Abstract
Excess salinity is a major environmental stress that limits growth and development of plants. Improving salt stress tolerance of plants is important in order to enhance land utilization and crop yield. Cold priming has been reported to trigger the protective processes in plants that increase their stress tolerance. Bermudagrass (Cynodon dactylon) is one of the most widely used turfgrass species around the world. However, the effect of cold priming on salt tolerance of bermudagrass is largely unknown. In the present study, wild bermudagrass was pre-treated with 4 °C for 6 h before 150 mM NaCl treatment for one week. The results showed that the cell membrane stability, ion homeostasis and photosynthesis process which are usually negatively affected by salt stress in bermudagrass were alleviated by short-term pre-cold treatment. Additionally, the gene expression profile also corresponded to the change of physiological indexes in bermudagrass. The results suggest that cold priming plays a positive role in improving salt stress tolerance of bermudagrass.
1. Introduction
Excess salinity is one of the main environmental stresses limiting growth and development, productivity and distribution of plants throughout the world. Salt stress causes changes in a series of cellular processes, photosynthesis process and gene expression in plants [1,2,3]. For instance, salt stress inhibits plant growth and damages the cell membrane [4,5]. Previous studies have revealed that electrolyte leakage (EL) increased dramatically in perennial ryegrass (Lolium perenne) [6]. The Na+ was accumulated, while K+ content decreased in Arabidopsis [7]. The change of ion accumulation will affect the metabolic process further [8]. Simultaneously, the chlorophyll content decreased in plant under salt stress [9]. In response, the plants have evolved several mechanisms such as: cell membrane system, antioxidant system and photosynthetic system, in order to protect themselves against salt stress [9,10,11]. Photosynthesis is one of the most essential physiological processes in plants that are sensitive to salt stress [12]. When a leaf is exposed to dark conditions, distinguishable changes of chlorophyll a fluorescence intensity can be detected, referred to the Kautsky effect [13]. Information about the different redox states of PSII and efficiencies of electron transfer in the photosynthetic electron transfer chain is contained in Chl a fluorescence transient [14]. For this reason, chlorophyll fluorescence analysis is widely used in plant physiology studies [15]. Generally, when Chl a fluorescence transient is plotted on a logarithmic time scale, four steps known as O-J-I-P could be clearly observed [13]. The four steps reflect different reactions in the photosynthetic pathway. Generally, the reaction centers (RCs) are totally open at O step, so the highest photochemistry yield and minimal Chl a fluorescence yield are measured. At the O-J step, the primary quinone electron acceptor QA was reduced to QA−; the J-I-P steps reflect the reduction of PQ pool; and the RCs are totally closed at P step, so the maximal fluorescence yield is measured. It was reported that the energy flux and electron transport were hindered in Brassica juncea by salt stress [16].
When plants are exposed to stress conditions, physiological and molecular changes occur in plant cells to counter the stresses. For example, the compatible solutes are accumulated as osmolytes, the membrane compositions are altered for enhanced fluidity, and the protective proteins are synthesized [17]. Cold acclimation is a natural physiological process in plants when treated with low but above freezing temperatures, enabling the plant to survive in freezing winter [18]. During this process, the cell wall is modified and the proteomic and metabolomic profilings of the plant change [19,20]. The freezing tolerance of alfalfa (Medicago sativa), eucalypt (Eucalyptus nitens) and tomato (Solanum lycopersicum) were improved after cold acclimation treatment [21,22,23]. It has been proven that CBF genes are involved in cold acclimation in Arabidopsis, and that the CBFs are also important for salt tolerance of plant [24]. Therefore, single short cold stress can improve cold tolerance of plants, and this process is called cold priming [25].
Advanced technologies such as high-throughput phenotyping and sequencing are contributed to identifying many stress response related genes in plant [26]. Transcriptomic analysis has showed that numerous genes are related to stress priming in plant [27]. For example, the genes of some crucial enzymes such as NADP-ME, SLP and Peroxidase were upregulated in peach after pre-cold treatment [28]. ECA4 is one of the Ca2+ ATPase genes, it is reported that the Ca2+ ATPase genes were remarkably upregulated by cold and salt stress in plant [29]. RAN is a small GTPase that plays important roles in nuclear transporting and assembling, it can affect plant response to cold and salt stress, and RAN1 gene is reported to be involved in cold resistance of rice (Oryza sativa) [30]. Moreover, the MHX gene which is a Mg2+/H+ exchanger gene [31] is regulated by cold and salt stress in plant according to our previous study of transcriptomic analysis [32]. In addition to the ion homeostasis-related genes, photosynthesis-related genes are also regulated by stress conditions in plants. The chloroplast encoded genes expression such as psbA, psbB and psbE are regulated by salt stress in plant [33]. Bermudagrass (Cynodon dactylon) is a warm-season turfgrass species. It is widely used in golf courses, gardens, sports fields and home lawns. However, the global change induced many environmental stresses, and it is very important for grass engineering to improve the stress tolerance [34]. The stress tolerance mechanisms of bermudagrass are largely unknown. Therefore, investigating the change in bermudagrass under stress conditions is necessary to understanding its stress tolerance mechanism. According to our previous study, numerous stress response genes were up-regulated after short-term cold acclimation [35]. Some of these genes are not only related to cold but also to other stress factors such as salt stress [36]. Hence, in the present study, the bermudagrass was treated with salt stress after pretreatment with low temperature to investigate whether short-term cold acclimation affects its salt stress tolerance.
2. Results
2.1. The Effect of Short-Term Pre-Cold Treatment on Cell Membrane Stability in Bermudagrass under Salt Stress
EL increase is usually induced by loss of membrane stability; to investigate the effect of cell membrane stability of pre-cold treatment under salt stress, relative EL was measured. The results showed that the relative EL under salt stress was 31.2% and 19.2% higher than that of the control in non-cold treatment and pre-cold treatment plants, respectively (Figure 1). There was no significant difference between non-cold treatment and pre-cold treatment plants under normal conditions. This suggests that short-term pre-cold treatment had a positive effect on bermudagrass tolerance to salt stress.
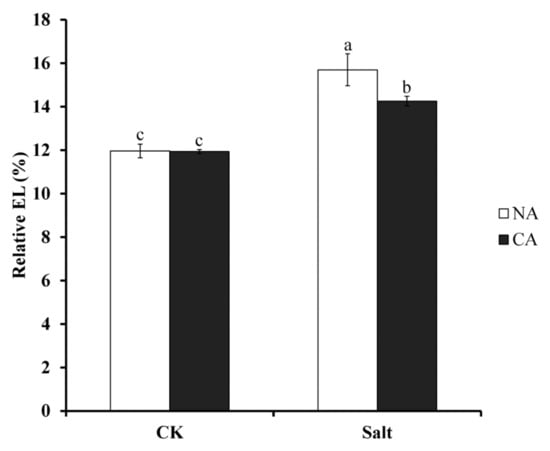
Figure 1.
Electrolyte leakage (EL) in bermudagrass under different treatments. Mean values and standard deviation (SD) were calculated from three independent experiments. Different letters indicated significant difference (p < 0.05) based on one-way analysis of variance (ANOVA), Duncan’s multiple range test. Bars show SD. CK = control; NA = non-cold treatment; CA = pre-cold treatment for 6 h.
2.2. The Effect of Short-Term Pre-Cold Treatment on Ion Homeostasis in Bermudagrass under Salt Stress
Ion homeostasis in plants is usually affected by salt stress. To investigate whether pre-cold treatment has an effect on ion homeostasis in bermudagrass under salt stress, K+ and Na+ concentrations were measured. The results showed that K+ content was remarkably decreased in the leaves of bermudagrass, 23.1% lower than that of control, when exposed to salt stress. However, after pre-cold treatment under salt stress, K+ content was 13.3% lower than that of control in bermudagrass (Figure 2A). Contrary to the result of K+ content, the concentration of Na+ increased significantly in bermudagrass leaf, 13.2% higher than that of control, under salt stress. After pretreatment with short-term low temperature, the Na+ content decreased to 11.2% higher than that of control under salt stress (Figure 2B). There was no significant difference between non-cold treatment and pre-cold treatment plants under control condition.
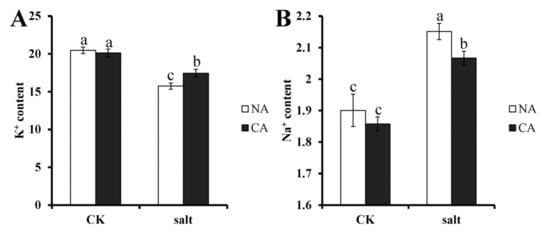
Figure 2.
Ion content in bermudagrass under different treatments. (A). K+ content; (B). Na+ content. Mean values and standard deviation (SD) were calculated from three independent experiments. Different letters indicated significant difference (p < 0.05) based on one-way analysis of variance (ANOVA), Duncan’s multiple range test. Bars show SD. CK = control; NA = non-cold treatment; CA = pre-cold treatment for 6 h.
2.3. The Effect of Short-Term Pre-Cold Treatment on Chlorophyll a/b (Chl a/b) in Bermudagrass under Salt Stress
As the pigment for photosynthesis, chlorophyll is very sensitive to stress conditions. To investigate the change of chlorophyll in bermudagrass after short-term pre-cold treatment under salt stress, Chl a/b ratio was measured. The results showed that this ratio was severely depressed to 46.9% lower than that of control when bermudagrass was exposed to salt stress. However, the ratio increased in the plants when pretreated by short-term low temperature, still under salt condition. Generally, although there was no statistical difference, the Chl a/b ratio was 14% higher in pre-cold treated plants compared with non-cold treated plants when exposed to salt stress (Figure 3).
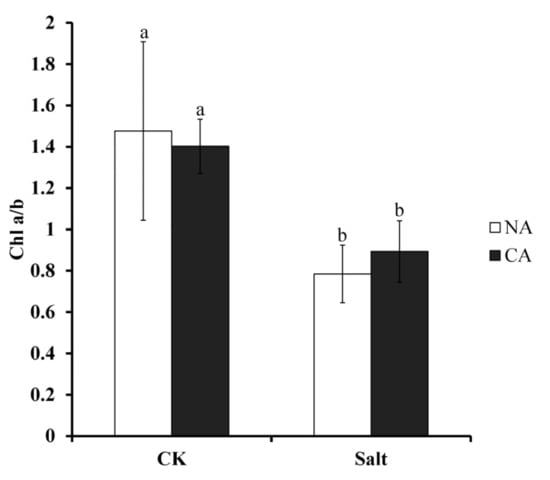
Figure 3.
Chlorophyll (Chl) a/b ratio of bermudagrass under different treatments. Mean values and standard deviation (SD) were calculated from three independent experiments. Different letters indicate significant difference (p < 0.05) based on one-way analysis of variance (ANOVA), Duncan’s multiple range test. Bars show SD. CK = control; NA = non-cold treatment; CA = pre-cold treatment for 6 h.
2.4. The Effect of Short-Term Pre-Cold Treatment on Chl a Fluorescence Transient (OJIP) in Bermudagrass under Salt Stress
To investigate the activity of photosystem II (PSII) in bermudagrass after short-term pre-cold treatment under salt stress, Chl a fluorescence transient was measured and JIP test was analyzed. The results showed that the OJIP curve declined remarkably as compared to control when the plants were exposed to salt stress. However, when pretreated with low temperature under salt stress, the OJIP curve was significantly higher than in the plants not pretreated with low temperature (Figure 4 and Figure S1). This result revealed that short-term pre-cold treatment could change the Chl a fluorescence transient in bermudagrass under salt stress.
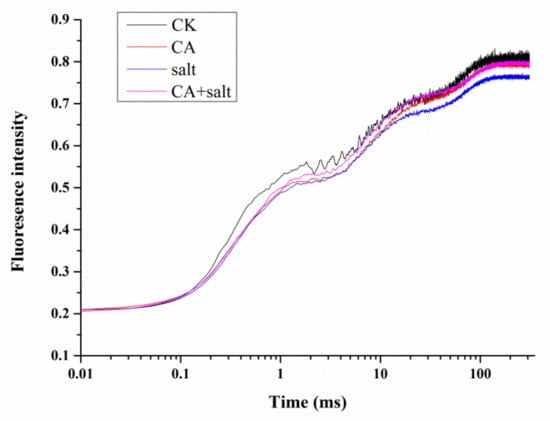
Figure 4.
Chlorophyll (Chl) a fluorescence transient in bermudagrass under different treatments. CK = control; CA = pre-cold treatment for 6 h; salt = 150 mM NaCl treatment for 1 week.
To further understand the change of PSII activity in bermudagrass after salt stress treatment, the JIP test was conducted. The result is shown in Figure 5 and Table S1. Parameters of specific energy fluxes; TRO/RC, ETO/RC and REO/RC decreased dramatically in PSII of bermudagrass when exposed to salt stress compared to control. TRO/RC was also decreased after pre-cold treatment. However, under salt stress conditions, TRO/RC and ETO/RC increased in the plants that were pretreated with cold. Contrary to the change of other parameters, DIO/RC increased significantly in bermudagrass under salt stress, while it dramatically declined in the plants pretreated with cold. This result implied that short-term pre-cold treatment could affect the energy fluxes in PSII of bermudagrass under salt stress. For the parameters of quantum yields and efficiencies, it was shown that φPo, ψEo, φEo, δRo, γRC and RC/ABS were found to be decreased in the plants that were treated with salt stress compared to control. These parameters increased in short-term pre-cold and salt-treated plants compared to only salt-treated plants. This result revealed that short-term pre-cold treatment had an effect on not only energy fluxes, but also quantum yields and efficiencies in PSII of bermudagrass under salt stress. The performance indexes (PI), including PIABS and PItoal, were important parameters, because they were composite indicators to reveal the responses of photosynthetic apparatus. The result showed that both of these two parameters decreased significantly in bermudagrass when exposed to salt stress. However, the performance index in plants that were treated with short-term pre-cold, increased remarkably compared to the plant treated with salt stress alone.
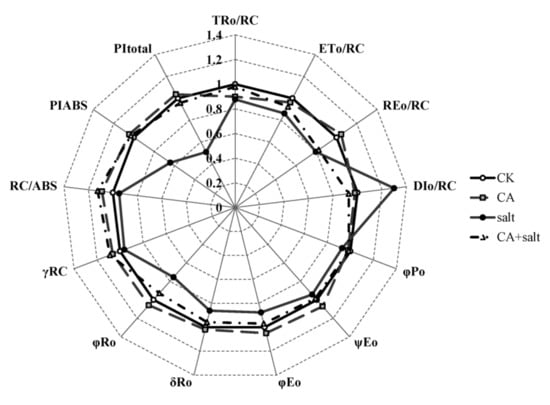
Figure 5.
Parameters of JIP test analysis in bermudagrass under different treatments. Mean values were calculated from three independent experiments. CK = control; NA = non-cold treatment; CA = pre-cold treatment for 6 h.
2.5. Gene Expression
To investigate the gene expression profile in bermudagrass under salt stress, expression of 7 candidate genes, including ECA4, RAN1, MHX1, psbA1, psbB1, psbP and psbY, were measured. The results showed that all of the genes were upregulated significantly in bermudagrass under different treatment compare to that in control. Generally, all of the genes were dramatically upregulated by short-term pre-cold treatment. However, ECA4 was downregulated by salt stress, and it was recovered after pre-cold treatment. For other genes, comparing between pre-cold treatment and salt stress, the ion transporting related genes such as RAN1 and MHX1 were more sensitive to salt stress, while the photosynthesis-related genes, including psbA1, psbB1, psbP and psbY, were more sensitive to pre-cold treatment (Figure 6). These results suggest that the genes expressions were regulated by pre-cold and salt treatment.
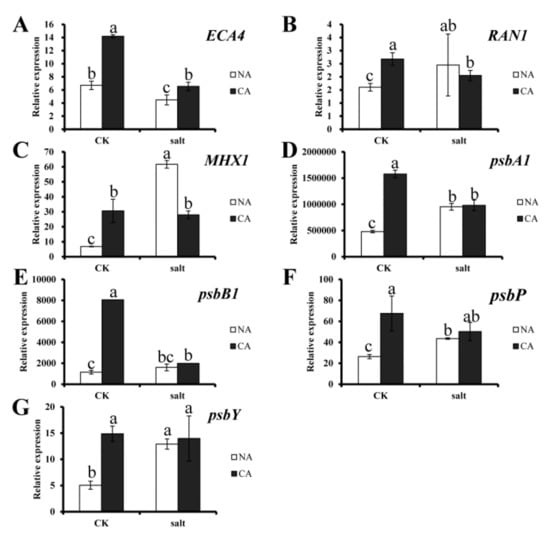
Figure 6.
Expression of candidate genes in bermudagrass under different treatments. (A) ECA4 gene expression; (B) RAN1 gene expression; (C) MHX1 gene expression; (D) psbA1 gene expression; (E) psbB1 gene expression; (F) psbP gene expression; (G) psbY gene expression. Quantitative real-time PCR was repeated three times. One-way analysis of variance (ANOVA), Duncan’s multiple range test was used to determine statistical differences. Bars show SD.
3. Discussion
Abiotic stresses such as salt, heat and drought can cause severe damages in plants. Salt stress can inhibit plant growth and decrease crop yield. When exposed to salt stress, as sessile organisms, plants have to respond and adapt to the adverse conditions. Investigating the mechanism of salt stress response and finding a way to improve the stress tolerance of plants is crucial. Recently, several studies have reported that not only chemical agents but also pre-acclimation can enhance abiotic stress tolerance of plant [37,38]. It was reported that low level salinity pre-treatment could increase salt tolerance of Zea mays [38], high blue light pre-treatment could improve UV stress tolerance of pepper (Capsicum annuum) [39], and cold acclimation pre-treatment could enhance cold tolerance of bermudagrass (Cynodon dactylon) [35]. However, the effect of cold priming on other stress responses in plants is largely unknown. The present study demonstrates that short-term pre-cold treatment could increase the salt tolerance of bermudagrass.
When exposed to salt stress, growth and development of plants is inhibited, and the metabolic processes in plant cells change [40]. Plenty of reactive oxygen species (ROS) accumulate in plant cells, which induces peroxidation of lipids, proteins and nucleic acids [41,42]. Hence, the cell membrane stability could be damaged leading to cell destruction [43]. The present study reveals that the relative EL of bermudagrass dramatically increases under salt stress. This implies that the cell membrane of bermudagrass is damaged by the adverse condition. Additionally, the growth rate of the plants under salt stress was significantly lower than that of control (data not shown). This result corresponds to that of sesame (Sesamum indicum) [44] and tomato (Lycopersicon esculentum) [5]. Many studies have shown that the activities of antioxidant enzymes would increase to scavenge the ROS induced by salt stress in plants [45,46,47].
As opposed to other stresses, salt stress can cause osmotic stress and ion toxicity in plants. The water loss and growth inhibition takes place first, a phenomenon termed ion-independent response. With continued salt stress, Na+ accumulates to toxic concentrations and premature senescence occurs then, referred to as ion-dependent response [48,49]. Ion homeostasis is easily disturbed by salt stress. Na+ and Cl− ions have been shown to significantly accumulate in cells of Arabidopsis and maize (Zea mays), while the concentration of K+ and Ca2+ decreased, which induced the Na+/K+ ratio increase [50,51]. The accumulation of Na+ and Cl- causes toxic effects on plant cell and induces cell death in the plant. The present study also shows that the Na+ content increased while K+ content decreased in bermudagrass under salt stress. This implies that ion homeostasis in cells of bermudagrass was affected by salt stress, which would have negative effects on the plant’s growth and nutrient element absorption [52].
Photosynthesis plays a crucial role in plants and it is sensitive to stress conditions [53]. As the infrastructure of photosynthesis, chlorophyll directly determines the photosynthetic efficiency of plants. Salt stress causes a decrease of chlorophyll synthesis and increase of chlorophyllase-mediated degradation [54]. It was reported that the accumulation of ROS in plant cells causes damage of Chl a, which induces a decrease in Chl a/b ratio in Hydrilla verticillata [55]. Consistent with this, the present study demonstrates that Chl a/b ratio decreased in bermudagrass after salt stress treatment. The damage to Chl a would result in malfunction of photosystem (PS), especially the PSII. The Chl a fluorescence transient and JIP test are usually applied to estimate change of PSII of plants under stress conditions [56]. The results in this study showed that the OJIP transient of bermudagrass was significantly changed by salt stress. This is in agreement with the results reported in Lemna gibba [57]. In addition, according to JIP test analysis, several parameters of energy flux and electron transport in PSII of bermudagrass were significantly changed. TRO/RC, ETO/RC and REO/RC reflect the flux of trapped energy, electron transport and the reduction of end electron acceptors at PSI acceptor side per RC, respectively [14]. The decrease of these parameters suggested that the RC activity was depressed by salt stress. The energy was dissipated as fluorescence which was reflected by increase of DIO/RC. In addition, the parameters of quantum yields and efficiencies were also changed in bermudagrass under salt stress. For instance, φPo, the maximum quantum yield of PSII, is usually used to evaluate the plant performance under stress conditions [58]. φPo dramatically declined in Lemna gibba under salt stress [57]. The present study also agrees with this result. The other parameters also detected a decrease in bermudagrass when it was exposed to salt stress. The parameters include: ψEo, which is the probability that the electrons move further than QA-; φEo, which represents the quantum yield for electron transport; δRo, which represents the probability of an electron transferred from carriers to acceptors at PSI side; γRC, which represents the probability of a Chl molecule functioning as RC; and RC/ABS, representing the probability of RCs reduced by QA per antenna Chl. This result is also consistent with a previous study in Ageratina adenophora [59]. The performance indexes, including PIABS and PItotal, are very sensitive to environmental stresses and are widely used to assess the photosynthetic functions of plant [60]. The performance indexes of bermudagrass were significantly decreased under salt stress, as shown in this study. A similar result was also reported in Arabidopsis under osmotic stress [61]. Above all, the results suggested that the salt stress negatively affected the electron transfer and energy transduction in PSII of bermudagrass.
When plants are exposed to a specific stress the tolerance to another stress could be improved, a phenomenon termed cross tolerance [62,63]. Previous studies have reported that short-term pre-cold treatment is involved in modifying the response to future stresses in plants, a process known as cold priming [64]. In plants, the cold-shock responses share the majority of pathways with heat-shock responses [65]. Many physiological traits such as antioxidant systems and osmolyte content are modulated [66]. The results of this study show that not only cell membrane stability, but also ion homeostasis and photosystem in bermudagrass, are all improved by cold priming under salt stress. In addition, gene expression is regulated by cold priming under stress condition [66]. Previous study showed that cold response related genes could be upregulated in bermudagrass when it was treated with cold stress [67]. The present study also shows that the candidate genes were remarkably upregulated by short-term pre-cold treatment. Among the genes, ECA4 is a Ca2+ transporter gene, RAN1 is a Cu2+ transporter gene, MHX1 is a sodium/calcium exchanger gene, and psbA1, psbB1, psbP and psbY are photosynthesis related genes in bermudagrass. Up-regulation of these genes implies that the ion transporting processes are triggered by cold priming and salt stress in bermudagrass. The photosynthesis related genes were significantly upregulated by short-term pre-cold treatment. This suggests that cold priming maybe related to alleviating the salt stress damage in bermudagrass through regulating the photosynthesis process.
4. Conclusions
Salt stress is an important environmental stress that negatively affects physiological processes in bermudagrass. The salt stress tolerance is enhanced by synthetic compounds [68]. The results of this study showed the salt-induced damage in bermudagrass can be alleviated by short-term pre-cold treatment.
It was shown that the relative EL was significantly decreased after short-term pre-cold treatment under salt stress. It is reasonable that the relative EL will increase under stress conditions because of the injury of cell membrane. The decrease of relative EL suggested the improvement of cell membrane stability after pre-cold treatment.
The results of ion homeostasis in bermudagrass revealed that Na+ concentration decreased under salt stress after cold priming treatment. The K+ concentration increased accordingly. This implied that the ion homeostasis in bermudagrass is improved by cold priming when the plants are exposed to salt stress.
The salt stress response of photosystem in bermudagrass is improved by cold priming. Under salt stress, the ratio of chlorophyll a and b slightly increased after short-term cold treatment. Additionally, Chl a fluorescence (OJIP) transient and JIP test analysis showed that the parameters of energy flux and electron transport in PSII of bermudagrass was improved by cold priming under salt stress. This implied that the salt tolerance of photosynthesis process in bermudagrass was enhanced by cold priming.
Furthermore, the expression of ion homeostasis and photosystem related genes were upregulated by cold priming. This result corresponded to the change of the physiological indexes.
5. Materials and Methods
5.1. Plant Materials and Growth Conditions
The bermudagrass (Cynodon dactylon) used in the present study was collected from wild field of Guangxi Province in China. It was sensitive to salt stress as demonstrated by the pre-experiment. To cultivate the seedlings, 30 stolons were planted in each pot (10 cm tall and 8 cm in diameter). All the pots were filled with brown coal soil and sand (1:1). The pots were kept in a greenhouse with temperature of 31/23 °C (day/night), the photoperiod was 12 h, the photosynthetically active radiation (PAR) on the canopy was 240 μmol·m−2·s−1 on average. The plants were cultivated in the pots for three weeks, which allowed the grass to be established. During the period, the plants were fertilized adequately with full-strength Hoagland nutrient solution every day.
5.2. Treatments
There are four different treatments—only salt, only cold, cold and salt treatment, and non-cold and salt treatment—in this study. Three repeats were set in each treatment. The established bermudagrass was pretreated with 4 °C for 6 h in a growth chamber (HP300GS-C; Wuhan Ruihua Instrument and Equipment Co., Wuhan, China). The PAR was 240 μmol·m−2·s−1, and the relative humidity was 60%. To avoid the thermal effect of the light, cold light illuminators were applied in this study. After cold acclimation, the bermudagrass was transferred to 25 °C, 14 h/10 h (day/night) photoperiod conditions and treated with 150 mM NaCl for 1 week. For salt treatment, 500 mL 150 mM NaCl solution was irrigated into the soil at the beginning of treatment. To avoid the solution draining out, a tray was placed under each pot. During the treatment, fully extended leaves after 12 h treatment were collected for RNA extraction. Other samples were collected after 7 days of salt stress treatment.
5.3. Measurements
5.3.1. Electrolyte Leakage
Electrolyte leakage (EL) was measured following Hu et al. (2012) [69]: 0.1 g of fully extended leaves was sampled and washed with deionized water three times. The leaves were cut into 0.5 cm fragments, and then transferred in 50 mL centrifuge tube which was contained 15 mL deionized water. After the fragments were shaken for 24 h at room temperature, the initial conductivity (EL1) was measured with a conductivity meter. The solution was then autoclaved at 121 °C for 10 min, the second conductivity (EL2) was detected after the solution was cooled to room temperature. The ratio of EL1/EL2 × 100 was the relative EL.
5.3.2. Ion Content
Total of 0.1 g leaves were dried in the oven and milled into fine powder. The powder was then solubilized in 5 mL nitric acid and 1 mL H2O2 with microwave digestion (Ethos One, Italy). The procedure was set as follows: 130 °C, 12 min; 160 °C, 38 min. After that, the ion contents, including K+ and Na+, were detected with inductively coupled plasma mass spectrometer (ICP-MS, X Series 2, Germany).
5.3.3. Chlorophyll Content
The chlorophyll content measurement was according to Hu et al. (2012) [6]. To extract the chlorophyll, a total of 0.1 g leaves was submerged into 10 mL dimethylsulfoxide (DMSO), which was contained in 15 mL centrifuge tubes. The tubes were incubated in the dark for 48 h until the leaves were totally faded. The absorbance at 645 nm and 663 nm of the extract solution was detected with the spectrophotometer (UV-2600, UNICO, Shanghai, China). The absorbance of DMSO was used as a blank. The chlorophyll content was calculated using the following formula:
OD645 and OD663 were the absorbance of the extract solution at 645 nm and 663 nm, respectively. Chl a/b is the ratio of Chl-a/Chl-b.
Chl-a content (mg·L−1) = 12.7 × OD663 − 2.59 × OD645,
Chl-b content (mg·L−1) = 22.9 × OD645 − 4.67 × OD663,
5.3.4. Chlorophyll (Chl) a Fluorescence Transient
Chlorophyll a fluorescence was measured after 7 days of salt stress treatment with a pulse-amplitude modulation fluorometer (PAM-2500, Heinz Walz GmbH) according to Chen et al. (2013) [70] with some modification. The plants were exposed to the dark for 30 min to ensure sufficient closure of PSII reaction centers. The third topmost expanded functional leaf was selected to be measured. The intensity of the saturating light pulse was 3000 μmol photons m−2 s−1. The Chl a fluorescence transient was then detected and digitized between 10 μs and 310 ms under room temperature.
5.3.5. The JIP-Test
The JIP-test was based on the theory of energy fluxes in bio-membranes and employed to the multi-parametric analysis of Chl a fluorescence transient [70]. For JIP-test, FO (fluorescence intensity at 20 μs); FM (the maximal fluorescence intensity); FJ (fluorescence intensity at 1.96 ms); FI (fluorescence intensity at 23.4 ms) were extracted from the original measurements. In addition, F300μs (fluorescence intensity at 300 μs) was also extracted for calculation of MO (the initial slope of the fluorescence kinetics). According to Srtasser et al. (2010) [14], the parameters for JIP-test include: (i) the specific energy fluxes per reaction center (RC), such as absorption (ABS/RC), trapping (TRO/RC), electron transport (ETO/RC), electron flux reducing end electron acceptors at the PSI acceptor side (REO/RC) and dissipation at the antenna chlorophyll level (DIO/RC); (ii) the quantum yields and efficiencies, such as maximum quantum yield for primary photochemistry (φPO), the efficiency that an electron moved further than QA− (ψEO), the efficiency that an electron was transferred from intersystem carriers to reduce end acceptors (δRO), the quantum yield for end electron acceptors reduction (φRO), probability that a PSII Chl functions as RC (γRC); (iii) performance indexes based on energy conservation such as PIABS and PItotal.
5.3.6. RNA Extraction and Gene Expression Analysis
For RNA extraction, 0.1 g bermudagrass leaves were sampled after 12 h treatment, and Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract the total RNA [67]. The RNA was then reversed to cDNA by M-MLV reverse transcriptase (Promega, Madison, WI) and oligo (dT) primer according to the operation manual. The gene expression was analyzed by two-step RT-PCR procedure with StepOnePlus Real-Time PCR Systems (Applied Biosystems, USA). SYBR Green (Toyobo, Osaka, Japan) was used as fluorescent dye and Real-time PCR Master Mix (Toyobo) was used for the reaction according to the operation manual. To quantify the gene expression, the Actin gene of bermudagrass was used as internal control, and the comparative Ct method was used to calculate the relative quantity of the target gene [71]. The primers of the specific genes are listed in Table 1.

Table 1.
Primers of candidate genes in bermudagrass.
5.4. Statistical Analysis
The statistical analysis used in this study was based on one-way analysis of variance (ANOVA), Duncan’s multiple range test. The statistically significant was considered when p < 0.05. Data were showed as mean ± standard deviation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/9/347/s1, Figure S1: O, J, I, P steps of bermudagrass under different treatments, Table S1: parameters of JIP test analysis in bermudagrass under different treatments.
Author Contributions
L.C. and D.L. designed the experiment; J.F. wrote the manuscript; M.A. revised the manuscript; W.Z. measured the physiological change of the plant; J.X. did the gene expression analysis.
Funding
This research was financially supported by National Natural Science Foundation of China (Grant No. 31702165, 31672482), General Financial Grant from the China Postdoctoral Science Foundation (Grant No. 2017M621838) and The Earmarked Fund for Jiangsu Agricultural Industry Technology System (Grant No. JATS(2018)314).
Acknowledgments
We thank Lipeng Zhou for helping us to collect the documents. We also thank the Public Laboratory Platform, Wuhan Botanical Garden, CAS, for providing techniques of ion content analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Xiong, L.; Ishitani, M.; Zhu, J.K. Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol. 1999, 119, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.G.; Toyota, M.; Kim, S.H.; Hilleary, R.; Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 2014, 111, 6497–6502. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Kanwar, P.; Singh, A.; Steinhorst, L.; Pandey, A.; Yadav, A.K.; Tokas, I.; Sanyal, S.K.; Kim, B.G.; Lee, S.C.; et al. Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 2015, 169, 780–792. [Google Scholar] [CrossRef] [PubMed]
- Demiral, T.; Türkan, İ. Comparative lipid peroxidation, antioxidant defense systems and proline content in roots of two rice cultivars differing in salt tolerance. Environ. Exp. Bot. 2005, 53, 247–257. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Ashraf, M.; Altunlu, H.; Yokas, I.; Yagmur, B. The effects of calcium sulphate on growth, membrane stability and nutrient uptake of tomato plants grown under salt stress. Environ. Exp. Bot. 2007, 59, 173–178. [Google Scholar] [CrossRef]
- Hu, L.; Hu, T.; Zhang, X.; Pang, H.; Fu, J. Exogenous glycine betaine ameliorates the adverse effect of salt stress on perennial ryegrass. J. Am. Soc. Hortic. Sci. 2012, 137, 38–46. [Google Scholar] [CrossRef]
- Kim, B.G.; Waadt, R.; Cheong, Y.H.; Pandey, G.K.; Dominguez-Solis, J.R.; Schültke, S.; Lee, S.C.; Kudla, J.; Luan, S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007, 52, 473–484. [Google Scholar] [CrossRef]
- Richter, J.A.; Behr, J.H.; Erban, A.; Kopka, J.; Zörb, C. Ion-dependent metabolic responses of Vicia faba L. to salt stress. Plant Cell Environ. 2019, 42, 295–309. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Sairam, R.K.; Srivastava, G.C. Changes in antioxidant activity in sub-cellular fractions of tolerant and susceptible wheat genotypes in response to long term salt stress. Plant Sci. 2002, 162, 897–904. [Google Scholar] [CrossRef]
- Regni, L.; Del Pino, A.M.; Mousavi, S.; Palmerini, C.A.; Baldoni, L.; Mariotti, R.; Mairech, H.; Gardi, T.; D’Amato, R.; Proietti, P. Behavior of four olive cultivars during salt stress. Front. Plant Sci. 2019, 10, 867. [Google Scholar] [CrossRef] [PubMed]
- Porcel, R.; Redondo-Gómez, S.; Mateos-Naranjo, E.; Aroca, R.; Garcia, R.; Ruiz-Lozano, J.M. Arbuscular mycorrhizal symbiosis ameliorates the optimum quantum yield of photosystem II and reduces non-photochemical quenching in rice plants subjected to salt stress. J. Plant Physiol. 2015, 185, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Stirbet, A. On the relation between the Kautsky effect (chlorophyll a fluorescence induction) and Photosystem II: Basics and applications of the OJIP fluorescence transient. J. Photochem. Photobiol. B Biol. 2011, 104, 236–257. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.J.; Tsimilli-Michael, M.; Qiang, S.; Goltsev, V. Simultaneous in vivo recording of prompt and delayed fluorescence and 820-nm reflection changes during drying and after rehydration of the resurrection plant Haberlea rhodopensis. BBA-Bioenerg. 2010, 1797, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.A.; Kumar, D.; Rajwanshi, R.; Strasser, R.J.; Tsimilli-Michael, M.; Sarin, N.B. Overexpression of γ-tocopherol methyl transferase gene in transgenic Brassica juncea plants alleviates abiotic stress: Physiological and chlorophyll a fluorescence measurements. BBA-Bioenerg. 2010, 1797, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.; Clement, C.; Barka, E.A. Physiological and molecular changes in plants grown at low temperatures. Planta 2012, 235, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Costa-Broseta, Á.; Perea-Resa, C.; Castillo, M.C.; Ruíz, M.F.; Salinas, J.; León, J. Nitric oxide deficiency decreases C-repeat bingding factor-dependent and -independent induction of cold acclimation. J. Exp. Bot. 2019, 70, 3283–3296. [Google Scholar] [CrossRef] [PubMed]
- Chai, F.; Liu, W.; Xiang, Y.; Meng, X.; Sun, X.; Cheng, C.; Liu, G.; Duan, L.; Xin, H.; Li, S. Comparative metabolic profiling of Vitis amurensis and Vitis vinifera during cold acclimation. Hortic. Res. 2019, 6, 8. [Google Scholar] [CrossRef]
- Takahashi, D.; Gorka, M.; Erban, A.; Graf, A.; Kopka, J.; Zuther, E.; Hincha, D.K. Both cold and sub-zero acclimation induce cell wall modification and changes in the extracellular proteome in Arabidopsis thaliana. Sci. Rep. 2019, 9, 2289. [Google Scholar] [CrossRef]
- Bertrand, A.; Bipfubusa, M.; Claessens, A.; Rocher, S.; Castonguay, Y. Effect of photoperiod prior to cold acclimation on freezing tolerance and carbohydrate metabolism in alfalfa (Medicago sativa L.). Plant Sci. 2017, 264, 122–128. [Google Scholar] [CrossRef]
- Gaete-Loyola, J.; Lagos, C.; Beltrán, M.F.; Valenzuela, S.; Emhart, V.; Fernández, M. Transcriptome profiling of Eucalyptus nitens reveals deeper insight into the molecular mechanism of cold acclimation and deacclimation process. Tree Genet. Genomes 2017, 13, 37. [Google Scholar] [CrossRef]
- Lv, X.; Li, H.; Chen, X.; Xiang, X.; Guo, Z.; Yu, J.; Zhou, Y. The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. J. Exp. Bot. 2018, 69, 4127–4139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [PubMed]
- Baier, M.; Bittner, A.; Prescher, A.; van Buer, J. Preparing plants for improved cold tolerance by priming. Plant Cell Environ. 2019, 42, 782–800. [Google Scholar] [CrossRef] [PubMed]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel—Dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Tanou, G.; Minas, I.S.; Scossa, F.; Belghazi, M.; Xanthopoulou, A.; Ganopoulos, I.; Madesis, P.; Fernie, A.; Molassiotis, A. Exploring priming responses involved in peach fruit acclimation to cold stress. Sci. Rep. 2017, 7, 11358. [Google Scholar] [CrossRef]
- Huda, K.M.K.; Yadav, S.; Banu, M.S.A.; Trivedi, D.K.; Tuteja, N. Genome-wide analysis of plant-type II Ca2+ATPases gene family from rice and Arabidopsis: Potential role in abiotic stresses. Plant Physiol. Biochem. 2013, 65, 32–47. [Google Scholar] [CrossRef]
- Xu, P.; Cai, W. RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J. Exp. Bot. 2014, 65, 3277–3287. [Google Scholar] [CrossRef]
- Pittman, J.K.; Hirschi, K.D. Phylogenetic analysis and protein structure modelling identifies distinct Ca 2+/Cation antiporters and conservation of gene family structure within Arabidopsis and rice species. Rice 2016, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liu, A.; Gitau, M.M.; Huang, X.; Chen, L.; Fu, J. Insights into the MicroRNA-regulated response of bermudagrass to cold and salt stress. Environ. Exp. Bot. 2018, 145, 64–74. [Google Scholar] [CrossRef]
- Chattopadhayay, M.K.; Tiwari, B.S.; Chattopadhyay, G.; Bose, A.; Sengupta, D.N.; Ghosh, B. Protective role of exogenous polyamines on salinity-stressed rice (Oryza sativa) plants. Physiol. Plant. 2002, 116, 192–199. [Google Scholar] [CrossRef]
- Shi, H.; Jiang, C.; Ye, T.; Yan, D.X.; Reiter, R.J.; Zhang, H.; Liu, R.; Chan, Z. Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 2015, 66, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fan, J.; Hu, L.; Hu, Z.; Xie, Y.; Zhang, Y.; Lou, Y.; Nevo, E.; Fu, J. A transcriptomic analysis of bermudagrass (Cynodon dactylon) provides novel insights into the basis of low temperature tolerance. BMC Plant Biol. 2015, 15, 216. [Google Scholar] [CrossRef]
- Liu, A.; Hu, Z.; Bi, A.; Fan, J.; Gitau, M.M.; Amombo, E.; Chen, L.; Fu, J. Photosynthesis, antioxidant system and gene expression of bermudagrass in response to low temperature and salt stress. Ecotoxicology 2016, 25, 1445–1457. [Google Scholar] [CrossRef]
- Savvides, A.; Ali, S.; Tester, M.; Fotopoulos, V. Chemical priming of plants against multiple abiotic stresses: Mission possible? Trends Plant Sci. 2016, 21, 329–340. [Google Scholar] [CrossRef]
- Pandolfi, C.; Azzarello, E.; Mancuso, S.; Shabala, S. Acclimation improves salt stress tolerance in Zea mays plants. J. Plant Physiol. 2016, 201, 1–8. [Google Scholar] [CrossRef]
- Hoffmann, A.M.; Noga, G.; Hunsche, M. High blue light improves acclimation and photosynthetic recovery of pepper plants exposed to UV stress. Environ. Exp. Bot. 2015, 109, 254–263. [Google Scholar] [CrossRef]
- Tang, X.; Mu, X.; Shao, H.; Wang, H.; Brestic, M. Global plant-responding mechanisms to salt stress: Physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 2015, 35, 425–437. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Koca, H.; Bor, M.; Özdemir, F.; Türkan, İ. The effect of salt stress on lipid peroxidation, antioxidative enzymes and proline content of sesame cultivars. Environ. Exp. Bot. 2007, 60, 344–351. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef] [PubMed]
- Farhangi-Abriz, S.; Torabian, S. Antioxidant enzyme and osmotic adjustment changes in bean seedlings as affected by biochar under salt stress. Ecotoxicol. Environ. Saf. 2017, 137, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Forni, C.; Duca, D.; Glick, B.R. Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 2017, 410, 335–356. [Google Scholar] [CrossRef]
- Sheldon, A.R.; Dalal, R.C.; Kirchhof, G.; Kopittke, P.M.; Menzies, N.W. The effect of salinity on plant-available water. Plant Soil 2017, 418, 477–491. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Egea, I.; Pineda, B.; Ortíz-Atienza, A.; Plasencia, F.A.; Drevensek, S.; García-Sogo, B.; Yuste-Lisbona, F.J.; Barrero-Gil, J.; Atarés, A.; Flores, F.B.; et al. The SlCBL10 calcineurin B-like protein ensures plant growth under salt stress by regulating Na+ and Ca2+ homeostasis. Plant Physiol. 2018, 176, 1676–1693. [Google Scholar] [CrossRef]
- El-Esawi, M.; Alaraidh, I.; Alsahli, A.; Alzahrani, S.; Ali, H.; Alayafi, A.; Ahmad, M. Serratia liquefaciens KM4 improves salt stress tolerance in maize by regulating redox potential, ion homeostasis, leaf gas exchange and stress-related gene expression. Int. J. Mol. Sci. 2018, 19, 3310. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, S.; Horie, T.; Hauser, F.; Deinlein, U.; Schroeder, J.I.; Uozumi, N. HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotechnol. 2015, 32, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.R. A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol. Plant. 1991, 81, 563–570. [Google Scholar] [CrossRef]
- Santos, C.V. Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Sci. Hortic. 2004, 103, 93–99. [Google Scholar] [CrossRef]
- Rout, N.P.; Shaw, B.P. Salt tolerance in aquatic macrophytes: Possible involvement of the antioxidative enzymes. Plant Sci. 2001, 160, 415–423. [Google Scholar] [CrossRef]
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Łukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 102. [Google Scholar] [CrossRef]
- Oukarroum, A.; Bussotti, F.; Goltsev, V.; Kalaji, H.M. Correlation between reactive oxygen species production and photochemistry of photosystems I and II in Lemna gibba L. plants under salt stress. Environ. Exp. Bot. 2015, 109, 80–88. [Google Scholar] [CrossRef]
- Zhang, C.J.; Lim, S.H.; Kim, J.W.; Nah, G.; Fischer, A.; Kim, D.S. Leaf chlorophyll fluorescence discriminates herbicide resistance in Echinochloa species. Weed Res. 2016, 56, 424–433. [Google Scholar] [CrossRef]
- Chen, S.; Yang, J.; Zhang, M.; Strasser, R.J.; Qiang, S. Classification and characteristics of heat tolerance in Ageratina adenophora populations using fast chlorophyll a fluorescence rise O-J-I-P. Environ. Exp. Bot. 2016, 122, 126–140. [Google Scholar] [CrossRef]
- Živčák, M.; Olšovská, K.; Slamka, P.; Galambošová, J.; Rataj, V.; Shao, H.B.; Brestič, M. Application of chlorophyll fluorescence performance indices to assess the wheat photosynthetic functions influenced by nitrogen deficiency. Plant Soil Environ. 2014, 60, 210–215. [Google Scholar] [CrossRef]
- Akilan, S.; Halima, T.H.; Sasi, S.; Kappachery, S.; Baniekal-Hiremath, G.; Venkatesh, J.; Gururani, A. Evaluation of osmotic stress tolerance in transgenic Arabidopsis plants expressing Solanum tuberosum D200 gene. J. Plant Interact. 2018, 14, 79–86. [Google Scholar] [CrossRef]
- Bowler, C.; Fluhr, R. The role of calcium and activated oxygen as signals for controlling cross-tolerance. Trends Plant Sci. 2000, 5, 241–246. [Google Scholar] [CrossRef]
- Faralli, M.; Lektemur, C.; Rosellini, D.; Gürel, F. Effects of heat shock and salinity on barley growth and stress-related gene transcription. Biol. Plant. 2015, 59, 537–546. [Google Scholar] [CrossRef]
- Hilker, M.; Schwachtje, J.; Baier, M.; Balazadeh, S.; Bäurle, I.; Geiselhardt, S.; Hincha, D.K.; Kunze, R.; Mueller-Roeber, B.; Rillig, M.C.; et al. Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 2016, 91, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Li, Z.G.; Hoque, T.S.; Burritt, D.J.; Fujita, M.; Munné-Bosch, S. Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: Key regulators and possible mechanisms. Protoplasma 2018, 255, 399–412. [Google Scholar] [CrossRef] [PubMed]
- van Buer, J.; Prescher, A.; Baier, M. Cold-priming of chloroplast ROS signalling is developmentally regulated and is locally controlled at the thylakoid membrane. Sci. Rep. 2019, 9, 3022. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ren, J.; Zhu, W.; Amombo, E.; Fu, J.; Chen, L. Antioxidant responses and gene expression in bermudagrass under cold stress. J. Am. Soc. Hortic. Sci. 2014, 139, 699–705. [Google Scholar] [CrossRef]
- Chan, Z.; Shi, H. Improved abiotic stress tolerance of bermudagrass by exogenous small molecules. Plant Signal. Behav. 2015, 10, e991577. [Google Scholar] [CrossRef]
- Hu, L.; Li, H.; Pang, H.; Fu, J. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. J. Plant Physiol. 2012, 169, 146–156. [Google Scholar] [CrossRef]
- Chen, K.; Chen, L.; Fan, J.; Fu, J. Alleviation of heat damage to photosystem II by nitric oxide in tall fescue. Photosynth. Res. 2013, 116, 21–31. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, H.; Ren, F.; Guo, Q.; Hu, X.P. A novel coldregulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant Cell Rep. 2011, 30, 463–471. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).