Early Detection of Sage (Salvia officinalis L.) Responses to Ozone Using Reflectance Spectroscopy
Abstract
1. Introduction
2. Results
2.1. Prediction of Leaf Traits
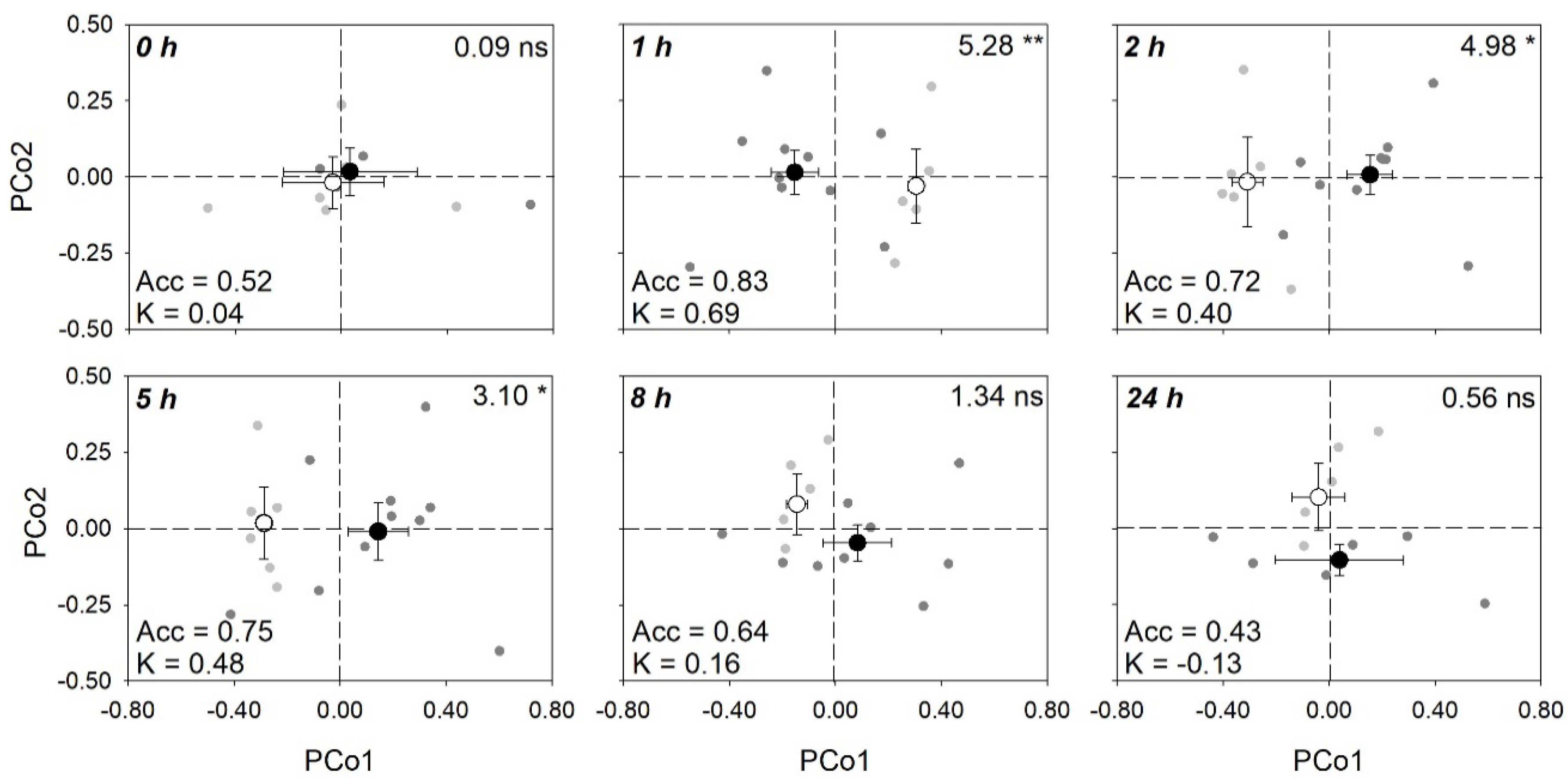
2.2. Analyses of Spectral Signatures
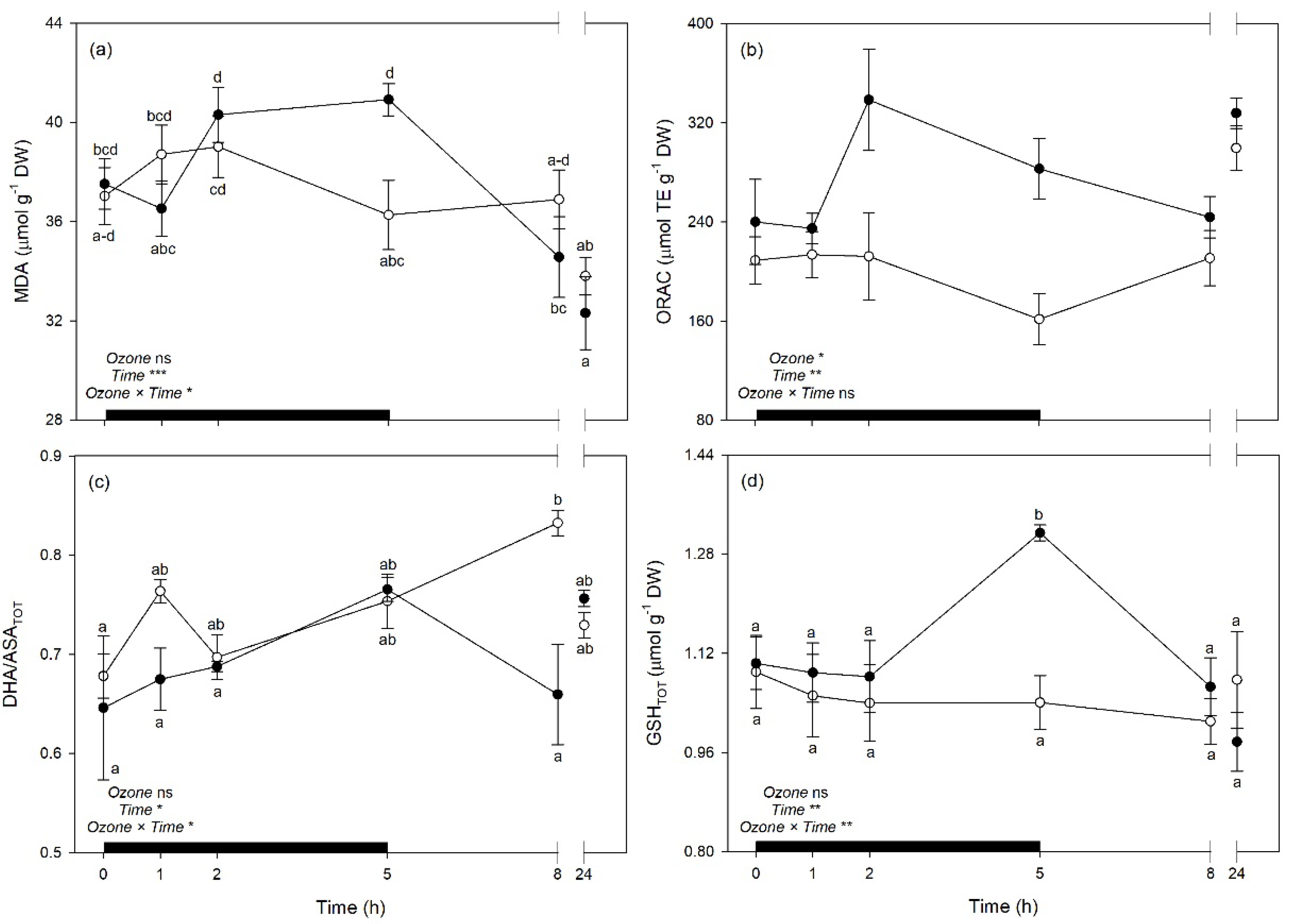
2.3. Variations of Vegetation Indices and Leaf Traits Derived from Spectra
3. Discussion
4. Materials and Methods
4.1. Plant Material and Experimental Design
4.2. Collection of Leaf Spectra
4.3. Foliar Gas-Exchange
4.4. Biochemical Analyses
4.5. PLSR-Model Calibration and Validation
4.6. Estimation of Leaf Traits by Spectral Indices and PLSR-Models
4.7. Analyses of Spectral Signatures
4.8. Other Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ainsworth, E.A. Understanding and improving global crop response to ozone pollution. Plant J. 2017, 90, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, M.A.; Khan, H.U.R.; Zaman, K.; Hishan, S.S. Measuring the impact of global tropospheric ozone, carbon dioxide and sulfur dioxide concentrations on biodiversity loss. Environ. Res. 2018, 160, 398–411. [Google Scholar] [CrossRef]
- Nuvolone, D.; Petri, D.; Voller, F. The effects of ozone on human health. Environ. Sci. Pollut. Res. 2018, 25, 8074–8088. [Google Scholar] [CrossRef]
- Lefohn, A.S.; Malley, C.S.; Smith, L.; Wells, B.; Hazucha, M.; Simon, H.; Naik, V.; Mills, G.; Schultz, M.G.; Paoletti, E.; et al. Tropospheric ozone assessment report: Global ozone metrics for climate change, human health, and crop/ecosystem research. Elementa 2018, 6, 28. [Google Scholar] [CrossRef]
- Ochoa-Hueso, R.; Munzi, S.; Alonso, R.; Arróniz-Crespo, M.; Avila, A.; Bermejo, V.; Bobbink, R.; Branquinho, C.; Concostrina-Zubiri, L.; Cruz, C.; et al. Ecological impacts of atmospheric pollution and interactions with climate change in terrestrial ecosystems of the Mediterranean Basin: Current research and future directions. Environ. Pollut. 2017, 227, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Sicard, P.; De Marco, A.; Troussier, F.; Renou, C.; Vas, N.; Paoletti, E. Decrease in surface ozone concentrations at Mediterranean remote sites and increase in the cities. Atmos. Environ. 2013, 79, 705–715. [Google Scholar] [CrossRef]
- Tonelli, M.; Pellegrini, E.; D’Angiolillo, F.; Petersen, M.; Nali, C.; Pistelli, L.; Lorenzini, G. Ozone-elicited secondary metabolites in shoot cultures of Melissa officinalis L. Plant Cell Tissue Organ Cult. 2015, 120, 617–629. [Google Scholar] [CrossRef]
- Pellegrini, E.; Campanella, A.; Cotrozzi, L.; Tonelli, M.; Nali, C.; Lorenzini, G. Ozone primes changes in phytochemical parameters in the medicinal herb Hypericum perforatum (St. John’s wort). Ind. Crop Prod. 2018, 126, 119–128. [Google Scholar] [CrossRef]
- Deacon, N.J.; Grossman, J.J.; Schweiger, A.K.; Armour, I.; Cavender-Bares, J. Genetic, morphological, and spectral characterization of relictual Niobrara River hybrid aspens (Populus × smithii). Am. J. Bot. 2017, 104, 1–13. [Google Scholar] [CrossRef]
- Cotrozzi, L.; Townsend, P.A.; Pellegrini, E.; Nali, C.; Couture, J.J. Reflectance spectroscopy: A novel approach to better understand and monitor the impact of air pollution on Mediterranean plants. Environ. Sci. Pollut. Res. 2018, 25, 8249–8267. [Google Scholar] [CrossRef]
- Gamon, J.A.; Field, C.B.; Goulden, M.L.; Griffin, K.L.; Hartley, A.E.; Joel, G.; Peñuelas, J.; Valentini, R. Relationships between NDVI, canopy structure, and photosynthesis in three Californian vegetation types. Ecol. Appl. 1995, 5, 28–41. [Google Scholar] [CrossRef]
- Gamon, J.A.; Serrano, L.; Surfus, J.S. The photochemical reflectance index: An optical indicator of photosynthetic radiation use efficiency across species, functional types, and nutrient levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef]
- Gao, B.C. NDWI-a normalized difference water index for remote sensing of vegetation liquid from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Quantitative estimation of chlorophyll a using reflectance spectra—Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Grossman, Y.L.; Ustin, S.L.; Jacquemond, S.; Sanderson, E.W.; Schmuck, G.; Verdebout, J. Critique of stepwise multiple linear regression for the extraction of leaf biochemistry information from leaf reflectance data. Remote Sens. Environ. 1996, 56, 182–193. [Google Scholar] [CrossRef]
- Couture, J.J.; Singh, A.; Rubert-Nason, K.F.; Serbin, S.P.; Lindroth, R.L.; Townsend, P.A. Spectroscopic determination of ecologically relevant plant secondary metabolites. Methods Ecol. Evol. 2016, 7, 1402–1412. [Google Scholar] [CrossRef]
- Bolster, K.L.; Martin, M.E.; Aber, J.D. Determination of carbon fraction and nitrogen concentration in tree foliage by near infrared reflectance: A comparison of statistical methods. Can. J. For. Res. 1996, 26, 590–600. [Google Scholar] [CrossRef]
- Atzberger, C.; Guerif, M.; Baret, F.; Werner, W. Comparative analysis of three chemometric techniques for the spectrometric assessment of canopy chlorophyll content in winter wheat. Comput. Electron. Agric. 2010, 73, 165–173. [Google Scholar] [CrossRef]
- Couture, J.J.; Lindroth, R.L. Atmospheric change alters performance of an invasive forest insect. Glob. Chang. Biol. 2012, 18, 3543–3557. [Google Scholar] [CrossRef]
- Petisco, C.; García-Criado, B.; Mediavilla, S.; Vázquez de Aldana, B.R.; Zabalgogeazcoa, I.; García-Ciudad, A. Near-infrared reflectance spectroscopy as a fast and non-destructive tool to predict foliar organic constituents of several woody species. Anal. Bioanal. Chem. 2006, 386, 1823–1833. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sens. Environ. 2008, 112, 3958–3970. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Tupayachi, R.; Emerson, R.; Marinez, P.; Sinca, F.; Powell, G.V.N.; Wright, S.J.; Lugo, A.E. Taxonomy and remote sensing of leaf mass per area (LMA) in humid tropical forests. Ecol. Appl. 2011, 21, 85–98. [Google Scholar] [CrossRef]
- Couture, J.J.; Serbin, S.P.; Townsend, P.A. Spectroscopic sensitivity of real-time, rapidly induced phytochemical change in response to damage. New Phytol. 2013, 198, 311–319. [Google Scholar] [CrossRef]
- Serbin, S.P.; Singh, A.; Desai, A.R.; Dubois, S.G.; Jablonski, A.D.; Kingdon, C.C.; Kruger, E.L.; Townsend, P.A. Remotely estimating photosynthetic capacity, and its response to temperature, in vegetation canopies using imaging spectroscopy. Remote Sens. Environ. 2015, 167, 78–87. [Google Scholar] [CrossRef]
- Cotrozzi, L.; Couture, J.J.; Cavender-Bares, J.; Kingdon, C.C.; Fallon, B.; Pilz, G.; Pellegrini, E.; Nali, C.; Townsend, P.A. Using foliar spectral properties to assess the effects of drought on plant water potential. Tree Physiol. 2017, 13, 1582–1591. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Meireles, J.E.; Couture, J.J.; Kaproth, M.A.; Kingdon, C.C.; Singh, A.; Serbin, S.P.; Center, A.; Zuniga, E.; Pilz, G.; et al. Associations of leaf spectra with genetic and phylogenetic variation in oaks: Prospects for remote detection of biodiversity. Remote Sens. 2016, 8, 201. [Google Scholar] [CrossRef]
- Runeckles, V.C.; Resh, H.M. The assessment of chronic ozone injury to leaves by reflectance spectrophotometry. Atmos. Environ. 1975, 9, 447–452. [Google Scholar] [CrossRef]
- Schutt, J.B.; Rowland, R.A.; Haggestad, H.E. Identification of injury resulting from atmospheric pollutants using reflectance measurements. J. Environ. Qual. 1984, 13, 605–608. [Google Scholar] [CrossRef]
- Grzegorczyk-Karolak, I.; Bilichowski, I.; Mikiciuk-Olasik, E.; Wysokińdka, H. In vitro cultures of Salvia officinalis L. as a source of antioxidant compounds. Acta Soc. Bot. Pol. 2005, 74, 17–21. [Google Scholar] [CrossRef]
- Pavić, V.; Jakovljević, M.; Molnar, M.; Jokić, S. Extraction of carnosic acid and carnosol from sage (Salvia officinalis L.) leaves by supercritical fluid extraction and their antioxidant and antibacterial activity. Plants 2019, 8, 16. [Google Scholar]
- Vergine, M.; Nicolì, F.; Negro, C.; Luvisi, A.; Nutricati, E.; Accogli, R.A.; Sabella, E.; Miceli, A. Phytochemical profiles and antioxidant activity of Salvia species from southern Italy. Rec. Nat. Prod. 2019, 13, 205–215. [Google Scholar] [CrossRef]
- Tounekti, T.; Abreau, M.E.; Khemira, H.; Munné-Bosch, S. Canopy position determines the photoprotective demand and antioxidant protection of leaves in salt-stressed Salvia officinalis L. Environ. Exp. Bot. 2012, 78, 146–156. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Drought-induced changes in the redox state of α-tocopherol, ascorbate and the diterpene carnosic acid in chloroplast of Labiatae species differing in carnosic acid contents. Plant Physiol. 2003, 131, 1816–1825. [Google Scholar] [CrossRef]
- Pellegrini, E.; Francini, A.; Lorenzini, G.; Nali, C. Ecophysiological and antioxidant traits of Salvia officinalis under ozone stress. Environ. Sci. Pollut. Res. 2015, 22, 13083–13093. [Google Scholar] [CrossRef]
- Marchica, A.; Lorenzini, G.; Papini, R.; Bernardi, R.; Nali, C.; Pellegrini, E. Signalling molecules responsive to ozone-induced oxidative stress in Salvia officinalis. Sci. Total Environ. 2019, 657, 568–576. [Google Scholar] [CrossRef]
- Peñuelas, J.; Garbulsky, M.F.; Filella, I. Photochemical reflectance index (PRI) and remote sensing of plant CO2 uptake. New Phytol. 2011, 191, 596–599. [Google Scholar] [CrossRef]
- Serbin, S.P.; Dillaway, D.N.; Kruger, E.L.; Townsend, P.A. Leaf optical properties reflect variation in photosynthetic metabolism and its sensitivity to temperature. J. Exp. Bot. 2012, 63, 489–502. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Serbin, S.P.; Skoneczka, J.A.; Townsend, P.A. Using leaf optical properties to detect ozone effects on foliar biochemistry. Phothosynth. Res. 2014, 119, 65–76. [Google Scholar] [CrossRef]
- Heckmann, D.; Schlüter, U.; Weber, A.P.M. Machine learning techniques for predicting crop photosynthetic capacity from leaf reflectance spectra. Mol. Plant 2017, 10, 878–890. [Google Scholar] [CrossRef]
- Yendrek, C.R.; Tomaz, T.; Montes, C.M.; Cao, Y.; Morse, A.M.; Brown, P.J.; McIntyre, L.M.; Leakey, A.D.B.; Ainsworth, E.A. High-throughput phenotyping maize leaf physiological and biochemical traits using hyperspectral reflectance. Plant Physiol. 2017, 173, 614–626. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Solovchenko, A.E.; Pogosyan, S.I. Application of reflectance spectroscopy for analysis of higher plant pigments. Russ. J. Plant Physiol. 2003, 50, 704–710. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Red edge shift and biochemical content in grass canopies. ISPRS J. Photogramm. Remote Sens. 2007, 62, 34–42. [Google Scholar] [CrossRef]
- Smith, K.L.; Steven, M.D.; Colls, J.J. Use of hyperspectral derivative ratios in red-edge region to identify plant stress response to gas leaks. Remote Sens. Environ. 2004, 92, 207–217. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Morales, A.; Berjón, A.; Agüera, J. Hyperspectral indices and model simulation for chlorophyll estimation in open-canopy tree crops. Remote Sens. Environ. 2004, 9, 463–476. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Cotrozzi, L.; Peron, R.; Tuinstra, M.R.; Mickelbart, M.V.; Couture, J.J. Spectroscopic determination of physiological and anatomical leaf traits related with water status in maize. Plant Physiol. (under review).
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Serbin, S.P.; Singh, A.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Spectroscopic determination of leaf morphological and biochemical traits for northern temperate and boreal tree species. Ecol. Appl. 2014, 24, 1651–1669. [Google Scholar] [CrossRef]
- Carter, G.A.; Mitchell, R.J.; Chappelka, A.H.; Brewer, C.H. Response of leaf spectral reflectance in loblolly pine to increased atmospheric ozone and precipitation acidity. J. Exp. Bot. 1992, 43, 577–584. [Google Scholar] [CrossRef]
- Couture, J.J.; Singh, A.; Charkowski, A.O.; Groves, R.L.; Gray, S.M.; Bethke, P.C.; Townsend, P.A. Integrating spectroscopy with potato disease management. Plant Dis. 2018, 10, 2233–2240. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The heart of the redox Hub1. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Kangasjärvi, J.; Jaspers, P.; Kollist, H. Signalling and cell death in ozone-exposed plants. Plant Cell Environ. 2005, 28, 1021–1036. [Google Scholar] [CrossRef]
- Vainonen, J.P.; Kangasjärvi, J. Plant signalling in acute ozone exposure. Plant Cell Environ. 2015, 38, 240–252. [Google Scholar] [CrossRef]
- Cotrozzi, L.; Pellegrini, E.; Guidi, L.; Landi, M.; Lorenzini, G.; Massai, R.; Remorini, D.; Tonelli, M.; Trivellini, A.; Vernieri, P.; et al. Losing the warning signal: Drought compromises the cross-talk of signaling molecules in Quercus ilex exposed to ozone. Front. Plant Sci. 2017, 8, 1020. [Google Scholar] [CrossRef]
- Guidi, L.; Remorini, D.; Cotrozzi, L.; Giordani, T.; Lorenzini, G.; Massai, R.; Nali, C.; Natali, L.; Pellegrini, E.; Trivellini, A.; et al. The harsh life of an urban tree: The effect of a single pulse of ozone in salt-stressed Quercus ilex saplings. Tree Physiol. 2016, 37, 246–260. [Google Scholar]
- Ou, B.; Hampsch-Woodill, M.; Prior, R.L. Development and validation of an improved oxygen radical absorbance capacity assay using fluorescein as fluorescent probe. J. Agric. Food Chem. 2001, 49, 4619–4626. [Google Scholar] [CrossRef]
- Ou, B.; Hampsch-Woodill, M.; Flanagan, J.; Deemer, E.K.; Prior, R.L.; Huang, D. Novel fluorometric assay for hydroxyl radical prevention capacity using fluorescein as the probe. J. Agric. Food Chem. 2002, 50, 2772–2777. [Google Scholar] [CrossRef]
- Kampfenkel, K.; Van Montagu, M.; Inzé, D. Extraction and determination of ascorbate and dehydroascorbate from plant tissue. Anal. Biochem. 1995, 225, 165–167. [Google Scholar] [CrossRef]
- Sgherri, C.L.M.; Navari-Izzo, F. Sunflower seedlings subjected to increasing water deficit stress: Oxidative stress and defence mechanisms. Physiol. Plant. 1995, 93, 25–30. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids, the pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and carotenoids–measurement and characterization by UV-VIS. Curr. Protoc. Food Anal. Chem. 2001, 1 (Suppl. 1), F431–F438. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Gillespie, K.M. Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin-Ciocalteau reagent. Nat. Protoc. 2007, 4, 875–877. [Google Scholar] [CrossRef]
- Chen, S.; Hong, X.; Harris, C.J.; Sharkey, P.M. Sparse modeling using orthogonal forest regression with PRESS statistic and regularization. IEEE Trans. Syst. Man Cybern. B Cybern. 2004, 34, 898–911. [Google Scholar] [CrossRef]
- Chong, I.-G.; Jun, C.-H. Performance of some variable selection methods when multicollinearity is present. Chemometr. Intell. Lab. Syst. 2005, 28, 103–112. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Dixon, P. VEGAN, A package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Chevallier, S.; Bertand, D.; Kohler, A.; Courcoux, P. Application of PLS-DA in multivariate analysis. J. Chemom. 2006, 20, 221–229. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 5. [Google Scholar] [CrossRef]
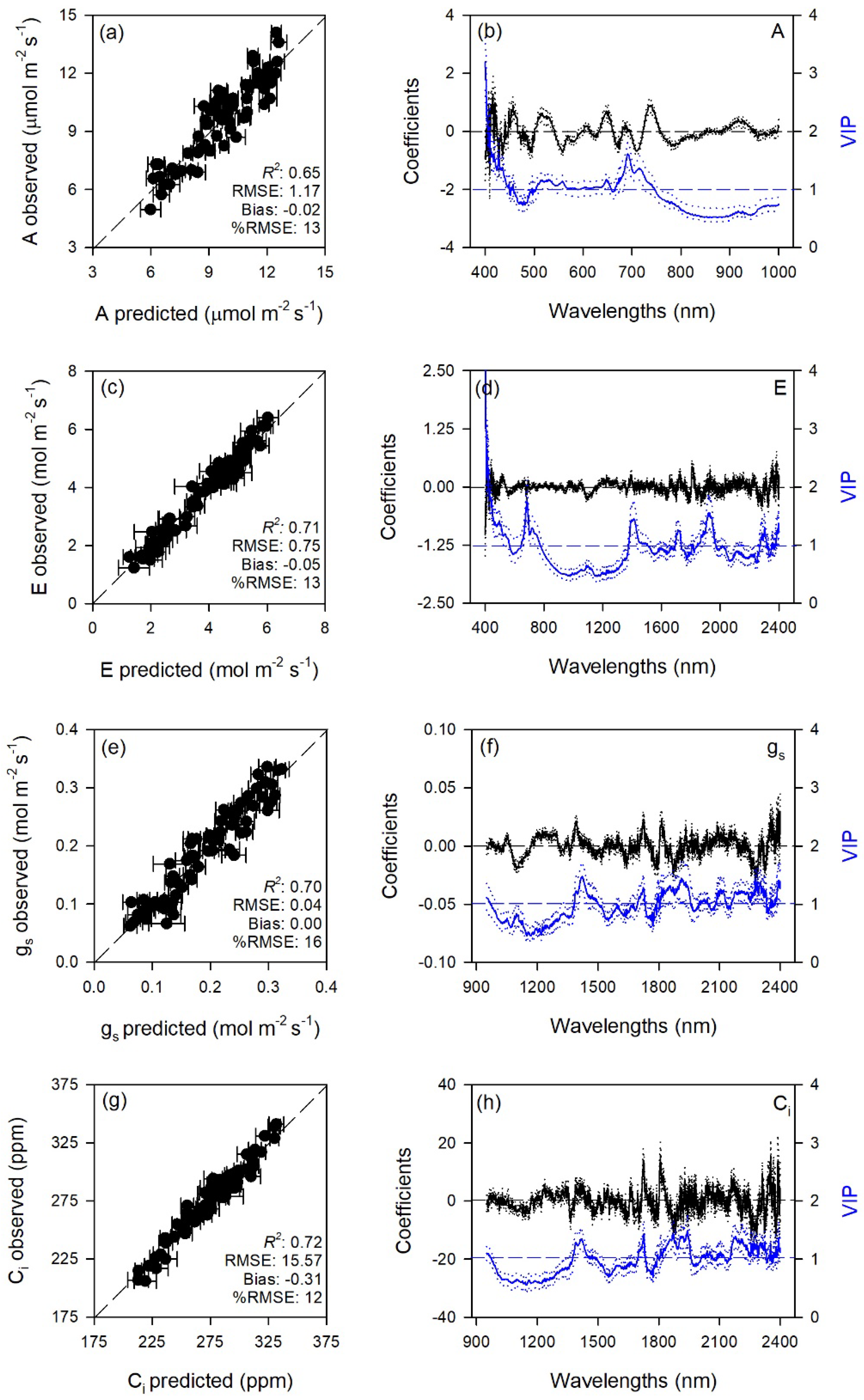
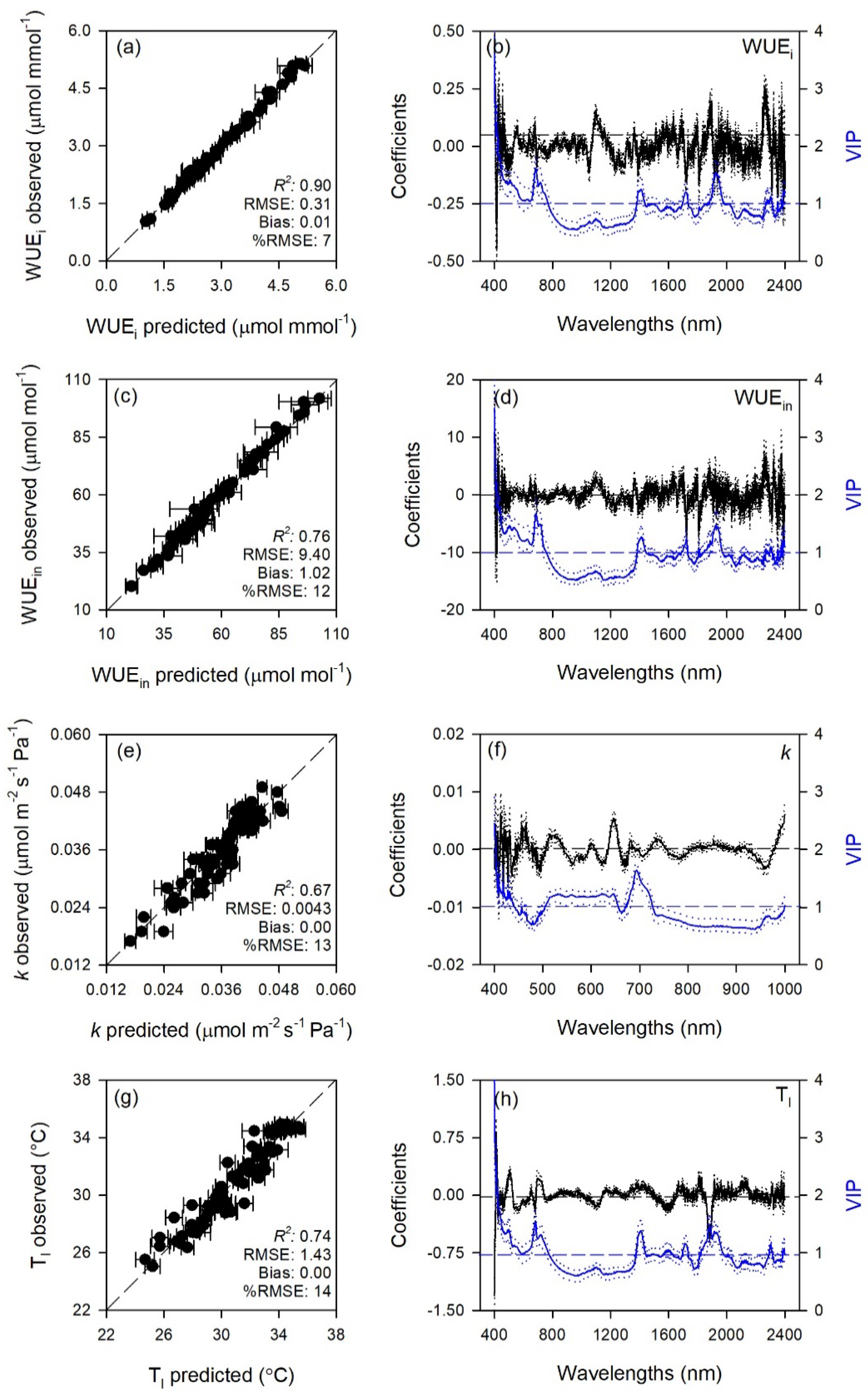
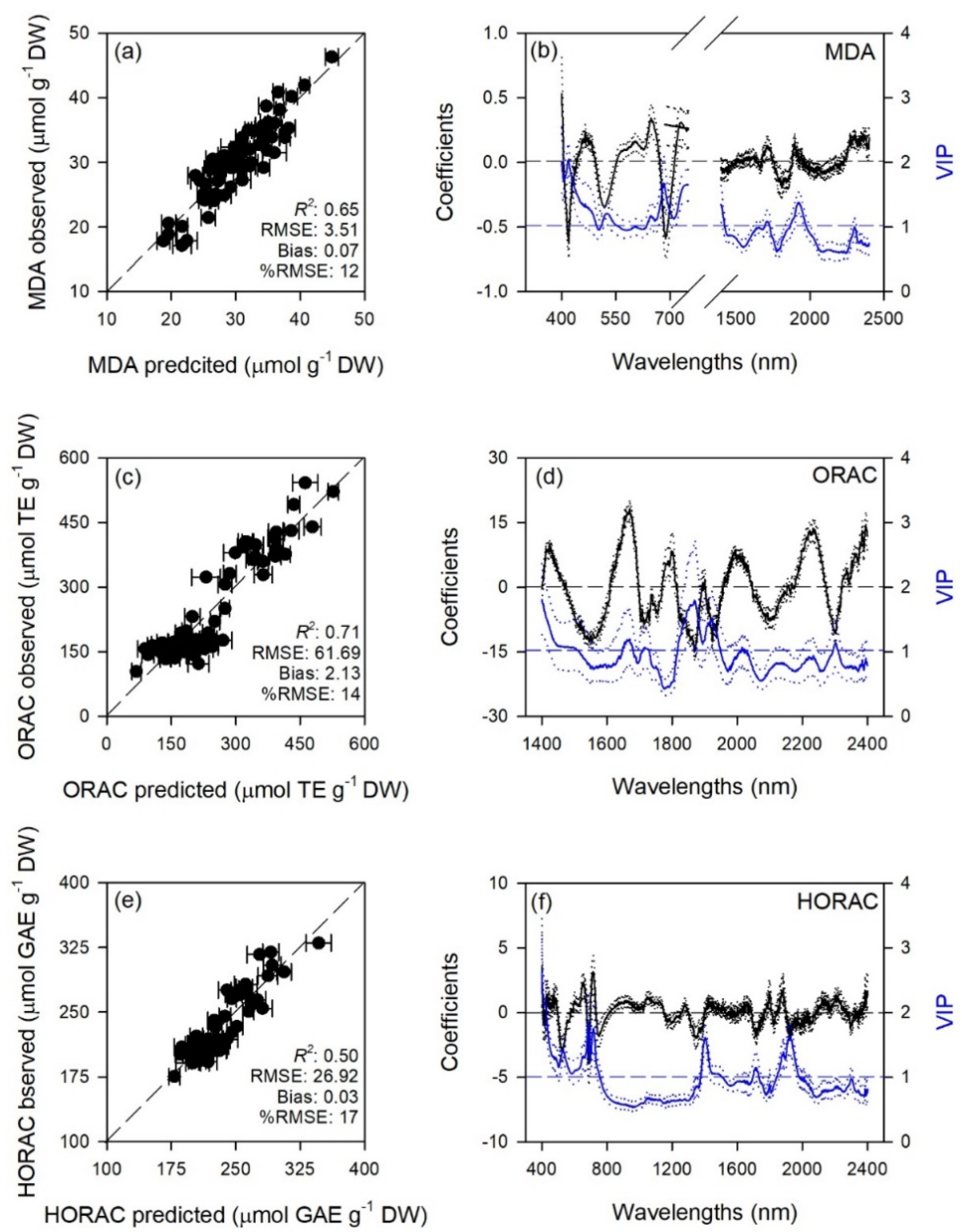
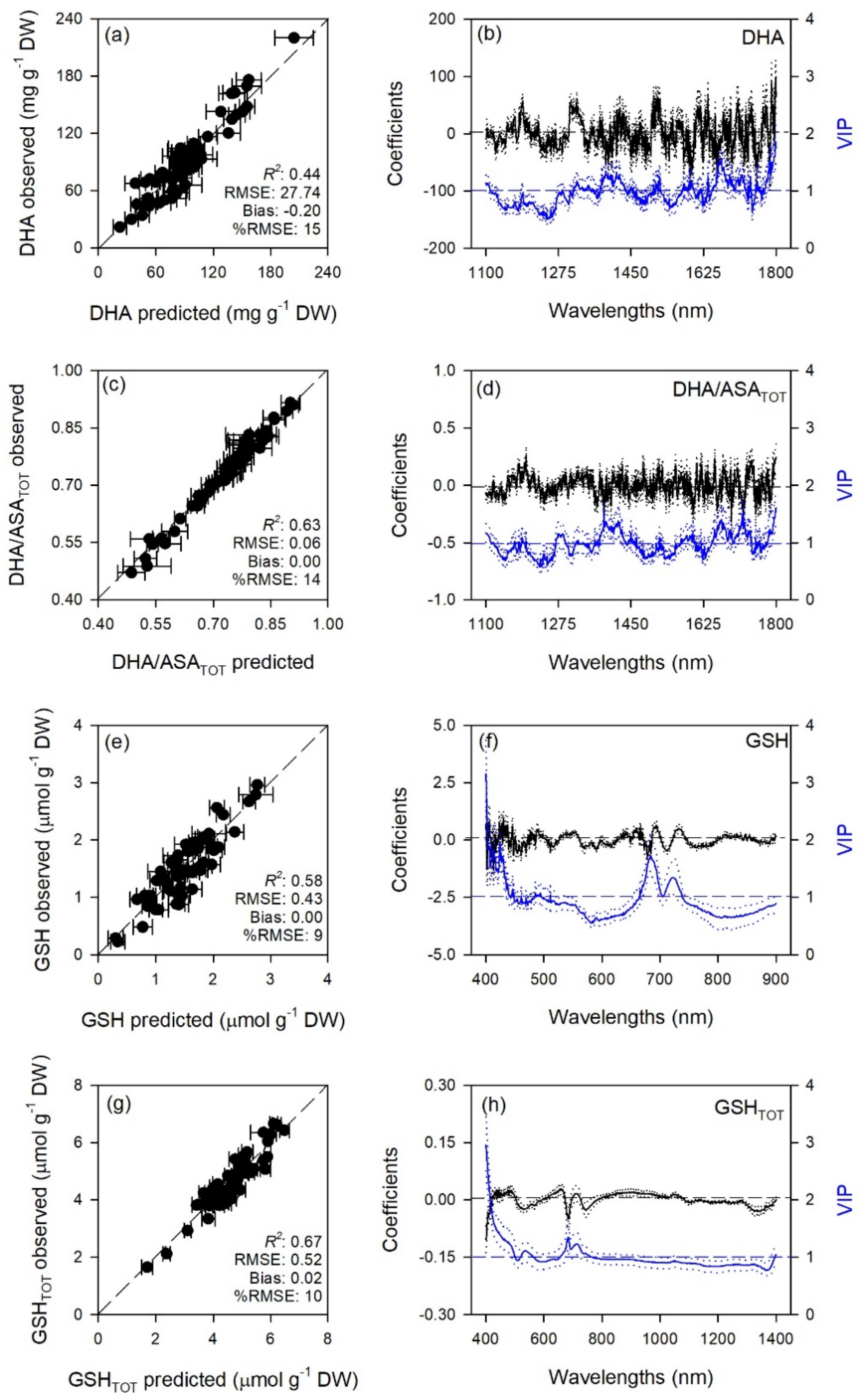
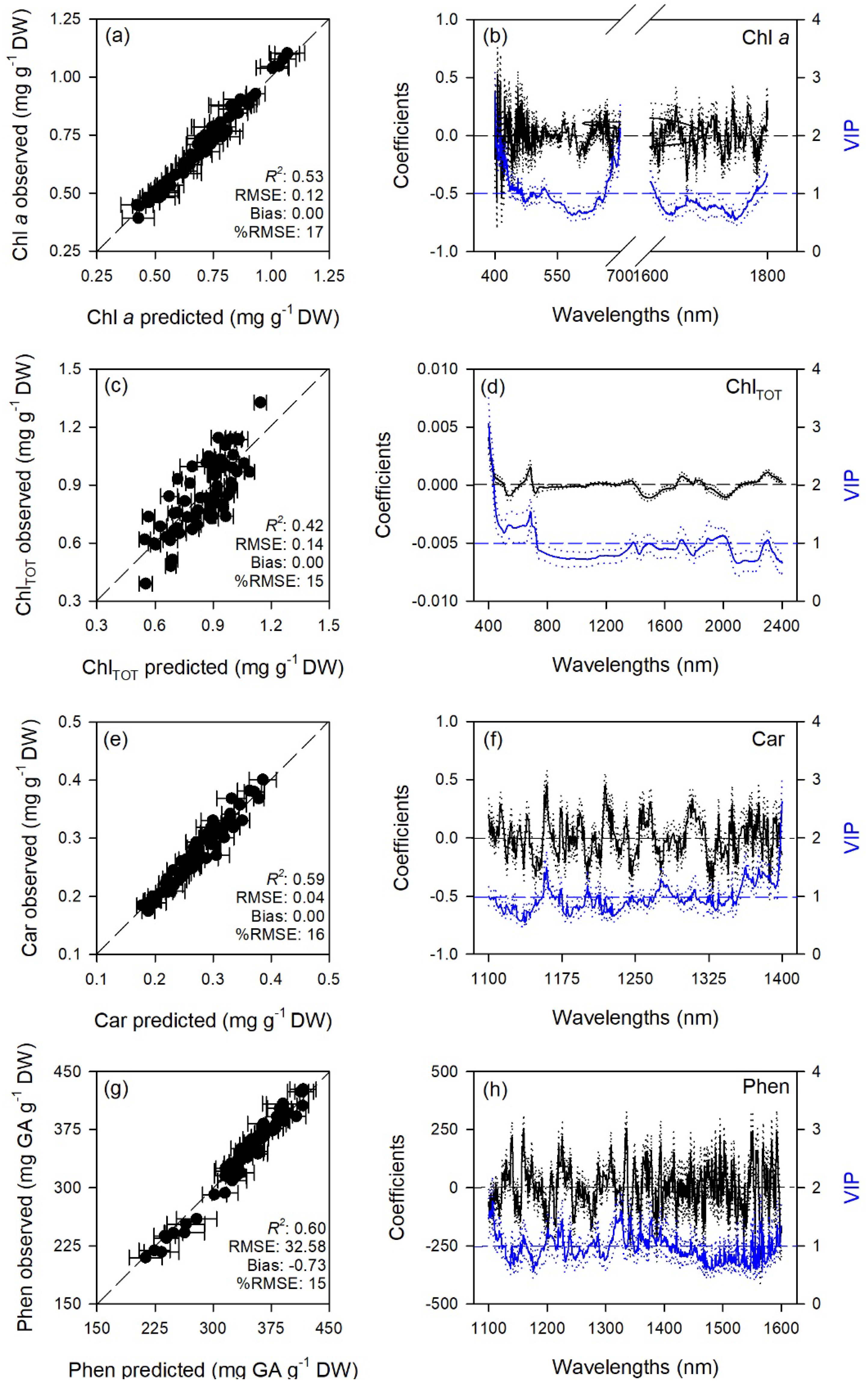

| Trait | Range | LV | Cal | Val | |||
|---|---|---|---|---|---|---|---|
| (nm) | R2 | RMSE | R2 | RMSE | Bias | ||
| A | 400–1000 | 11 | 0.84 ± 0.02 | 0.81 ± 0.04 | 0.65 ± 0.13 | 1.17 ± 0.19 | −0.02 ± 0.34 |
| E | 400–2400 | 20 | 0.97 ± 0.01 | 0.21 ± 0.02 | 0.71 ± 0.11 | 0.75 ± 0.14 | −0.05 ± 0.22 |
| gs | 950–2400 | 13 | 0.92 ± 0.01 | 0.02 ± 0.00 | 0.70 ± 0.12 | 0.04 ± 0.01 | 0.00 ± 0.01 |
| Ci | 950–2400 | 15 | 0.96 ± 0.01 | 5.88 ± 0.51 | 0.72 ± 0.13 | 15.57 ± 2.95 | −0.31 ± 4.54 |
| WUEi | 400–2400 | 22 | 1.00 ± 0.00 | 0.05 ± 0.01 | 0.90 ± 0.06 | 0.31 ± 0.08 | 0.01 ± 0.09 |
| WUEin | 400–2400 | 25 | 1.00 ± 0.00 | 0.59 ± 0.09 | 0.76 ± 0.14 | 9.40 ± 2.58 | 1.02 ± 2.67 |
| k | 400–1000 | 13 | 0.88 ± 0.02 | 2 × 10−4 ± 2 × 10−4 | 0.67 ± 0.14 | 4 × 10−4 ± 7 × 10−4 | 0.00 ± 0.00 |
| Tl | 400–2400 | 15 | 0.92 ± 0.01 | 0.78 ± 0.05 | 0.74 ± 0.10 | 1.43 ± 0.25 | −0.00 ± 0.43 |
| MDA | 400–750 + 1400–2400 | 10 | 0.84 ± 0.02 | 2.33 ± 0.13 | 0.65 ± 0.15 | 3.51 ± 0.58 | 0.07 ± 1.16 |
| ORAC | 1400–2400 | 8 | 0.85 ± 0.02 | 45.68 ± 2.51 | 0.71 ± 0.13 | 61.69 ± 10.44 | 2.13 ± 18.18 |
| HORAC | 400–2400 | 12 | 0.82 ± 0.02 | 15.36 ± 0.94 | 0.50 ± 0.19 | 26.92 ± 4.36 | 0.03 ± 8.18 |
| DHA | 1100–1800 | 13 | 0.91 ± 0.02 | 10.7 ± 1.04 | 0.44 ± 0.19 | 27.74 ± 4.02 | −0.20 ± 8.31 |
| DHA/ASATOT | 1100–1800 | 16 | 1.00 ± 0.00 | 0.01 ± 0.00 | 0.63 ± 0.15 | 0.06 ± 0.01 | 0.00 ± 0.02 |
| GSH | 400–900 | 13 | 0.90 ± 0.03 | 0.22 ± 0.01 | 0.58 ± 0.24 | 0.43 ± 0.07 | 0.00 ± 0.13 |
| GSHTOT | 400–2400 | 11 | 0.88 ± 0.02 | 0.32 ± 0.02 | 0.67 ± 0.16 | 0.52 ± 0.09 | 0.02 ± 0.16 |
| Chl a | 400–700 + 1600–1800 | 20 | 1.00 ± 0.00 | 0.01 ± 0.00 | 0.53 ± 0.20 | 0.12 ± 0.02 | 0.00 ± 0.04 |
| ChlTOT | 400–2400 | 6 | 0.62 ± 0.04 | 0.11 ± 0.00 | 0.42 ± 0.17 | 0.14 ± 0.02 | 0.00 ± 0.04 |
| Car | 1100–1400 | 11 | 0.96 ± 0.01 | 0.01 ± 0.00 | 0.59 ± 0.15 | 0.04 ± 0.01 | 0.00 ± 0.01 |
| Phen | 1100–1600 | 13 | 0.98 ± 0.00 | 7.14 ± 0.90 | 0.60 ± 0.19 | 32.58 ± 7.57 | −0.73 ± 10.40 |
| Effect | df | F | p |
|---|---|---|---|
| Ozone | 1 | 11.10 | *** |
| Time | 5 | 1.05 | ns |
| Ozone × Time | 5 | 0.43 | ns |
| Time (h) | LV | Acc | 95% CI Acc | K |
|---|---|---|---|---|
| 0 | 3 | 0.52 | 0.04–0.96 | 0.04 ± 0.64 |
| 1 | 1 | 0.83 | 0.20–0.99 | 0.69 ± 0.35 |
| 2 | 8 | 0.72 | 0.15–0.97 | 0.40 ± 0.57 |
| 5 | 1 | 0.75 | 0.16–0.99 | 0.48 ± 0.44 |
| 8 | 1 | 0.64 | 0.10–0.97 | 0.16 ± 0.41 |
| 24 | 2 | 0.43 | 0.02–0.96 | −0.13 ± 0.56 |
| Trait | Ozone (df: 1) | Time (df: 5) | Ozone × Time (df: 5) |
|---|---|---|---|
| PRI | 2.46 ns | 4.54 ** | 3.51 * |
| NDVI | 0.15 ns | 1.11 ns | 0.42 ns |
| CI | 0.30 ns | 1.46 ns | 0.51 ns |
| sPSRI | 0.36 ns | 1.09 ns | 0.81 ns |
| WUEi | 2.53 ns | 76.27 *** | 16.96 *** |
| WUEin | 1.99 ns | 31.66 *** | 18.24 *** |
| Ci | 0.92 ns | 68.21 *** | 0.54 ns |
| MDA | 0.00 ns | 11.15 *** | 3.67 * |
| ORAC | 12.54 * | 5.29 ** | 2.23 ns |
| DHA/ASATOT | 3.28 ns | 3.51 * | 3.61 * |
| GSHTOT | 0.95 ns | 3.98 ** | 5.01 ** |
| Phen | 0.30 ns | 4.59 ** | 0.35 ns |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchica, A.; Loré, S.; Cotrozzi, L.; Lorenzini, G.; Nali, C.; Pellegrini, E.; Remorini, D. Early Detection of Sage (Salvia officinalis L.) Responses to Ozone Using Reflectance Spectroscopy. Plants 2019, 8, 346. https://doi.org/10.3390/plants8090346
Marchica A, Loré S, Cotrozzi L, Lorenzini G, Nali C, Pellegrini E, Remorini D. Early Detection of Sage (Salvia officinalis L.) Responses to Ozone Using Reflectance Spectroscopy. Plants. 2019; 8(9):346. https://doi.org/10.3390/plants8090346
Chicago/Turabian StyleMarchica, Alessandra, Silvia Loré, Lorenzo Cotrozzi, Giacomo Lorenzini, Cristina Nali, Elisa Pellegrini, and Damiano Remorini. 2019. "Early Detection of Sage (Salvia officinalis L.) Responses to Ozone Using Reflectance Spectroscopy" Plants 8, no. 9: 346. https://doi.org/10.3390/plants8090346
APA StyleMarchica, A., Loré, S., Cotrozzi, L., Lorenzini, G., Nali, C., Pellegrini, E., & Remorini, D. (2019). Early Detection of Sage (Salvia officinalis L.) Responses to Ozone Using Reflectance Spectroscopy. Plants, 8(9), 346. https://doi.org/10.3390/plants8090346






