuge tubes were used. Reactions were performed in triplicate and incubated at 37 °C, 280 RPM for either 90 or 150 min as expressly noted. Fluorescence was measured with the Biotek (Winooski, VT, USA) SynergyMx plate reader.
2.2. Paper-Based Colorimetric Cell-Free Protein Synthesis
The paper-based, lyophilized sensor format was prepared according to previously reported protocols [42,43]. First, 12 μL liquid solutions containing all cell-free reagents besides glutamine were pipetted onto chromatography paper (Thick Chromatography Paper Grade 238, VWR 28342-036, VWR International, Radnor, PA, USA) and then lyophilized for 24 h. Chromatography paper was chosen as the paper substrate based on favorable results from prior work comparing the compatibility of cellulose, nylon, chromatography paper, and other paper substrates with cell-free protein synthesis biosensors [56]. DNA encoding for β-galactosidase and its substrate CRPG were used for this sensor format. The assay was performed by rehydrating the papers with 12 μL aqueous solutions containing glutamine at the indicated concentrations. The papers were then covered with clear tape to prevent dehydration before being placed in an incubator at 37 °C. The assay was photographed using an iPhone XR smartphone every 3 min for 150 min followed by every 15 min until t = 225 min. Each condition was run in triplicate.
Photos were cropped using Adobe Lightroom and then aligned chronologically using Microsoft PowerPoint. No other modifications were made to the original images. Figure S1 is an example of an original image before any cropping was applied. Quantification of the color change was produced using ImageJ version 1.54, where the integrated density of the images at each timepoint was calculated, normalized, and plotted [57].
2.3. Use of Artificial Intelligence
Claude by Anthropic was used to generate text in the Introduction section of this publication.
3. Results
In this work, we engineered a novel cell-free protein synthesis (CFPS) system that utilized alternative salts to produce a paper-based, colorimetric glutamine biosensor that overcomes prior background limitations while maintaining a sufficient yield for the sensor to function (Figure 1). It was hypothesized that metabolic enzymes such as glutaminases converted the high concentrations of glutamate into glutamine, resulting in the significant unwanted protein synthesis of our previous sensor [43,44]. Here, we quantitatively examine three alternative cell extract compositions and four alternative PANOxSP compositions to determine an optimal composition for glutamine sensing.
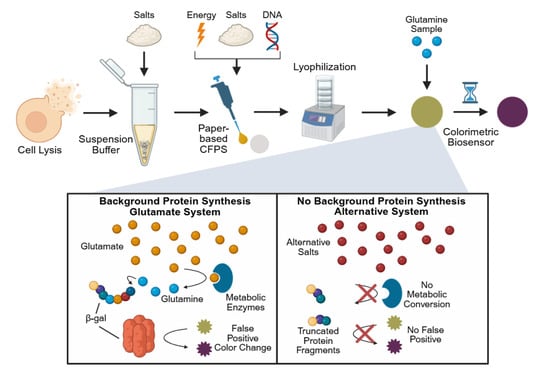
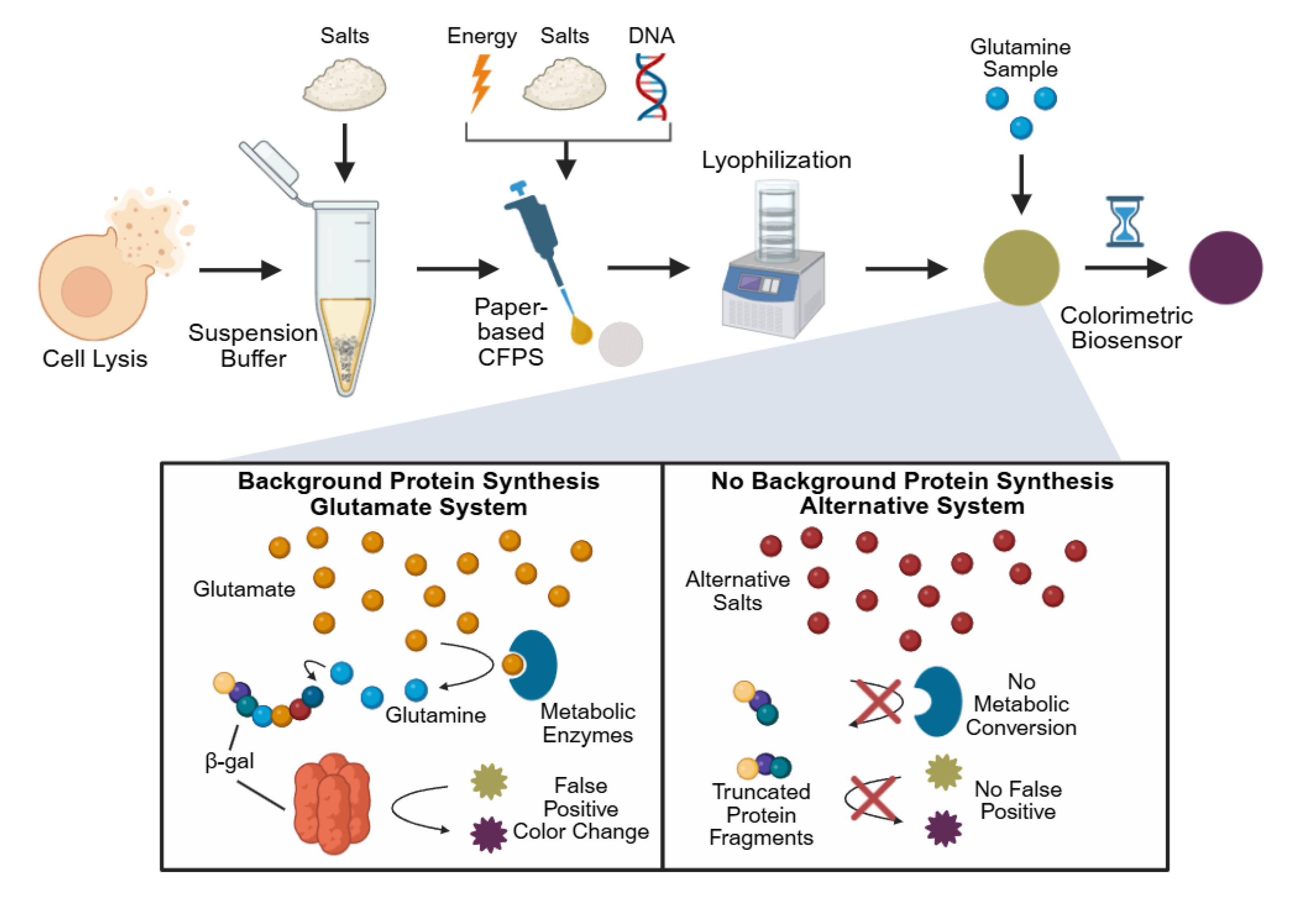
Figure 1.
Overview of paper-based, cell-free glutamine biosensor development and background reduction strategy. Cells are first lysed and then resuspended in Buffer S30, which typically contains glutamate-based salts. This buffer, along with additional components for protein synthesis, including additional glutamate-based salts, is pipetted onto chromatography paper where it is subsequently lyophilized. Rehydration with a glutamine sample is then performed to run the assay, where the protein β-galactosidase is produced, converting the substrate CRPG from yellow to purple, generating a visible response. Glutamate is hypothesized to contribute to the production of glutamine by metabolic enzymes, leading to a false positive color change even in glutamine-free samples. Replacing the glutamate-based salts with alternative salts is believed to overcome this limitation.
3.1. Alternative Cell Extract Buffers
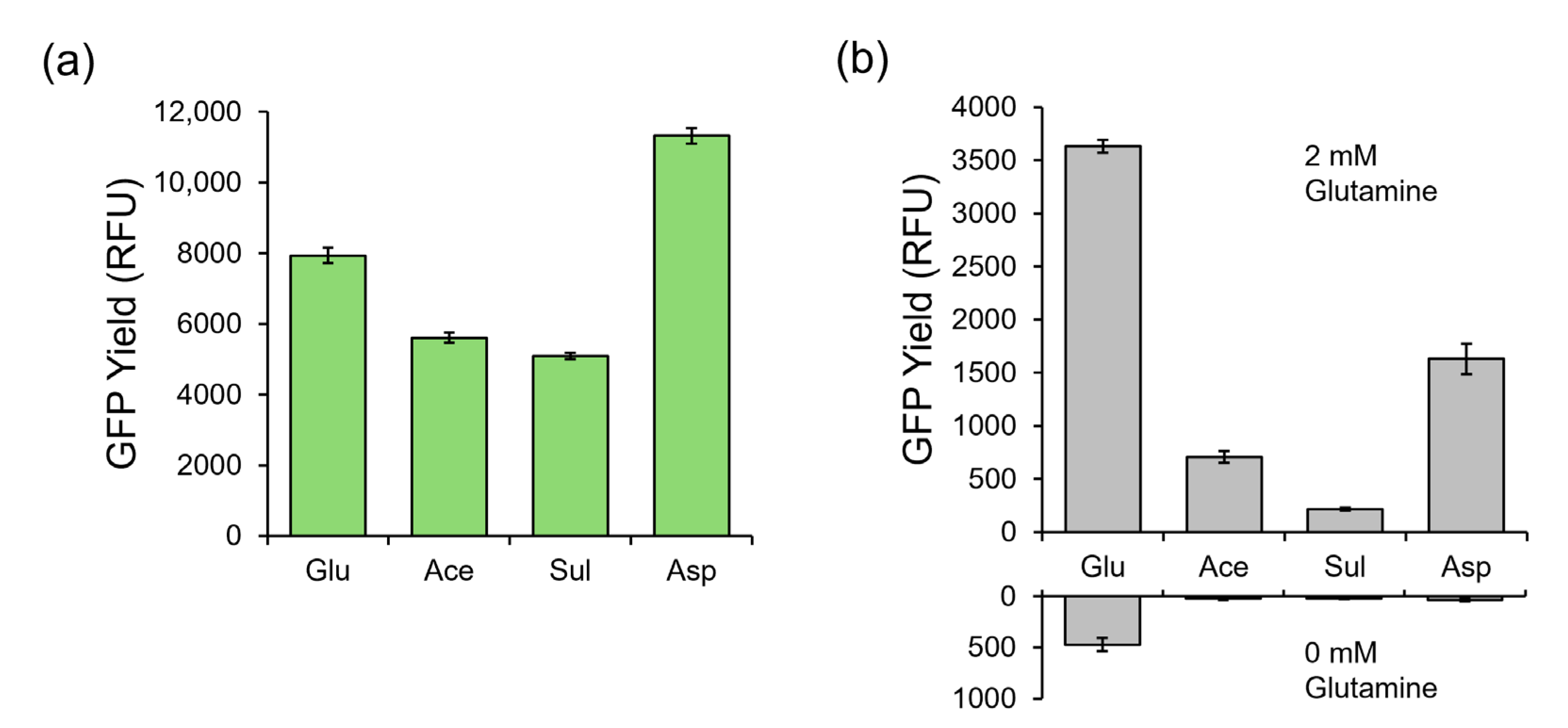
In E. coli-based CFPS reactions, glutamate, in the form of potassium glutamate, is the most abundant salt and is found in the cell extract buffer, Buffer S30. E. coli cells are washed and resuspended in this buffer after harvest. The cells are subsequently lysed in this buffer, which is then added as a primary reagent in the CFPS reaction. Buffer S30 contributes 15 mM of potassium glutamate to the CFPS reaction. For reference, canonical amino acids are provided at a concentration of 2 mM to the CFPS reaction, so potassium glutamate in Buffer S30 provides a 7.5× excess of glutamate relative to other amino acids present. It was thus hypothesized that excess potassium glutamate could drive metabolic conversion to glutamine with the active enzymes in the clarified lysate. To test this hypothesis, we composed three alternative Buffer S30 mixtures, replacing potassium glutamate with either potassium acetate, potassium sulfate, or potassium aspartate. E. coli extracts for CFPS were prepared from the same fermentation batch of E. coli but using different aliquots that were washed and suspended with either the traditional glutamate-based Buffer S30 or the three alternative Buffer S30 mixtures without glutamate. These extracts were used in CFPS reactions producing a model GFP using a glutamate-based PANOxSP small molecule supplementation recipe. The CFPS reactions were incubated for 150 min at 37 °C to compare the protein synthesis capabilities of each of these extracts against a glutamate-based control extract. Relative GFP yields are found in Figure 2a where the yields are compared against the glutamate-based extract. Glutamate-based yields were 409 μg/mL. Acetate-based and sulfate-based extracts performed similarly to each other with protein yields reaching approximately 71 and 64% of the glutamate control yields, respectively. It was observed that the aspartate-based extract had approximately 43% higher yields than the glutamate control. All extracts were treated with MSO and protease inhibitors, as those components were previously reported to improve the signal-to-noise ratio of the glutamine biosensor [42].
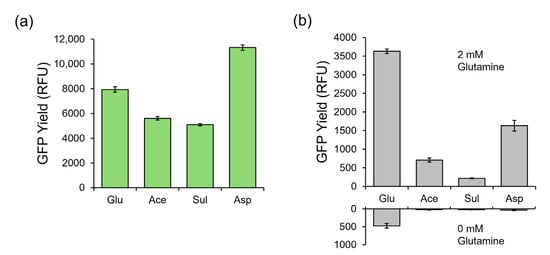
Figure 2.
Performance of alternative salt-based CFPS systems in comparison with the standard glutamate-based system. (a) The GFP protein yield in relative fluorescence units of glutamate-, acetate-, sulfate-, and aspartate-based cell extracts using a standard glutamate-based PANOxSP formula. (b) The GFP protein yield in relative fluorescence units of glutamate-, acetate-, sulfate-, and aspartate-based CFPS systems where each extract was paired with its respective PANOxSP formula under saturating glutamine (upper) and no glutamine (lower) conditions. The protein yield of acetate-, sulfate-, and aspartate-based systems under no glutamine conditions was within the range of background of the plate reader. Error bars represent one standard deviation of n = 3 replicates.
3.2. Alternative PANOxSP Compositions
The above replacement of potassium glutamate in the S30 Buffer reduced the concentration of glutamate in the CPFS reaction by 15 mM. However, a traditionally used small molecule mixture (PANOxSP which contains phosphoenylpyruvate as an energy source, amino acids, tRNA, buffer molecules, and nucleic acid stabilizing polymers) is also a major source of glutamate. PANOxSP contributes approximately 185 mM concentrations of glutamate to CFPS reactions in the forms of ammonium glutamate and potassium glutamate. To remove glutamate from PANOxSP, we composed four alternative PANOxSP mixtures, replacing both salts with their acetate, sulfate, citrate, or aspartate counterparts. CFPS reactions producing GFP using each PANOxSP mixture with their respective cell extract counterparts were incubated for 90 min at 37 °C in zero and saturated (2 mM) glutamine conditions to determine signal strength and background noise levels in comparison to a glutamate-based system (Figure 2b). Additionally, it should be noted that magnesium glutamate is traditionally used to provide magnesium for the system. In this work, magnesium sulfate was used as a replacement in each of the systems. As shown in Figure 2b, the glutamate-based system had the highest signal strength; however, it also had a significantly higher background signal than the rest of the systems. The other systems all had background levels that were within the range of background of the plate reader, suggesting that there was zero or near zero protein that was produced in these systems. Citrate and sulfate had the lowest signal strengths out of the systems. It is important to note that the citrate-based PANOxSP was paired with an acetate-based extract. The acetate system had the third highest signal that was approximately 20% of the glutamate-based signal. Aspartate had the highest non-glutamate-based signal that was approximately 45% of the glutamate-based signal.
3.3. Lyophilized, Paper-Based, Colorimetric Biosensor
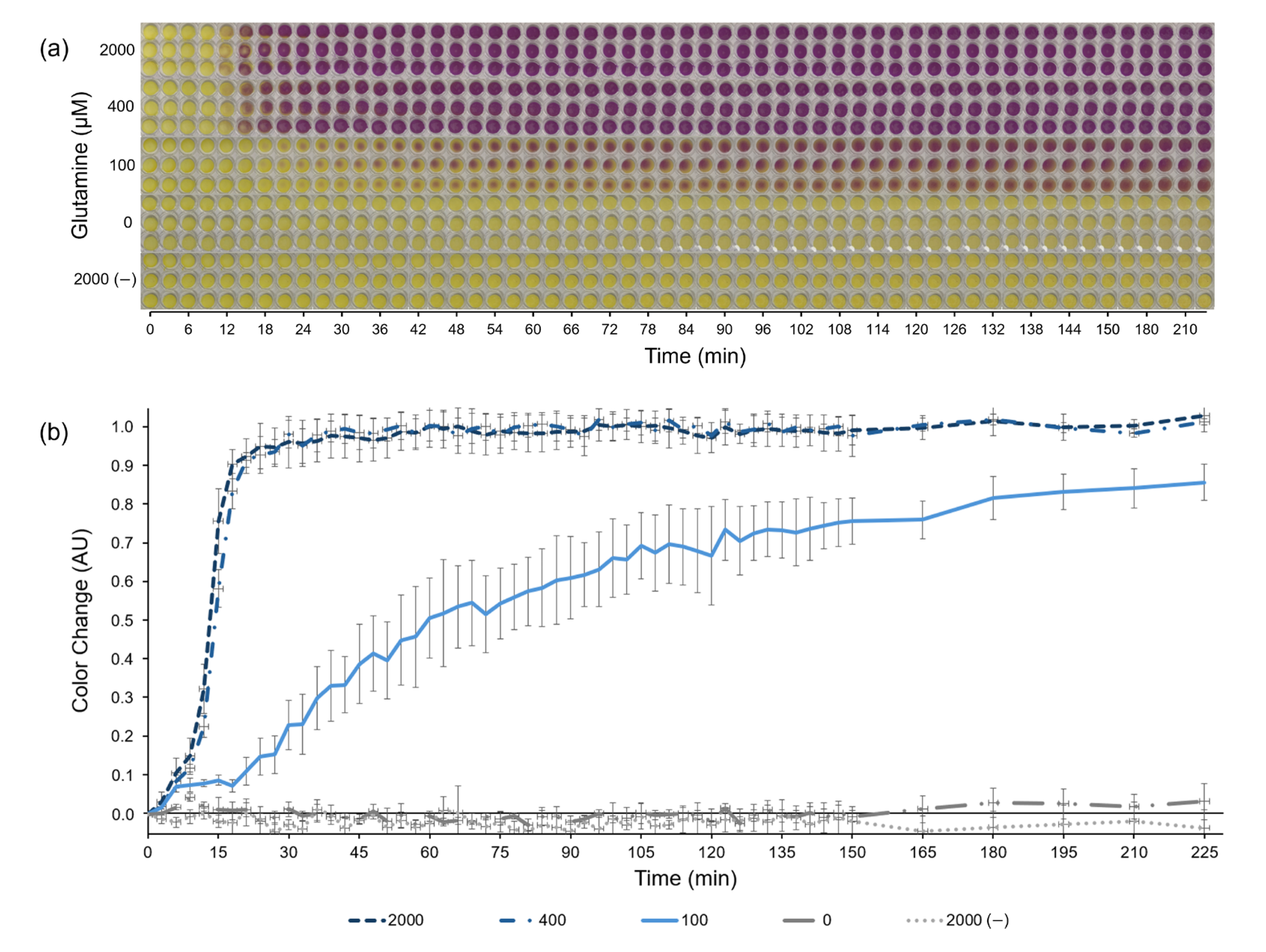
The novel aspartate-based CFPS system (combined aspartate-based S30 buffer and aspartate-based PANOxSP) described above was selected for use as a low-cost paper-based CFPS glutamine biosensor due to its combination of high signal strength in the presence of glutamine and undetectable signal without glutamine (signal was not statistically significant relative to background, p < 0.05). With this system, we developed a lyophilized, paper-based format for the sensor that utilizes the reporter protein β-galactosidase and its substrate CRPG, which changes color from yellow to purple in the presence of β-galactosidase (Figure 3). Glutamine concentrations of 0, 100, 400, and 2000 μM were tested by the sensor with a control lacking the DNA template encoding for β-galactosidase and saturating glutamine levels (2000 μM). The aspartate-based CFPS system led to a colorimetric sensor with significantly improved background reduction and sensitivity range compared with prior reported CFPS sensors. No color change was observed in the zero-glutamine sample after 225 min, indicating an achievement of the initial goal to eliminate background in the sensor. Additionally, there was a distinct difference in the color change between 0, 100, and 400 μM concentrations. Under saturating glutamine concentrations, a signal response was observed after around 15 min. This incubation period before a response is expected due to the time required to transcribe and translate the reporter protein β-galactosidase.
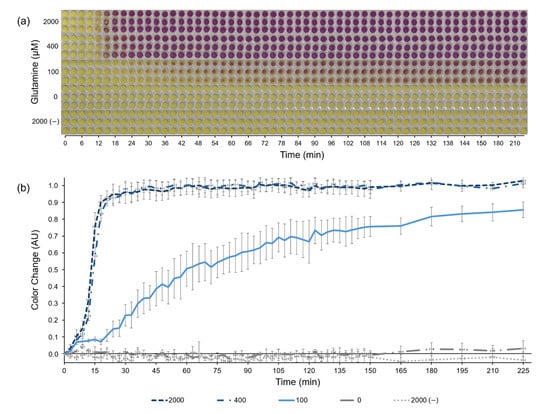
Figure 3.
Paper-based, colorimetric glutamine biosensor. (a) Chronological images of the paper-based, colorimetric glutamine biosensor using the fully aspartate-based cell-free protein synthesis system over 225 min. The reactions produce the protein β-galactosidase, which converts the substrate chlorophenol red-β-D-galactopyranoside (CRPG) from yellow to purple. All necessary protein synthesis reagents besides glutamine were pipetted onto the papers before lyophilization. Papers were rehydrated with aqueous samples containing either 0, 100, 400, or 2000 μM of glutamine, as indicated on the vertical axis. A negative control lacking the DNA encoding β-galactosidase is listed as 2000 (−). (b) The color change as quantified from (a) based off of ImageJ analysis. Error bars represent one standard deviation of n = 3 replicates.
4. Discussion
This work takes advantage of the open reaction environment of cell-free protein synthesis to explore and optimize the composition of the reaction, enabling the production of a paper-based glutamine biosensor. While other glutamine sensing technology suffers from slow response times, the need for specialized equipment and lab technicians, as well as high production costs, our cell-free biosensor provides an affordable and rapid way to determine glutamine concentrations in a sample. Additionally, to our knowledge, this represents the first time an aspartate salt-based CFPS system has been developed or successfully reported.
4.1. Alternative Cell Extract Buffers
Glutamate and acetate are the two most commonly used major anions in Buffer S30 for E. coli cell extracts [42,43,58,59,60,61,62]. For high yielding systems, glutamate is preferred due to its involvement in energy regeneration [63,64] and stabilizing effect on proteins [65,66]. However, the presence of glutamate in our cell-free sensor was hypothesized to be contributing to the presence of background glutamine levels, likely through glutaminase enzymes, which under normal conditions catalyze the conversion of glutamine to glutamate [67], but under the high glutamate concentrations of CFPS systems (100× higher concentrations of glutamate than glutamine), likely work in reverse. To validate that hypothesis, it became necessary to replace glutamate with a different anion while still maintaining a sufficiently high protein yield.
It was observed that the aspartate-based cell extract had the highest yields when paired with a glutamate-based PANOxSP mixture even in comparison to a glutamate-based extract. This could in part be because it is involved in the TCA cycle through its ability to be converted into oxaloacetate and activity as a nitrogen donor. Additionally, it also works to stabilize proteins. Acetate- and sulfate-based extracts performed the worst. Acetate, while also being involved in the TCA cycle [68], does not act as a nitrogen donor, and higher concentrations can inhibit protein production [69]. Sulfate is not directly involved in energy regeneration and therefore likely only contributes to the ionic strength of the system.
4.2. Alternative PANOxSP Compositions
PANOxSP was designed as an energy regeneration system for CFPS while also providing necessary salts and amino acids to the system [70]. For this reason, glutamate and acetate are the most commonly used major anions for this mixture [42,43,59,60,61,70]. In addition to composing PANOxSP mixtures using acetate, sulfate, and aspartate, a citrate-based PANOxSP mixture was also tested due to its involvement in the TCA cycle [71,72].
We observed that all non-glutamate-based systems resulted in background protein yields that were indistinguishable from the noise observed by the plate reader, whereas the glutamate-based system had significantly higher background yields, validating our hypothesis that metabolic enzymes, likely glutaminase, were contributing to the noise of the system. It is important to note that glutamine synthetase had previously been knocked out of the E. coli cell strain used in this work and is thus not a contributing metabolic enzyme. The selection criteria for determining the optimal sensor configuration focused on which system facilitated production of the highest concentration of visual reporter protein. Relative to the traditional glutamate-based systems, the aspartate, acetate, and sulfate-based systems achieved 40%, 15%, and 8%, of the synthesized reporter protein concentrations, respectively. While these reporter protein production yields of alternative PANOxSP systems are less than the yields of the traditional glutamate-based PANOxSP system, these findings provide support for the assumption that glutamate plays important roles in stabilization and energy regeneration, as the aspartate-based system had the next highest protein yields at approximately 40% of the glutamate-based system. Interestingly, the aspartate cell extract produced more reporter protein than the glutamate extract with a glutamate-based PANOxSP mixture; however, the fully glutamate-based system outperformed the fully aspartate-based system. It is important to note that the glutamate-based extract was tested with the aspartate-based PANOxSP, where the protein yield did not improve beyond the fully aspartate-based system. This demonstrates that aspartate does not provide the same energy regeneration capabilities as glutamate; however, it may result in a healthier and more active cell extract as seen by its improved yields. This also highlights the capabilities of CFPS to tease out nuances in cellular metabolism and chemical interactions. Although outside the scope of this work, the aspartate-based extract could be explored further in high-yielding systems to potentially improve synthesis yields.
Acetate had the third highest yielding system, which was likely due to its involvement in the TCA cycle, as mentioned previously. Sulfate had the next lowest system for reasons previously mentioned. The citrate-based system had the lowest yields out of all the systems tested. It is important to note that both the citrate- and aspartate-based PANOxSP mixtures had to be titrated with potassium hydroxide to a pH of 7, since they were both highly acidic under normal conditions. Additionally, HEPES free acid was added as a buffer to both these systems since its pKa value is ~7.5, meaning it is a good buffer in the range needed for protein synthesis. Without titration, both systems yielded zero protein. The citrate-based system likely had the lowest yields due in part to it being a strong magnesium chelator. Under normal magnesium concentrations, no protein was produced in this system. Protein synthesis was only observed after the addition of approximately five times the normal amount of magnesium. This highlights how the open reaction environment of cell-free systems allows for the facile addition of different components to recover and optimize protein yields.
4.3. Lyophilized, Paper-Based, Colorimetric Biosensor
While acetate, citrate, and aspartate-based CFPS glutamine sensors each resulted in undetectable levels of reporter protein production when glutamate was not added to the system, the aspartate-based system was chosen to develop the paper-based colorimetric format of this sensor due to its ability to produce more reporter protein when 2 mM glutamate was added—150% and 300% of the acetate and citrate systems, respectively (Figure 2). As previously stated under saturating conditions, an assay response was observed after approximately 15 min, which is similar to our previous sensor, indicating that signal strength was sufficient for sensor functionality. Additionally, whereas in our previous sensor, a background signal in the zero-glutamine sample was observed around 75 min after sensor rehydration with a near total color change after 144 min [42], no color change was observed in this work even after 225 min. Given the intended application of this technology in a rapid, at-home test, the scope of this work was limited to designing assays with response times under 3 h. It should be noted that the reaction chambers were sealed with tape to prevent evaporative dehydration, which would prematurely end colorimetric reactions. After the reaction, the chambers were visually inspected to confirm that chambers were still sealed and hydrated. The prior sensor’s response at glutamine concentrations of 20 μM concentrations mimics the color response of this sensor at glutamine concentrations of 100 μM, demonstrating a drastic improvement to the sensitivity range of the biosensor.
Normal physiological serum glutamine levels are between 400 and 900 μM, which is above the sensitivity range of this biosensor [73,74]. Prior work has shown that the dilution of samples that contain high glutamine concentrations to a range detectable by the sensor is a viable option for this technology [43]. Additionally, prior work has proven that human samples such as urine, blood, and saliva are compatible with cell-free biosensors with the addition of RNase inhibitors, paving the way for this sensor to move forward in the healthcare industry [56,75,76]. Urea, a common protein denaturant, has previously been reported to be compatible with cell-free biosensors under low to moderate concentrations [77].
5. Conclusions
Cancer treatments targeting glutamine metabolism have the potential to advance the fight against cancer; however, a lack of rapid, portable, low-cost glutamine sensors is preventing this type of treatment from moving forward. This work addresses that need by producing a rapid, portable, low-cost, glutamine biosensor using cell-free protein synthesis. Additionally, this work overcomes previous, significant background limitations by exploring alternative non-glutamate-based systems, leading to the discovery of an aspartate-based system that provides a sufficiently high signal response for sensing purposes and a potentially high yielding cell extract. Presented in this work is a paper-based, colorimetric biosensor with no visible background color change over 225 min and a fast response time.
This sensor has the potential to push forward cancer treatments involving glutamine metabolism. Additionally, this work highlights how the open environment of cell-free systems allows for the optimization of biosensors by altering the metabolic landscapes of the reactions. This approach could also be applied to other cell-free biosensors as we work to maximize signal-to-background ratios and eliminate background, which is essential for reliable usage of at-home colorimetric paper-based CFPS biosensors.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/chemosensors13060206/s1, Figure S1: Original Image from Figure 3a.
Author Contributions
Conceptualization, J.P.T., T.J.F. and B.C.B.; methodology, J.P.T., T.J.F. and T.P.G.; validation, J.P.T., T.J.F., T.P.G. and D.M.C.; formal analysis, J.P.T. and B.C.B.; investigation, J.P.T., T.J.F., T.P.G. and D.M.C.; resources, B.C.B.; data curation, J.P.T.; writing—original draft preparation, J.P.T. and T.P.G.; writing—review and editing, J.P.T., T.J.F., T.P.G., D.M.C. and B.C.B.; visualization, J.P.T. and B.C.B.; supervision, J.P.T., T.J.F. and B.C.B.; project administration, J.P.T., T.J.F. and B.C.B.; funding acquisition, J.P.T. and B.C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Simmons Center for Cancer Research at Brigham Young University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article and Supplementary Information.
Acknowledgments
The authors gratefully acknowledge the generous support from the Simmons Center for Cancer Research at Brigham Young University. The authors gratefully acknowledge Landon Bundy for help in analyzing the colorimetric sensor. During the preparation of this manuscript, the authors used Claude Sonnet 3.7 by Anthropic for the purposes of text generation of the introduction. The authors have reviewed and edited the output and take full responsibility for the content of this publication. We acknowledge the use of BioRender.com to create Figure 1.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| TCA | tricarboxylic acid |
| LC-MS | liquid chromatography-mass spectrometry |
| CFPS | cell-free protein synthesis |
| MSO | L-methionine sulfoximine |
| RCF | relative centrifugal force |
| GFP | green fluorescent protein |
| CRPG | chlorophenol red-β-D-galactopyranoside |
References
- Sikaris, K.A. Enhancing the Clinical Value of Medical Laboratory Testing. Clin. Biochem. Rev. 2017, 38, 107–114. [Google Scholar] [PubMed]
- Hawkes, N. Cancer survival data emphasise importance of early diagnosis. BMJ 2019, 364, l408. [Google Scholar] [CrossRef]
- Mani, K.; Deng, D.; Lin, C.; Wang, M.; Hsu, M.L.; Zaorsky, N.G. Causes of death among people living with metastatic cancer. Nat. Commun. 2024, 15, 1519. [Google Scholar] [CrossRef]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef]
- Peng, B.; Li, H.; Peng, X.X. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. [Google Scholar] [CrossRef]
- Li, T.; Copeland, C.; Le, A. Glutamine Metabolism in Cancer. Adv. Exp. Med. Biol. 2021, 1311, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Yang, L.F.; Venneti, S.; Nagrath, D. Glutaminolysis: A Hallmark of Cancer Metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Invest. 2013, 123, 3678–3684. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Burgess, D.J. Metabolism: Glutamine connections. Nat. Rev. Cancer 2013, 13, 293. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zeng, H.; Fan, J.; Wang, F.; Xu, C.; Li, Y.; Tu, J.; Nephew, K.P.; Long, X. Glutamine metabolism in breast cancer and possible therapeutic targets. Biochem. Pharmacol. 2023, 210, 115464. [Google Scholar] [CrossRef] [PubMed]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 2017, 3, 169–180. [Google Scholar] [CrossRef]
- De Vitto, H.; Perez-Valencia, J.; Radosevich, J.A. Glutamine at focus: Versatile roles in cancer. Tumour Biol. 2016, 37, 1541–1558. [Google Scholar] [CrossRef]
- Jin, J.; Byun, J.K.; Choi, Y.K.; Park, K.G. Targeting glutamine metabolism as a therapeutic strategy for cancer. Exp. Mol. Med. 2023, 55, 706–715. [Google Scholar] [CrossRef]
- Halama, A.; Suhre, K. Advancing Cancer Treatment by Targeting Glutamine Metabolism-A Roadmap. Cancers 2022, 14, 553. [Google Scholar] [CrossRef]
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef]
- Yang, W.H.; Qiu, Y.J.; Stamatatos, O.; Janowitz, T.; Lukey, M.J. Enhancing the Efficacy of Glutamine Metabolism Inhibitors in Cancer Therapy. Trends Cancer 2021, 7, 790–804. [Google Scholar] [CrossRef]
- Yang, L.; Moss, T.; Mangala, L.S.; Marini, J.; Zhao, H.; Wahlig, S.; Armaiz-Pena, G.; Jiang, D.; Achreja, A.; Win, J.; et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol. 2014, 10, 728. [Google Scholar] [CrossRef]
- Lukey, M.J.; Wilson, K.F.; Cerione, R.A. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med. Chem. 2013, 5, 1685–1700. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.W. The Essence of Modern HPLC: Advantages, Limitations, Fundamentals, and Opportunities. LCGC N. Am. 2013, 31, 472–479. [Google Scholar]
- Monge-Acuña, A.A.; Fornaguera-Trías, J. A high performance liquid chromatography method with electrochemical detection of gamma-aminobutyric acid, glutamate and glutamine in rat brain homogenates. J. Neurosci. Meth 2009, 183, 176–181. [Google Scholar] [CrossRef]
- Lim, Y.; Kim, J.Y.; Jung, Y.H.; Lee, J.H.; Baek, M.S.; Jung, J.H.; Kim, H.Y.; Lee, W.; Park, K.; Seo, M.H. Q-SHINE: A versatile sensor for glutamine measurement via ligand-induced dimerization. Sens. Actuators B Chem. 2023, 390, 133951. [Google Scholar] [CrossRef]
- Lam, H.; Kostov, Y.; Rao, G.; Tolosa, L. Low-cost optical lifetime assisted ratiometric glutamine sensor based on glutamine binding protein. Anal. Biochem. 2008, 383, 61–67. [Google Scholar] [CrossRef]
- Liu, B.J.; Zhao, Z.J.; Wang, P.C.; Aihemaiti, K.; Zhu, L.X.; Wei, Q.P.; Li, W.Z.; Yuan, X.; Wu, J.; Jiang, C.T.; et al. GlutaR: A High-Performance Fluorescent Protein-Based Sensor for Spatiotemporal Monitoring of Glutamine Dynamics In Vivo. Angew. Chem. Int. Edit 2025, 64, e202416608. [Google Scholar] [CrossRef]
- Albayrak, D.; Karakus, E. A novel glutamine biosensor based on zinc oxide nanorod and glutaminase enzyme from Hypocria jecorina. Artif. Cell Nanomed. B 2016, 44, 92–97. [Google Scholar] [CrossRef]
- Devi, P.; Kukkar, D.; Kaur, M.; Thakur, A.; Kim, K.H.; Kukkar, P.; Kaur, K.; Kaur, H. Conjugate of graphene quantum dots and glutaminase for the sensing of L-glutamine: Electrochemical vs. fluorescent sensing approaches. Inorg. Chem. Commun. 2021, 130, 108745. [Google Scholar] [CrossRef]
- Luck, L.A.; Moravan, M.J.; Garland, J.E.; Salopek-Sondi, B.; Roy, D. Chemisorptions of bacterial receptors for hydrophobic amino acids and sugars on gold for biosensor applications: A surface plasmon resonance study of genetically engineered proteins. Biosens. Bioelectron. 2003, 19, 249–259. [Google Scholar] [CrossRef]
- Chen, J.F.; Ding, L.Y.; Zhao, J.; Jiang, X.D.; Ma, F.; Li, H.J.; Zhang, Y.M. A L-glutamine binding protein modified MNM structured optical fiber biosensor based on surface plasmon resonance sensing for detection of L-glutamine metabolism in vitro embryo culture. Biosens. Bioelectron. 2023, 237, 115537. [Google Scholar] [CrossRef]
- Takamatsu, S.; Lee, J.; Asano, R.; Tsugawa, W.; Ikebukuro, K.; Sode, K. Continuous electrochemical monitoring of L-glutamine using redox-probe-modified L-glutamine-binding protein based on intermittent pulse amperometry. Sensors Actuators B Chem. 2021, 346, 130554. [Google Scholar] [CrossRef]
- Tanigawa, M.; Yamamoto, K.; Nagatoishi, S.; Nagata, K.; Noshiro, D.; Noda, N.N.; Tsumoto, K.; Maeda, T. A glutamine sensor that directly activates TORC1. Commun. Biol. 2021, 4, 1093. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Ma, D.; Shen, L.; Wu, K.; Diehnelt, C.W.; Green, A.A. Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment. Synth. Biol. 2018, 3, ysy018. [Google Scholar] [CrossRef]
- McNerney, M.P.; Zhang, Y.; Steppe, P.; Silverman, A.D.; Jewett, M.C.; Styczynski, M.P. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 2019, 5, eaax4473. [Google Scholar] [CrossRef]
- Soltani, M.; Davis, B.R.; Ford, H.; Nelson, J.A.D.; Bundy, B.C. Reengineering cell-free protein synthesis as a biosensor: Biosensing with transcription, translation, and protein-folding. Biochem. Eng. J. 2018, 138, 165–171. [Google Scholar] [CrossRef]
- Hunt, J.P.; Yang, S.O.; Wilding, K.M.; Bundy, B.C. The growing impact of lyophilized cell-free protein expression systems. Bioengineered 2017, 8, 325–330. [Google Scholar] [CrossRef]
- Soltani, M.; Hunt, J.P.; Bundy, B.C. Rapid RNase inhibitor production to enable low-cost, on-demand cell-free protein synthesis biosensor use in human body fluids. Biotechnol. Bioeng. 2021, 118, 3973–3983. [Google Scholar] [CrossRef] [PubMed]
- Soltani, M.; Bundy, B.C. Streamlining cell-free protein synthesis biosensors for use in human fluids: RNase inhibitor production during extract preparation. Biochem. Eng. J. 2022, 177, 108158. [Google Scholar] [CrossRef]
- commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015, 478, 107–120. [CrossRef]
- Lopreside, A.; Wan, X.Y.; Michelini, E.; Roda, A.; Wang, B.J. Comprehensive Profiling of Diverse Genetic Reporters with Application to Whole-Cell and Cell-Free Biosensors. Anal. Chem. 2019, 91, 15284–15292. [Google Scholar] [CrossRef]
- Free, T.J.; Talley, J.P.; Hyer, C.D.; Miller, C.J.; Griffitts, J.S.; Bundy, B.C. Engineering the Signal Resolution of a Paper-Based Cell-Free Glutamine Biosensor with Genetic Engineering, Metabolic Engineering, and Process Optimization. Sensors 2024, 24, 3073. [Google Scholar] [CrossRef]
- Free, T.J.; Tucker, R.W.; Simonson, K.M.; Smith, S.A.; Lindgren, C.M.; Pitt, W.G.; Bundy, B.C. Engineering At-Home Dilution and Filtration Methods to Enable Paper-Based Colorimetric Biosensing in Human Blood with Cell-Free Protein Synthesis. Biosensors 2023, 13, 104. [Google Scholar] [CrossRef]
- Hunt, J.P.; Barnett, R.J.; Robinson, H.; Soltani, M.; Nelson, J.A.D.; Bundy, B.C. Rapid sensing of clinically relevant glutamine concentrations in human serum with metabolically engineered -based cell-free protein synthesis. J. Biotechnol. 2021, 325, 389–394. [Google Scholar] [CrossRef]
- Laohakunakorn, N. Cell-Free Systems: A Proving Ground for Rational Biodesign. Front. Bioeng. Biotech. 2020, 8, 788. [Google Scholar] [CrossRef] [PubMed]
- Aw, R.; Polizzi, K.M. Biosensor-assisted engineering of a high-yield cell-free protein synthesis platform. Biotechnol. Bioeng. 2019, 116, 656–666. [Google Scholar] [CrossRef]
- Holecek, M. Roles of malate and aspartate in gluconeogenesis in various physiological and pathological states. Metabolism 2023, 145, 155614. [Google Scholar] [CrossRef]
- Lefin, N.; Miranda, J.; Munhoz Costa, I.; Pedroso Reynaldo, A.; Monteiro, G.; Zamorano, M.; Pessoa, A.; Farias, J.G. Optimized Amino Acid-Enhanced Medium for Efficient L-Asparaginase II Production in E. coli: From Shake Flask to Bioreactor. Fermentation 2025, 11, 239. [Google Scholar] [CrossRef]
- Schubert, C.; Zedler, S.; Strecker, A.; Unden, G. L-Aspartate as a high-quality nitrogen source in: Regulation of L-aspartase by the nitrogen regulatory system and interaction of L-aspartase with GlnB. Mol. Microbiol. 2021, 115, 526–538. [Google Scholar] [CrossRef]
- Reidman, S.; Cohen, A.; Kupiec, M.; Weisman, R. The cytosolic form of aspartate aminotransferase is required for full activation of TOR complex 1 in fission yeast. J. Biol. Chem. 2019, 294, 18244–18255. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, J.W. Cell-free protein synthesis: The state of the art. Biotechnol. Lett. 2013, 35, 143–152. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Infantino, V. Citrate—New functions for an old metabolite. Biol. Chem. 2014, 395, 387–399. [Google Scholar] [CrossRef]
- Zhao, H. Effect of ions and other compatible solutes on enzyme activity, and its implication for biocatalysis using ionic liquids. J. Mol. Catal. B Enzym. 2005, 37, 16–25. [Google Scholar] [CrossRef]
- Failmezger, J.; Rauter, M.; Nitschel, R.; Kraml, M.; Siemann-Herzberg, M. Cell-free protein synthesis from non-growing, stressed. Sci. Rep. 2017, 7, 16524. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Michael, H.T.; Bundy, B.C. Streamlined Extract Preparation for Escherichia Coli-Based Cell-Free Protein Synthesis by Sonication or Bead Vortex Mixing. Biotechniques 2012, 53, 163–174. [Google Scholar] [CrossRef]
- Hunt, J.P.; Zhao, E.L.; Free, T.J.; Soltani, M.; Warr, C.A.; Benedict, A.B.; Takahashi, M.K.; Griffitts, J.S.; Pitt, W.G.; Bundy, B.C. Towards detection of SARS-CoV-2 RNA in human saliva: A paper-based cell-free toehold switch biosensor with a visual bioluminescent output. New Biotechnol. 2022, 66, 53–60. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Katsura, K.; Seki, E.; Takemoto, C.; Shirouzu, M.; Terada, T.; Mukai, T.; Sakamoto, K.; Yokoyama, S. Cell-free protein synthesis using S30 extracts from Escherichia coli RFzero strains for efficient incorporation of non-natural amino acids into proteins. Int. J. Mol. Sci. 2019, 20, 492. [Google Scholar] [CrossRef] [PubMed]
- Des Soye, B.J.; Gerbasi, V.R.; Thomas, P.M.; Kelleher, N.L.; Jewett, M.C. A Highly Productive, One-Pot Cell-Free Protein Synthesis Platform Based on Genomically Recoded Escherichia coli. Cell Chem. Biol. 2019, 26, 1743–1754.e9. [Google Scholar] [CrossRef]
- Levin, R.; Löhr, F.; Karakoc, B.; Lichtenecker, R.; Dötsch, V.; Bernhard, F.E. coli “Stablelabel” S30 lysate for optimized cell-free NMR sample preparation. J. Biomol. NMR 2023, 77, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Krüger, A.; Mueller, A.P.; Rybnicky, G.A.; Engle, N.L.; Yang, Z.K.; Tschaplinski, T.J.; Simpson, S.D.; Köpke, M.; Jewett, M.C. Development of a clostridia-based cell-free system for prototyping genetic parts and metabolic pathways. Metab. Eng. 2020, 62, 95–105. [Google Scholar] [CrossRef]
- Wu, K.; Xu, G.; Tian, Y.; Li, G.; Yi, Z.; Tang, X. Synthesis and Evaluation of Aquatic Antimicrobial Peptides Derived from Marine Metagenomes Using a High-Throughput Screening Approach. Mar. Drugs 2025, 23, 178. [Google Scholar] [CrossRef]
- Jewett, M.C.; Calhoun, K.A.; Voloshin, A.; Wuu, J.J.; Swartz, J.R. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 2008, 4, 220. [Google Scholar] [CrossRef]
- Cai, Q.; Hanson, J.A.; Steiner, A.R.; Tran, C.; Masikat, M.R.; Chen, R.; Zawada, J.F.; Sato, A.K.; Hallam, T.J.; Yin, G. A simplified and robust protocol for immunoglobulin expression in Escherichia coli cell-free protein synthesis systems. Biotechnol. Prog. 2015, 31, 823–831. [Google Scholar] [CrossRef]
- Cheng, X.; Guinn, E.J.; Buechel, E.; Wong, R.; Sengupta, R.; Shkel, I.A.; Record, M.T. Basis of Protein Stabilization by K Glutamate: Unfavorable Interactions with Carbon, Oxygen Groups. Biophys. J. 2016, 111, 1854–1865. [Google Scholar] [CrossRef]
- Izzi, G.; Campanile, M.; Del Vecchio, P.; Graziano, G. On the Stabilizing Effect of Aspartate and Glutamate and Its Counteraction by Common Denaturants. Int. J. Mol. Sci. 2024, 25, 9360. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Aplin, C.; Nguyen, T.-T.T.; Milano, S.K.; Cerione, R.A. Filament formation drives catalysis by glutaminase enzymes important in cancer progression. Nat. Commun. 2024, 15, 1971. [Google Scholar] [CrossRef]
- Arnold, P.K.; Finley, L.W.S. Regulation and function of the mammalian tricarboxylic acid cycle. J. Biol. Chem. 2023, 299, 102838. [Google Scholar] [CrossRef]
- Leone, S.; Sannino, F.; Tutino, M.L.; Parrilli, E.; Picone, D. Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Fact. 2015, 14, 106. [Google Scholar] [CrossRef] [PubMed]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004, 86, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.C.; O’Neill, L.A.J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef]
- Icard, P.; Coquerel, A.; Wu, Z.; Gligorov, J.; Fuks, D.; Fournel, L.; Lincet, H.; Simula, L. Understanding the Central Role of Citrate in the Metabolism of Cancer Cells and Tumors: An Update. Int. J. Mol. Sci. 2021, 22, 6587. [Google Scholar] [CrossRef]
- Bergström, J.; Fürst, P.; Noree, L.; Vinnars, E. Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol. 1974, 36, 693–697. [Google Scholar] [CrossRef]
- Rodas, P.C.; Rooyackers, O.; Hebert, C.; Norberg, Å.; Wernerman, J. Glutamine and glutathione at ICU admission in relation to outcome. Clin. Sci. 2012, 122, 591–597. [Google Scholar] [CrossRef]
- Voyvodic, P.L.; Bonnet, J. Cell-free biosensors for biomedical applications. Curr. Opin. Biomed. Eng. 2020, 13, 9–15. [Google Scholar] [CrossRef]
- Wen, K.Y.; Cameron, L.; Chappell, J.; Jensen, K.; Bell, D.J.; Kelwick, R.; Kopniczky, M.; Davies, J.C.; Filloux, A.; Freemont, P.S. A Cell-Free Biosensor for Detecting Quorum Sensing Molecules in P. aeruginosa-Infected Respiratory Samples. Acs Synth. Biol. 2017, 6, 2293–2301. [Google Scholar] [CrossRef]
- Salehi, A.S.M.; Yang, S.O.; Earl, C.C.; Tang, M.J.S.; Hunt, J.P.; Smith, M.T.; Wood, D.W.; Bundy, B.C. Biosensing estrogenic endocrine disruptors in human blood and urine: A RAPID cell-free protein synthesis approach. Toxicol. Appl. Pharm. 2018, 345, 19–25. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).