CRISPR-Cas9-Mediated Knockout of MLO3 Confers Enhanced Resistance to Reniform Nematode in Upland Cotton
Abstract
1. Introduction
2. Results
2.1. Identification and Sequence Validation of MLO3 Knockout Lines in the F2 and F3 Generations
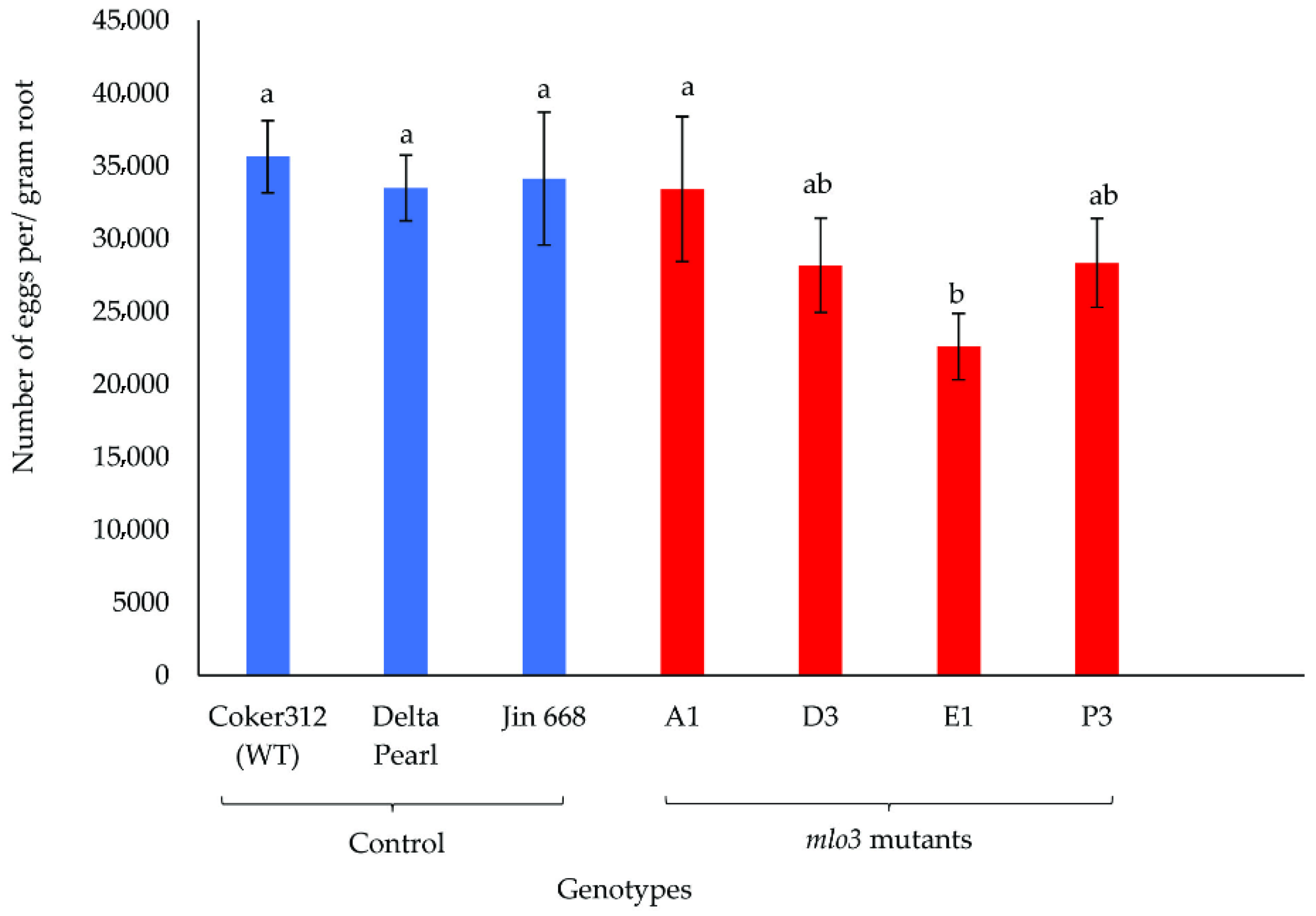
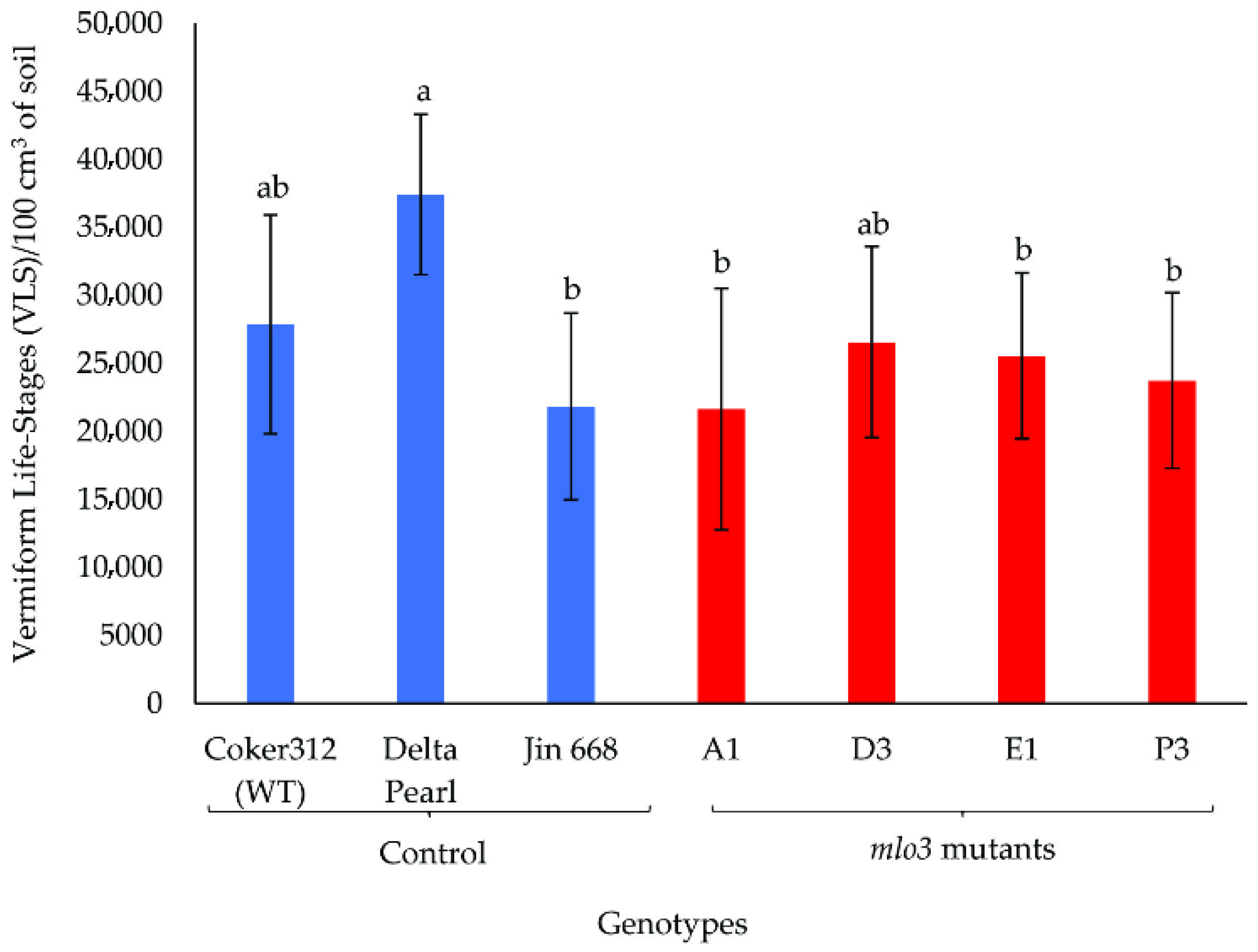
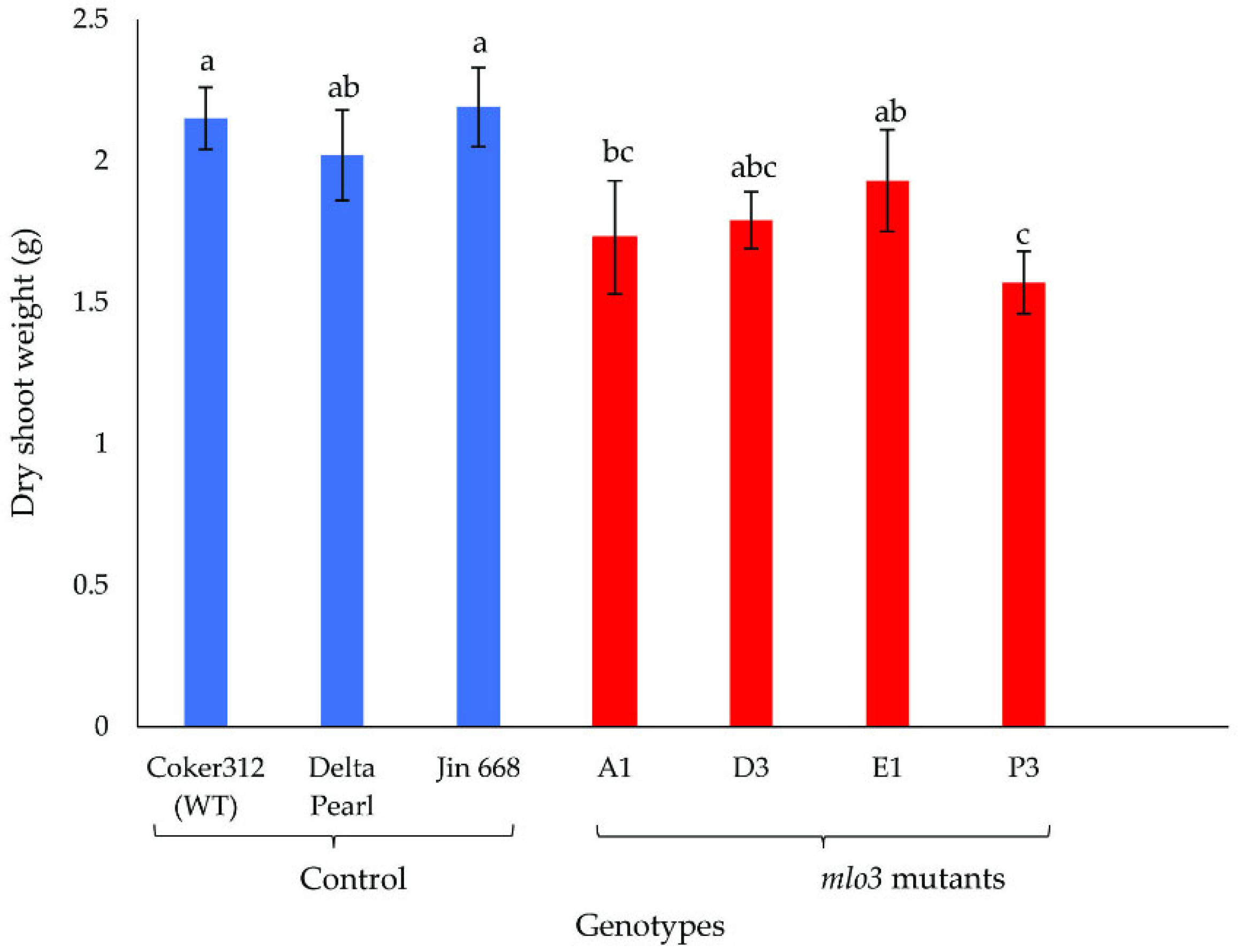
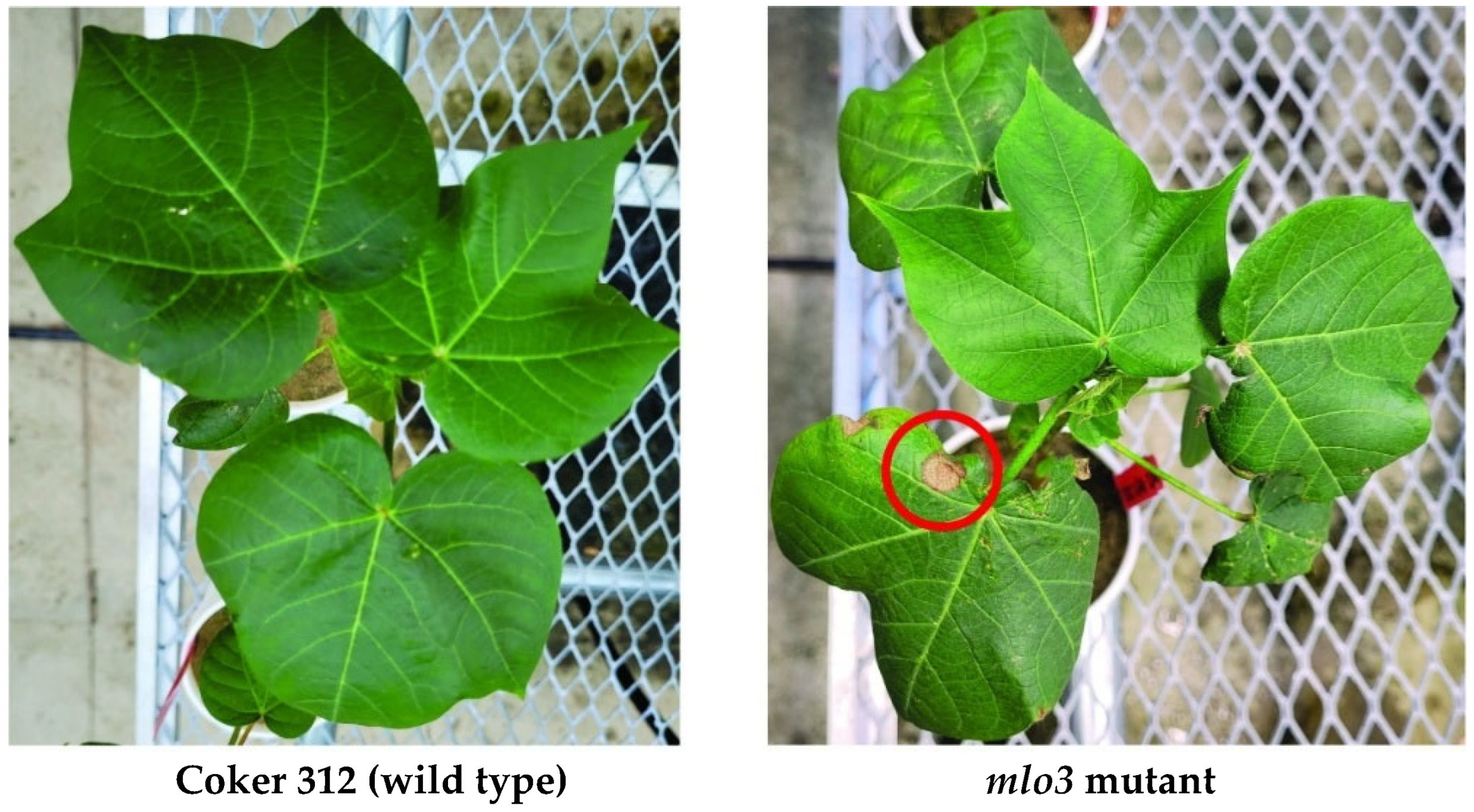
2.2. Phenotyping
3. Discussion
4. Materials and Methods
4.1. Generation and Characterization of Mlo3 Knockouts
4.2. Reniform Nematode Resistance Assessment of Mlo3 Knockout Lines
4.3. Establishment of Experiments
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MLO | Mildew Resistance Locus O |
| GhiMLO3 | Mildew Resistance Locus O from G. hirsutum |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats (CRIPSR)–CRISPR-associated protein 9 |
| gRNAs | Guide RNAs |
| AtMLO | Arabidopsis thaliana Mildew Resistance Locus O |
| WT | Wild type |
| VLS | Vermiform life-stage |
References
- Meyer, L.A. Cotton and Wool Outlook, CWS-21; U.S. Department of Agriculture, Economic Research Service: Washington, DC, USA, 2021; pp. 1–8.
- Colyer, P.; Kirkpatrick, T.; Caldwell, W.; Vernon, P. Plant Pathology and Nematology Root-Knot Nematode Reproduction and Root Galling Severity on Related Conventional and Transgenic Cotton Cultivars. J. Cotton Sci. 2000, 4, 232–236. [Google Scholar]
- Galbieri, R.; Davis, R.F.; Kobayasti, L.; Albuquerque, M.C.F.; Belot, J.L.; Echer, F.R.; Boldt, A.S. Influence of Cotton Root System Size on Tolerance to Rotylenchulus reniformis. Plant Dis. 2018, 102, 2473–2479. [Google Scholar] [CrossRef] [PubMed]
- Koenning, S.R.; Wrather, J.A.; Kirkpatrick, T.L.; Walker, N.R.; Starr, J.L.; Mueller, J.D. Plant-Parasitic Nematodes Attacking Cotton in the United States: Old and Emerging Production Challenges. Plant Dis. 2004, 88, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Faske, T.R.; Kandel, Y.; Allen, T.W.; Grabau, Z.J.; Hu, J.; Kemerait, R.C.; Lawrence, G.W.; Lawrence, K.S.; Mehl, H.L.; Overstreet, C.; et al. Meta-Analysis of the Field Efficacy of Seed- and Soil-Applied Nematicides on Meloidogyne incognita and Rotylenchulus reniformis Across the U.S. Cotton Belt. Plant Dis. 2022, 106, 2228–2238. [Google Scholar] [CrossRef] [PubMed]
- Dyer, D.R.; Groover, W.; Lawrence, K.S. Yield Loss of Cotton Cultivars Due to Rotylenchulus reniformis and the Added Benefit of a Nematicide. Plant Health Prog. 2020, 21, 113–118. [Google Scholar] [CrossRef]
- Robinson, A.F. Reniform in U.S. cotton: When, where, why, and some remedies. Annu. Rev. Phytopathol. 2007, 45, 263–288. [Google Scholar] [CrossRef]
- Popp, J.; Pető, K.; Nagy, J. Pesticide productivity and food security. A review. Agron. Sustain. Dev. 2013, 33, 243–255. [Google Scholar] [CrossRef]
- Smith, A.L.; Taylor, A.L. Nematode distribution in the 1940 regional cotton-wilt plots. Phytopathology 1940, 31, 771. [Google Scholar]
- Dasgupta, D.R.; Raski, D.J.; Sher, S.A. A revision of the genus Rotylenchulus Linford and Oliveira, 1940 (Nematoda: Tylenchiclae). Proc. Helminthol. Soc. Wash. 1968, 35, 169–182. [Google Scholar]
- Weaver, D.B. Cotton Nematodes. Cotton 2015, 57, 547–570. [Google Scholar]
- Fábia, S.O.L.; Valdir, R.C.; Sônia Regina, N.; Patrícia, R.R.S. Nematodes Affecting Soybean and Sustainable Practices for Their Management. In Soybean; Minobu, K., Ed.; IntechOpen: Rijeka, Croatia, 2017; Chapter 6. [Google Scholar]
- Wilson, B.R.; Allen, T.W.; Catchot, A.L.; Krutz, L.J.; Dodds, D.M. Determining the Profitability of Reniform Nematode Control Practices in the Mississippi Cotton Production System. Plant Health Prog. 2020, 21, 105–112. [Google Scholar] [CrossRef]
- Arias, R.S.; Stetina, S.R.; Tonos, J.L.; Scheffler, J.A.; Scheffler, B.E. Microsatellites reveal genetic diversity in Rotylenchulus reniformis populations. J. Nematol. 2009, 41, 146–156. [Google Scholar]
- Stetina, S.R.; Sciumbato, G.L.; Young, L.D.; Blessitt, J.A. Cotton Cultivars Evaluated for Tolerance to Reniform Nematode. Plant Health Prog. 2009, 10, 25. [Google Scholar] [CrossRef]
- Van Den Berg, E.; Palomares-Rius, J.E.; Vovlas, N.; Tiedt, L.R.; Castillo, P.; Subbotin, S.A. Morphological and molecular characterisation of one new and several known species of the reniform nematode, Rotylenchulus Linford & Oliveira, 1940 (Hoplolaimidae: Rotylenchulinae), and a phylogeny of the genus. Nematology 2016, 18, 67–107. [Google Scholar] [CrossRef]
- Weaver, D.B.; Lawrence, K.S.; van Santen, E. Reniform nematode resistance in upland cotton germplasm. Crop Sci. 2007, 47, 19–24. [Google Scholar] [CrossRef]
- McCarty, J.C.; Jenkins, J.N.; Wubben, M.J.; Hayes, R.W.; Callahan, F.E.; Deng, D. Registration of Six Germplasm Lines of Cotton with Resistance to the Root-Knot and Reniform Nematodes. J. Plant Regist. 2017, 11, 168–171. [Google Scholar] [CrossRef]
- Khanal, C.; McGawley, E.C.; Overstreet, C.; Stetina, S.R. The Elusive Search for Reniform Nematode Resistance in Cotton. Phytopathology 2018, 108, 532–541. [Google Scholar] [CrossRef]
- Holguin, C.M.; Gerard, P.; Mueller, J.D.; Khalilian, A.; Agudelo, P. Spatial Distribution of Reniform Nematode in Cotton as Influenced by Soil Texture and Crop Rotations. Phytopathology 2015, 105, 674–683. [Google Scholar] [CrossRef]
- Singh, B.; Chastain, D.; Kaur, G.; Snider, J.L.; Stetina, S.R.; Bazzer, S.K. Reniform nematode impact on cotton growth and management strategies: A review. Agron. J. 2023, 115, 2140–2158. [Google Scholar] [CrossRef]
- Abd El-Aal, E.M.; Shahen, M.; Sayed, S.; Kesba, H.; Ansari, M.J.; El-Ashry, R.M.; Aioub, A.A.A.; Salma, A.S.A.; Eldeeb, A.M. In Vivo and In Vitro management of Meloidogyne incognita (Tylenchida: Heteroderidae) using rhizosphere bacteria, Pseudomonas spp. and Serratia spp. compared with oxamyl. Saudi J. Biol. Sci. 2021, 28, 4876–4883. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M. Optimizing Safe Approaches to Manage Plant-Parasitic Nematodes. Plants 2021, 10, 1911. [Google Scholar] [CrossRef] [PubMed]
- Khanal, C.; McGawley, E.; Overstreet, C.; Stetina, S.; Myers, G.; Kularathna, M.; McInnes, B.; Godoy, F. Reproduction and pathogenicity of endemic populations of rotylenchulus reniformis on cotton. Nematropica 2018, 48, 68–81. [Google Scholar]
- Selosse, M.-A.; Baudoin, E.; Vandenkoornhuyse, P. Symbiotic microorganisms, a key for ecological success and protection of plants. Comptes Rendus Biol. 2004, 327, 639–648. [Google Scholar] [CrossRef]
- Li, R.; Rashotte, A.M.; Singh, N.K.; Lawrence, K.S.; Weaver, D.B.; Locy, R.D. Transcriptome Analysis of Cotton (Gossypium hirsutum L.) Genotypes That Are Susceptible, Resistant, and Hypersensitive to Reniform Nematode (Rotylenchulus reniformis). PLoS ONE 2015, 10, e0143261. [Google Scholar] [CrossRef] [PubMed]
- Yik, C.; Birchfield, W. Resistant Germplasm in Gossypium Species and Related Plants to Rotylenchulus reniformis. J. Nematol. 1984, 16, 146–153. [Google Scholar]
- Robinson, A.F.; Bell, A.A.; Dighe, N.D.; Menz, M.A.; Nichols, R.L.; Stelly, D.M. Introgression of Resistance to Nematode Rotylenchulus reniformis into Upland Cotton (Gossypium hirsutum) from Gossypium longicalyx. Crop Sci. 2007, 47, 1865–1877. [Google Scholar] [CrossRef]
- Kate Turner, A.; Graham, S.H.; Potnis, N.; Brown, S.M.; Donald, P.; Lawrence, K.S. Evaluation of Meloidogyne Incognita and Rotylenchulus reniformis Nematode-resistant Cotton Cultivars with Supplemental Corteva Agriscience Nematicides. J. Nematol. 2023, 55, 20230001. [Google Scholar] [CrossRef]
- Han, X.; Li, S.; Zeng, Q.; Sun, P.; Wu, D.; Wu, J.; Yu, X.; Lai, Z.; Milne, R.J.; Kang, Z.; et al. Genetic engineering, including genome editing, for enhancing broad-spectrum disease resistance in crops. Plant Commun. 2025, 6, 101195. [Google Scholar] [CrossRef]
- Khanal, C.; Rutter, W.; Alam, M.S.; Alarcon-Mendoza, I. Meloidogyne floridensis has a unique virulence profile against root-knot nematode resistant and susceptible pepper (Capsicum annuum) lines. J. Nematol. 2025, 57, 20250007. [Google Scholar] [CrossRef]
- Davis, T.C.; Jones, D.S.; Dino, A.J.; Cejda, N.I.; Yuan, J.; Willoughby, A.C.; Kessler, S.A. Arabidopsis thaliana MLO genes are expressed in discrete domains during reproductive development. Plant Reprod. 2017, 30, 185–195. [Google Scholar] [CrossRef]
- Kim, M.C.; Lee, S.H.; Kim, J.K.; Chun, H.J.; Choi, M.S.; Chung, W.S.; Moon, B.C.; Kang, C.H.; Park, C.Y.; Yoo, J.H.; et al. Mlo, a Modulator of Plant Defense and Cell Death, Is a Novel Calmodulin-binding Protein: Isolation and Characterization of a Rice Mlo Homologue. J. Biol. Chem. 2002, 277, 19304–19314. [Google Scholar] [CrossRef]
- Dreiseitl, A. Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations. Plants 2025, 14, 2091. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; Lee, T.A.J.; Diergaarde, P.; Groenendijk, J.; et al. The Barley Mlo Gene: A Novel Control Element of Plant Pathogen Resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Gruner, K.; Reinstädler, A.; Kemen, A.; Kemen, E.; Cao, L.; Takken, F.L.W.; Reitz, M.U.; Schäfer, P.; O’Connell, R.J.; et al. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 2017, 7, 9319. [Google Scholar] [CrossRef] [PubMed]
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Valè, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 2016, 3, 16016. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Wang, M.; He, H.-X.; Xiao, H.-X.; Zhang, Y.; Wang, L.-F. Identification and Characterization of a Potential Candidate Mlo Gene Conferring Susceptibility to Powdery Mildew in Rubber Tree. Phytopathology 2019, 109, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Walden, K.; Leissing, F.; Baumgarten, K.; Drwiega, K.; Kwaaitaal, M.; Reinstädler, A.; Freh, M.; Dong, X.; James, G.V.; et al. Barley Ror1 encodes a class XI myosin required for mlo-based broad-spectrum resistance to the fungal powdery mildew pathogen. Plant J. 2022, 112, 84–103. [Google Scholar] [CrossRef]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef]
- Wang, X.; Ma, Q.; Dou, L.; Liu, Z.; Peng, R.; Yu, S. Genome-wide characterization and comparative analysis of the MLO gene family in cotton. Plant Physiol. Biochem. 2016, 103, 106–119. [Google Scholar] [CrossRef]
- Kumar, R.; Das, J.; Puttaswamy, R.K.; Kumar, M.; Balasubramani, G.; Prasad, Y.G. Targeted genome editing for cotton improvement: Prospects and challenges. Nucleus 2024, 67, 181–203. [Google Scholar] [CrossRef]
- Lei, J.; Li, Y.; Dai, P.; Liu, C.; Zhao, Y.; You, Y.; Qu, Y.; Chen, Q.; Liu, X. Efficient virus-mediated genome editing in cotton using the CRISPR/Cas9 system. Front. Plant Sci. 2022, 13, 1032799. [Google Scholar] [CrossRef]
- Sheri, V.; Mohan, H.; Jogam, P.; Alok, A.; Rohela, G.K.; Zhang, B. CRISPR/Cas genome editing for cotton precision breeding: Mechanisms, advances, and prospects. J. Cotton Res. 2025, 8, 4. [Google Scholar] [CrossRef]
- Kangben, F.; Kumar, S.; Li, Z.; Sreedasyam, A.; Dardick, C.; Jones, D.; Saski, C.A. Phylogenetic and functional analysis of tiller angle control homeologs in allotetraploid cotton. Front. Plant Sci. 2024, 14, 1320638. [Google Scholar] [CrossRef]
- Pepin, N.; Hebert, F.O.; Joly, D.L. Genome-Wide Characterization of the MLO Gene Family in Cannabis sativa Reveals Two Genes as Strong Candidates for Powdery Mildew Susceptibility. Front. Plant Sci. 2021, 12, 729261. [Google Scholar] [CrossRef]
- Gong, F.; Lan, Y.; Zhang, T.; Li, C.; Li, Y.; Xia, F.; Liu, X.; Liu, D.; Liang, G.; Cai, P.; et al. Genome-Wide Analysis of Mlo Genes and Functional Characterization of Cm-mlo38 and Cm-mlo44 in Regulating Powdery Mildew Resistance in Melon. Horticulturae 2025, 11, 509. [Google Scholar] [CrossRef]
- Kusch, S.; Frantzeskakis, L.; Lassen, B.D.; Kümmel, F.; Pesch, L.; Barsoum, M.; Walden, K.D.; Panstruga, R. A fungal plant pathogen overcomes mlo-mediated broad-spectrum disease resistance by rapid gene loss. New Phytol. 2024, 244, 962–979. [Google Scholar] [CrossRef]
- Chen, X.; Lu, X.; Shu, N.; Wang, S.; Wang, J.; Wang, D.; Guo, L.; Ye, W. Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci. Rep. 2017, 7, 44304. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Z.; Bai, W.; Huang, S.; Chen, Y.; Sun, J. Heterotrimeric G protein interacts with MLO1 to regulate a trade-off between disease resistance and growth in wheat. Physiol. Plant. 2025, 177, e70100. [Google Scholar] [CrossRef] [PubMed]
- Kusch, S.; Panstruga, R. mlo-Based Resistance: An Apparently Universal “Weapon” to Defeat Powdery Mildew Disease. Mol. Plant Microbe Interact. 2017, 30, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.; Somerville, S.C. MLO, a novel modulator of plant defenses and cell death, binds calmodulin. Trends Plant Sci. 2002, 7, 379–380. [Google Scholar] [CrossRef]
- Reinstädler, A.; Müller, J.; Czembor, J.H.; Piffanelli, P.; Panstruga, R. Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley MLO protein. BMC Plant Biol. 2010, 10, 31. [Google Scholar] [CrossRef]
- Hilbert, M.; Novero, M.; Rovenich, H.; Mari, S.; Grimm, C.; Bonfante, P.; Zuccaro, A. MLO Differentially Regulates Barley Root Colonization by Beneficial Endophytic and Mycorrhizal Fungi. Front. Plant Sci. 2019, 10, 1678. [Google Scholar] [CrossRef]
- Pessina, S.; Pavan, S.; Catalano, D.; Gallotta, A.; Visser, R.G.F.; Bai, Y.; Malnoy, M.; Schouten, H.J. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genom. 2014, 15, 618. [Google Scholar] [CrossRef]
- Ingvardsen, C.R.; Massange-Sánchez, J.A.; Borum, F.; Füchtbauer, W.S.; Bagge, M.; Knudsen, S.; Gregersen, P.L. Highly effective mlo-based powdery mildew resistance in hexaploid wheat without pleiotropic effects. Plant Sci. 2023, 335, 111785. [Google Scholar] [CrossRef]
- Li, P.; Xiao, S. Diverse Functions of Plant MLO Proteins: From Mystery to Elucidation. Annu. Rev. Phytopathol. 2025, 63, 147–173. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Moolhuijzen, P.; Hickey, L.; Wentzel, E.; Deng, W.; Dinglasan, E.G.; Ellwood, S.R. Physiological Changes in Barley mlo-11 Powdery Mildew Resistance Conditioned by Tandem Repeat Copy Number. Int. J. Mol. Sci. 2020, 21, 8769. [Google Scholar] [CrossRef] [PubMed]
- Gruner, K.; Esser, T.; Acevedo-Garcia, J.; Freh, M.; Habig, M.; Strugala, R.; Stukenbrock, E.; Schaffrath, U.; Panstruga, R. Evidence for Allele-Specific Levels of Enhanced Susceptibility of Wheat mlo Mutants to the Hemibiotrophic Fungal Pathogen Magnaporthe oryzae pv. Triticum. Genes 2020, 11, 517. [Google Scholar] [CrossRef]
- Kumar, S.; Ruggles, A.; Logan, S.; Mazarakis, A.; Tyson, T.; Bates, M.; Grosse, C.; Reed, D.; Li, Z.; Grimwood, J.; et al. Comparative Transcriptomics of Non-Embryogenic and Embryogenic Callus in Semi-Recalcitrant and Non-Recalcitrant Upland Cotton Lines. Plants 2021, 10, 1775. [Google Scholar] [CrossRef]
- Kokalis-Burelle, N.; Chellemi, D.O.; Périès, X. Effect of Soils from Six Management Systems on Root-knot Nematodes and Plant Growth in Greenhouse Assays. J. Nematol. 2005, 37, 467–472. [Google Scholar] [PubMed]
- Hussey, R.S.; Barker, K.R. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis. Report. 1973, 57, 1025–1028. [Google Scholar]
- Jenkins, W.R. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Report. 1964, 48, 692–695. [Google Scholar]
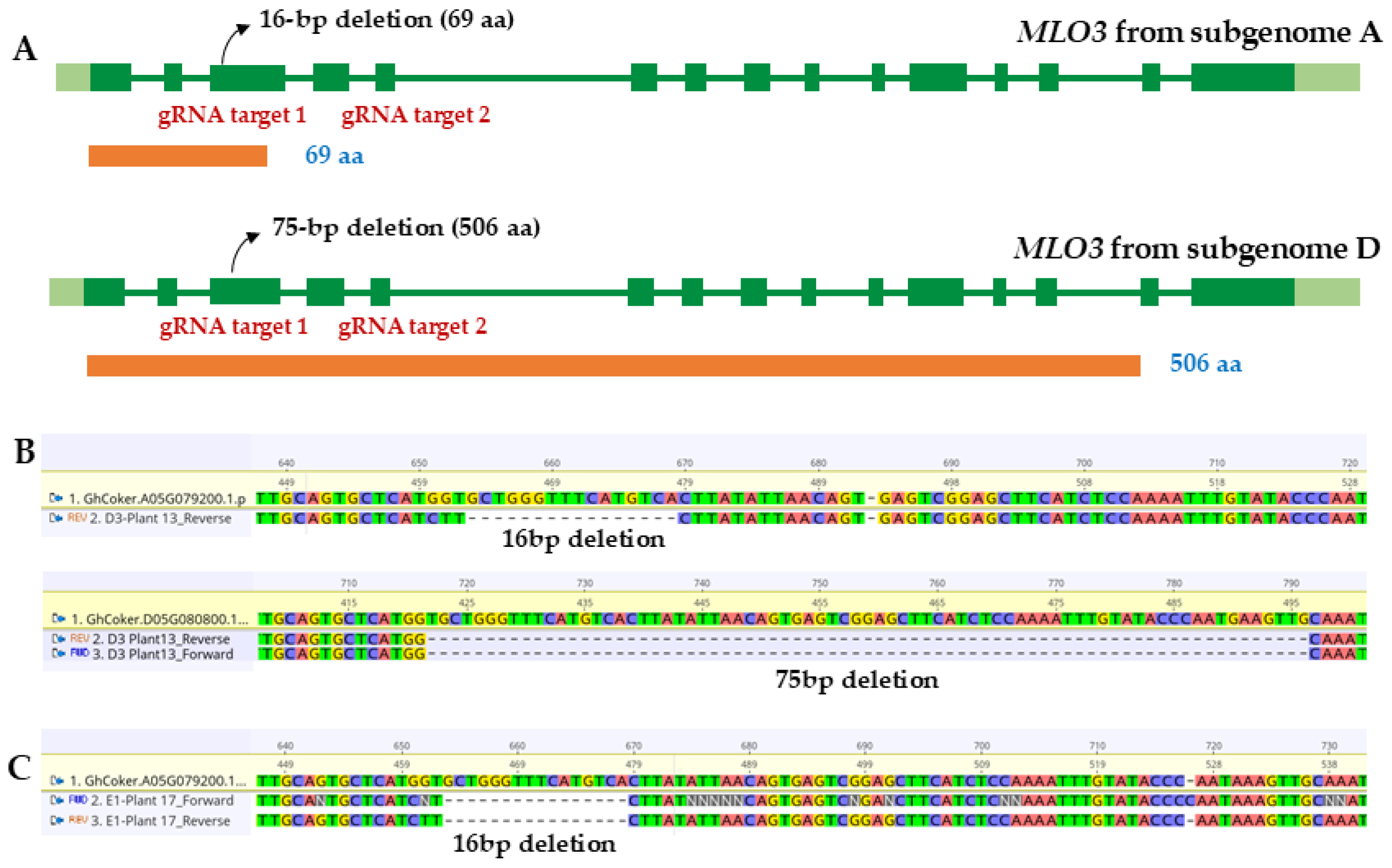
| Transgenic Individual | Mutations in MLO3 (A) | Mutations in MLO3 (D) | ||
|---|---|---|---|---|
| gRNA1 | gRNA2 | gRNA1 | gRNA2 | |
| A1: 1 | A insertion (HM) | CT deletion (HM) | T insertion (HM) | TT insertion (HM) |
| A1: 2–4 | No PCR band | GG deletion (HM) | TGCTCACTATGGCTT deletion (HM) | |
| A1: 5 | A insertion (HM) | CT deletion (HT) | T insertion (HM) | TT insertion (HM) |
| D3: 1–3 | A insertion; 16-bp insertion | 60-bp deletion (HT) | 75-bp deletion (HM) | 147-bp deletion (HM) |
| D3: 4 | A insertion; 16-bp insertion | 60-bp deletion (HT) | No PCR band | |
| D3: 5 | A insertion; 16-bp insertion | 25-bp deletion (HT) | 75-bp deletion (HM) | 147-bp deletion (HM) |
| E1: 1 | A insertion (HM) | CT deletion (HM) | T insertion (HM) | TT insertion (HM) |
| E1: 2 | 16-bp deletion (HM) | 60-bp deletion (HM) | No PCR band | |
| E1: 3–5 | 16-bp deletion (HM) | 60-bp deletion (HM) | T insertion (HM) | TT insertion (HM) |
| P3: 1 | 404-bp deletion (HM) | 507-bp deletion (HM) | ||
| P3: 2 & 3 | A insertion (HM) | CT deletion (HM) | 507-bp deletion (HM) | |
| P3: 4 | A insertion (HM) | CT deletion (HM) | No PCR band | |
| P3: 5 | 404-bp deletion (HM) | 507-bp deletion (HM) | ||
| Transgenic Individual | Mutations in MLO3 (A) | Mutations in MLO3 (D) | ||
|---|---|---|---|---|
| Guide RNA1 | Guide RNA2 | Guide RNA1 | Guide RNA2 | |
| D3: 13 | 16-bp deletion | replaced and substituted | 75-bp deletion (HM) | replaced and substituted |
| E1: 17 | 16-bp deletion and A insertion | A insertion | extra A insertion | extra AA insertion |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kangben, F.; Kumar, S.; Xing, A.; Wen, L.; Li, W.; Parris, S.; Lawson, J.; Li, Z.; Carneal, L.; Cobb, M.; et al. CRISPR-Cas9-Mediated Knockout of MLO3 Confers Enhanced Resistance to Reniform Nematode in Upland Cotton. Plants 2025, 14, 3491. https://doi.org/10.3390/plants14223491
Kangben F, Kumar S, Xing A, Wen L, Li W, Parris S, Lawson J, Li Z, Carneal L, Cobb M, et al. CRISPR-Cas9-Mediated Knockout of MLO3 Confers Enhanced Resistance to Reniform Nematode in Upland Cotton. Plants. 2025; 14(22):3491. https://doi.org/10.3390/plants14223491
Chicago/Turabian StyleKangben, Foster, Sonika Kumar, Anqi Xing, Li Wen, Wei Li, Stephen Parris, John Lawson, Zhigang Li, Lauren Carneal, Meredith Cobb, and et al. 2025. "CRISPR-Cas9-Mediated Knockout of MLO3 Confers Enhanced Resistance to Reniform Nematode in Upland Cotton" Plants 14, no. 22: 3491. https://doi.org/10.3390/plants14223491
APA StyleKangben, F., Kumar, S., Xing, A., Wen, L., Li, W., Parris, S., Lawson, J., Li, Z., Carneal, L., Cobb, M., Nichols, R. L., Wells, C., Agudelo, P., Khanal, C., & Saski, C. A. (2025). CRISPR-Cas9-Mediated Knockout of MLO3 Confers Enhanced Resistance to Reniform Nematode in Upland Cotton. Plants, 14(22), 3491. https://doi.org/10.3390/plants14223491






