Abstract
Various kinds of reproductive barriers have been reported in intraspecific and interspecific crosses between the AA genome Oryza species, to which Asian rice (O. sativa) and African rice (O. glaberrima) belong. A hybrid seed sterility phenomenon was found in the progeny of the cross between O. sativa and O. meridionalis, which is found in Northern Australia and Indonesia and has diverged from the other AA genome species. This phenomenon could be explained by an egg-killer model. Linkage analysis using DNA markers showed that the causal gene was located on the distal end of chromosome 1. Because no known egg-killer gene was located in that chromosomal region, this gene was named HYBRID SPIKELET STERILITY 57 (abbreviated form, S57). In heterozygotes, the eggs carrying the sativa allele are killed, causing semi-sterility. This killer system works incompletely: some eggs carrying the sativa allele survive and can be fertilized. The distribution of alleles in wild populations of O. meridionalis was discussed from the perspective of genetic differentiation of populations.
1. Introduction
Reproductive barriers have been observed in the intraspecific and interspecific cross between the AA genome Oryza species, to which Asian rice (O. sativa) and African rice (O. glaberrima) belong. We identified some causal genes of these reproductive barriers, such as hybrid weakness [1,2,3] and hybrid chlorosis [4] in Oryza sativa. These barriers are obstacles to the genetic improvement of rice, on which more than half of all people depend for staple food. Reproductive barriers have also been studied in the context of differentiation and speciation [5,6,7]. Detailed studies on reproductive barriers contribute to both rice breeding and evolutionary biology [8,9]. Among reproductive barriers, hybrid seed (spikelet) sterility has been intensively studied because it is related to hybrid rice breeding. Asian rice O. sativa comprises two major subspecific groups, indica and japonica. The hybrid between indica and japonica shows hybrid vigor, but also shows hybrid seed sterility [10]. The major gene causing hybrid seed sterility is S5 [11]. The hybrid seed sterility caused by the S5 gene is explained by an egg-killer model: In heterozygotes of S5-i derived from indica and S5-j derived from japonica, S5-i and S5-j respectively act as killer allele and abortive allele, leading to semi-sterility, with about half of the seeds being sterile. Other genes, such as S7 and S9, also follow the same egg-killer model [12,13]. The interspecific crosses between O. sativa and the African rice O. glaberrima show hybrid vigor but are highly sterile both in pollen and seeds [14]. Some causes of this hybrid sterility are explained by a gametic eliminator model [14,15,16]: In a heterozygous state of killer allele and abortive allele, both the eggs and pollen carrying the abortive allele are killed in the gametic eliminator model, causing semi-sterility in both spikelet and pollen. In the selfed progeny of the egg-killer model, the ratio of homozygotes of killer allele and heterozygotes are expected to be 1:1, and no homozygotes of the abortive allele are expected to appear. On the other hand, in the selfed progeny of the gametic eliminator model, only homozygotes of the killer allele are expected to appear. Mapping and characterization of the causal genes of hybrid seed and pollen sterility could contribute to hybrid rice breeding.
O. meridionalis is an AA genome Oryza species from Northern Australia and Indonesia [17]. Several types of molecular data indicate that this species has diverged from the other AA genome species and that it is distantly related to them [18,19]. This divergence is reflected by the low pollen fertility of the hybrid between O. meridionalis and the other AA genome species [20,21,22]. Li et al. [23] identified five hybrid pollen sterility genes, all of which follow the pollen-killer model [15,16]. No genes conferring hybrid seed sterility have been reported in the cross between O. meridionalis and the other AA genome species before our recent study [24], in which we reported seed abortion after fertilization in the progeny from the cross between O. sativa and O. meridionalis was controlled by a SEED DEVELOPMENT 1 (SDV1) gene and a SEED DEVELOPMENT 2 (SDV2) gene. This gene model is not an egg-killer model but a duplication and loss of a functional gene for seed development—a new finding in rice genetics. However, seed sterility is not completely explained by these genes in the cross between an O. meridionais strain Jpn2 and O. sativa. In the present study, we report that an egg-killer gene also contributes to the sterility in the above cross combination. Judging from the gene location and gene action, this gene is thought to be a new finding in rice genetics.
2. Results
2.1. Finding of a Hybrid Sterility Phenomenon
The BC4F2 population using Jpn2 as a donor parent and an Oryza sativa cultivar Taichung 65 (T65) as the recurrent parent showed diverse seed fertility, which was not explained only by the SDV1 gene [24].
To remove the genetic noise, we backcrossed further to produce a BC5F2. However, the situation was not improved (Figure 1). The SDV1 gene was linked closely to HD1, a major heading-time gene [24,25]. Therefore, early heading plants for which days to heading ranged from 98 to 106 were expected to be homozygotes of the Sdv1-s allele, the functional allele for seed development, and to be segregating for the other gene(s) causing seed fertility. Ten plants varying in seed fertility were selected, and their selfed progenies were checked for seed fertility (Table 1). All the plants in the BC5F3 lines headed as early as the recurrent parent T65, suggesting that they were homozygotes of T65 at the HD1 locus (data not shown). Three BC5F3 lines derived from plants with fertility ranging from 80.3% to 88.7% were skewed toward high fertility. Three BC5F3 lines derived from plants with fertility ranging from 57.3% to 79.3% showed a wider range of seed fertility. The other four BC5F3 lines derived from plants with fertility ranging from 44.0% to 54.0% showed bimodal seed fertility with one peak around 50% and the other peak around 90%. Among them, BC5F3 line 8 showed clear bimodal distributions comprising 16 semi-sterile plants with fertility ranging from 0.45 to 0.65 and 18 fertile plants with fertility ranging from 0.85 to 1.00.
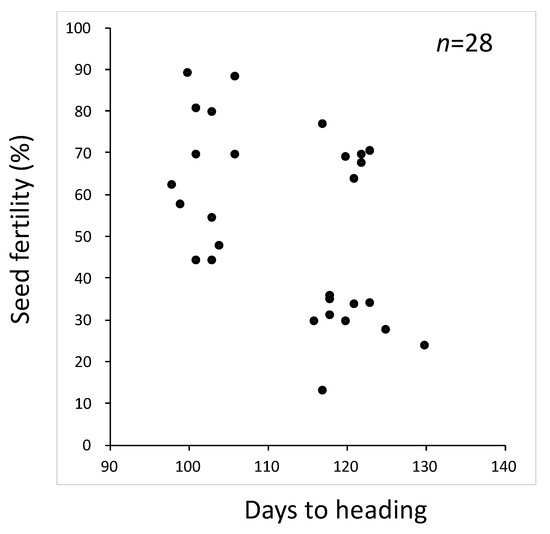
Figure 1.
Scatter diagram of days to heading and seed fertility in the BC5F2 population using T65 as the recurrent parent and Jpn2 as the donor parent.

Table 1.
Frequency distribution of seed fertility in BC5F3 generation using T65 as the recurrent parent and Jpn2 as the donor parent.
Bulked DNA composed of 10 randomly chosen plants per BC5F3 line were subjected to genotyping of 76 DNA markers covering the whole 12 rice chromosomes, with the result that the genotype of S13374 (Table 2), located on the distal end of the short arm of chromosome 1, showed four homozygotes of the Jpn2 allele and six heterozygotes, though these lines underwent backcross five times using T65 as the recurrent parent. All the homozygotes of Jpn2 were derived from BC5F2 plants with high seed fertility, and all the heterozygotes were derived from BC5F2 plants with semi-sterility. These experimental results could be explained by the hypothesis that eggs carrying a gene linked with S13374 from T65 are selectively eliminated during egg development in heterozygotes, following an egg-killer model. Deformed pollen and weakly stained pollen were observed in some low seed-fertility plants. However, the degree of deformation and stain varied highly and was difficult to quantify (data not shown). All plants of BC5F3 line 8 showed high pollen fertility (more than 90%). Therefore, the cause of seed fertility could be confined to the egg. A BC5F4 line derived from a semi-sterile BC5F3 line 8 was used for further analysis.

Table 2.
Primer sequences of Indel markers designed or redesigned for mapping the S57 gene.
2.2. Linkage Analysis of the Causal Gene of the Hybrid Sterility
The distribution of the BC5F4 line showed a clear bimodal distribution of seed fertility (Figure 2). When divided by the dotted line in Figure 2, the ratio of the semi-sterile group and fertile group was 37:39, very close to 1:1. Most semi-sterile plants were heterozygotes, and most fertile plants were homozygotes of the Jpn2 allele at S13374. These experimental results could be explained by the above hypothesis.
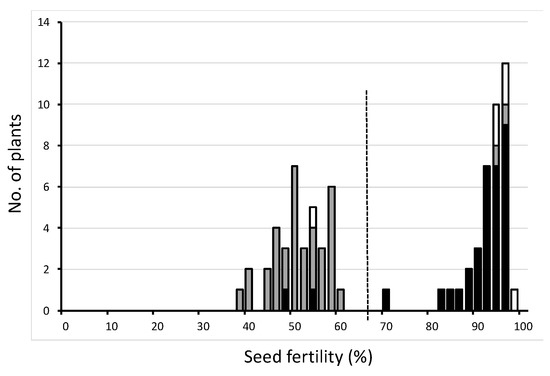
Figure 2.
Frequency distribution of seed fertility of one BC5F4 population using T65 as the recurrent parent and Jpn2 as the donor parent. Three classified genotypes were assessed for S13374 as indicated: white, homozygous for T65; gray, heterozygous; black, homozygous for Jpn2.
Then, we examined the BC5F5 lines shown in Table 3. We designed five PCR-based DNA markers located close to S13374 (Table 2) and checked the bulk DNA of each BC5F5 line for the genotypes of these DNA markers. Then we genotyped the six DNA markers in Table 2 of a total of 506 plants composed of BC5F5 line 1–9 to find recombinants between the DNA markers.

Table 3.
Genotypes of selected BC5F4 and seed fertility of BC5F5 generation classified by KGC1_1.24.
The seed fertility and genotypes of the DNA markers of the recombinants suggested that the causal gene of semi-sterility is located between KGC1_1.24 and KGC1_1.67 (Table 4). Heterozygotes of KGC1_1.24 showed semi-sterility and homozygotes of Jpn2 showed high fertility, except that line 7 individual 61 (hereafter abbreviated as 7-61) showed high fertility, though it was heterozygous at KGC1_1.24. Then 15 non-recombinants between the two markers of each BC5F5 line were evaluated for seed fertility. Hence, all data support the evidence that the causal gene was located close to KGC1_1.24 (Table 3).

Table 4.
Segregation of DNA markers of the progeny of the recombinants between KGC1_1.24 and KGC1_1.67 in BC5F5 and BC5F6 generation.
To confirm the location of the causal gene, the progeny of the above recombinants were genotyped for KGC1_1.24 or KGC1_1.67. Segregation distortion occurred in the upper three BC5F6 lines while it did not in the lower six BC5F6 lines in Table 4, which is consistent with the seed fertility of the previous generation. The genotypes of the causal sterility gene were the same as those of KGC1_1.24 for all the recombinants in Table 4 except 7-61 in that the genotype was the same as that of KGC1_1.67. About 30 plants of BC5F6 lines from 3-21, 7-61 and 3-57 were checked for seed fertility (Table 4). The distribution of seed fertility of each line was consistent with the inferred genotypes of the causal gene.
Based on the inference that the genotype of the causal gene was the same as KGC1_1.24 except 7-61 (Table 4), the haplotypes of 506 plants composed of BC5F5 line 1-9 (Table 3) surrounding the causal gene were classified, as shown in Table 5. As will be described in the discussion section, the causal gene was named S57. A total of 17 homozygotes of the T65 allele confirmed that the selective elimination of the T65 allele during egg development is incomplete. The linkage map of the causal gene was constructed based on Table 6 and genotypes of S13374, KGC1_0.61, KGC1_2.24 and KGC1_2.50 (Figure 3). The S57 gene was closely linked with KGC1_1.24, with a genetic distance of 0.1cM calculated by the Kosambi function.

Table 5.
Haplotypes around the segregation distortion region on chromosome 1 of BC5F5 using Jpn2 as the donor parent and T65 as the recurrent parent.

Table 6.
Segregation of progeny of BC5F5 heterozygous for KGC1_1.24 genotype.
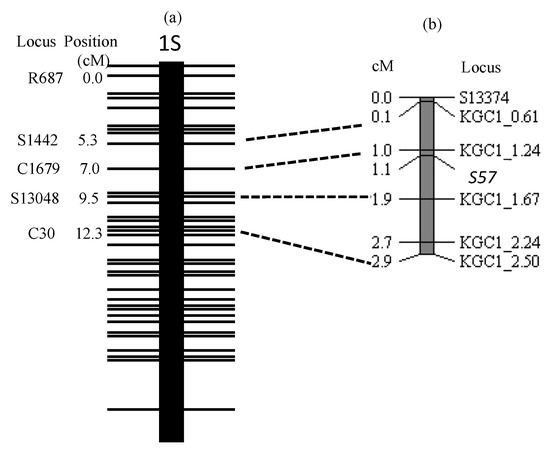
Figure 3.
Linkage map showing S57 gene on the short arm of chromosome 1. (a) RFLP framework map of the short arm of chromosome 1 modified from Harushima et al. [26]. (b) linkage map of S57 gene constructed from the BC5F5 population using Jpn2 as donor parent and T65 as the recurrent parent (n = 506). DNA markers near each other on Nipponbare pseudomolecules are connected by dotted lines.
2.3. Confirmation of Egg-Killer Phenomenon by Testcross
Apart from the BC5F5 lines for linkage analysis of the S57 gene, we separately planted about 15 plants each from the BC5F5 line 1-9. The three plants heterozygous at S13374 were reciprocally backcrossed with the recurrent parent to produce BC6F1 generation and selfed to produce BC5F6 generation. When T65 was used as a pollen donor, the genotype ratio of KGC1_1.24 deviated significantly from the expected ratio of 1:1 (Table 6). The BC5F6 generation also showed segregation distortion. On the other hand, when T65 was used as an egg donor, the genotype ratio of KGC1_1.24 of BC6F1 generation fitted the expected ratio. This result confirms that S57 is an egg-killer gene.
2.4. Distribution of the Egg-Killer Gene
In our previous paper [24], seed sterility caused by genes other than SDV1 was observed only in the cross with Jpn2. We inferred the genotype of the causal sterility gene in this study of another O. meridionalis strain W1297 and an Australian O. rufipogon strain Jpn1 [24] by the seed fertility pollinated by T65 pollen of the BC1F1, BC2F1 and BC3F1 generation. When using Jpn1 as the donor parent, average seed fertility remained above 69%, indicating that Jpn1 does not carry a causal gene of semi-sterility (Figure 4). When W1297 was used as donor parent, average seed fertility elevated as backcross times increased, and in the BC3F1 generation, the average seed fertility was 71% with the seed fertility of few plants less than 50%. This is in contrast to the case of Jpn2: where seed fertility of BC1F1 was 26%, and that of BC3F1 was 49% with 29 BC3F1plants showing seed fertility less than 50%. The S57 gene found in Jpn2 almost completely eliminates the egg carrying the T65 allele. Therefore, when backcrossed using T65 as the pollen donor, the Jpn2 allele tends to be transmitted preferentially and remains the cause of semi-sterility. Because no similar tendency as Jpn2 was observed in W1297, W1297 is thought to not carry a causal gene for hybrid seed sterility.
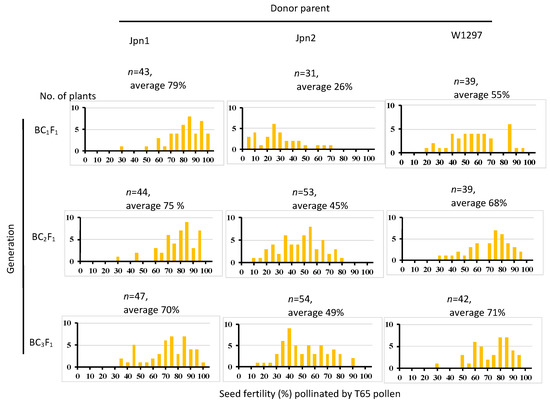
Figure 4.
Seed fertility pollinated by T65 of backcross generation using Jpn1, Jpn2 and W1297 as donor parents and T65 as the recurrent parent.
3. Discussion
In this study, seed semi-sterility observed in the backcross progeny using an O. meridionalis strain Jpn2 as donor parent and T65 as a recurrent parent was genetically analyzed. A gene from Jpn2 was the cause of semi-sterility. As shown in Figure 5, in heterozygous form, eggs carrying the T65 allele were incompletely sterilized. This gene exerts no effect on pollen sterilization. Therefore, this phenomenon can be explained by an egg-killer gene model. Linkage analysis indicated that this gene is located on the distal end of the short arm of chromosome 1 (Figure 3). Genes with similar functions were observed in intersubspecific crosses in Oryza sativa, such as S5 [11], S7 [12] and S9 [13]. According to [27], in the cross between O. sativa and O. meridionalis, only pollen-killer genes have been reported. Li et al. [23] performed linkage analysis of five pollen-killer genes: S51 located on chromosome 1, S52 and S53 located on chromosome 2, S54 and S55 on chromosome 7. Yu et al. [28] also identified the pollen-killer gene qSHM7 in the cross between O. sativa and O. meridionalis and located it on the same chromosomal location as S55. Furthermore, O. meridionalis alleles behaved as pollen-killer alleles at the S51 and S54 loci and behaved as abortive alleles at the S52, S53 and S55 (qHMS7) loci. To our knowledge, there have been no reports of egg-killer genes in this chromosomal region, and no report of those found in the cross between O. sativa and O. meridionalis. Two egg-killer genes were reported to be located on chromosome 1: ESA was detected in the cross between O. sativa and O. rufipogon. This gene was tightly linked with RM24, located at 18.9 Mb on chromosome 1 of IRGSP 1.0 pseudomolecule [29]. S40 was detected in the cross between O. sativa and O. longistaminata. This gene was tightly linked with RM575, located at 8.1 Mb [30]. Therefore, the egg-killer gene in this study is a new gene.
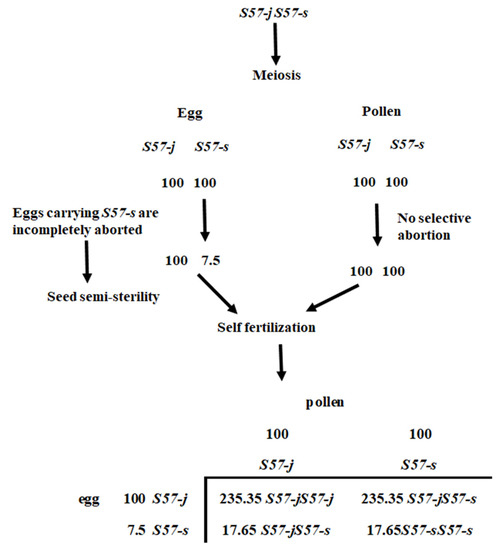
Figure 5.
Genetic model explaining the seed semi-sterility and segregation distortion in the genotype of S57-j S57-s at S57 locus. The transmission ratio of S57-s is inferred from observed data in Table 5.
In the history of rice gene nomenclature, S with digits have been applied to the gene symbols of both pollen sterility genes and egg sterility genes. According to a database of rice, Oryzabase (Oryzabase.Available online: https://shigen.nig.ac.jp/rice/oryzabase/locale/change?lang=en accessed on 31 December 2021) [31], and a recent review on hybrid sterility by Li et al. [27], S56(t) is the latest gene symbol, which was found in the cross between O. sativa and O. glumaepatula, and caused pollen semi-sterility [32]. Therefore, we name the gene found in this study as HYBRID SPIKELET STERILITY 57, with the gene symbol of S57, following Yoshimura and Nagato [33] and McCouch and CGSNL [34]. In the intraspecific crosses among O. sativa, there have been no reports of hybrid spikelet sterility phenomena caused by a gene located on the distal end of the short arm of chromosome 1. Therefore, all O. sativa is thought to share the same allele found in T65. This allele was called S57-s (s for sativa). The allele of Jpn2 was not shared by another O. meridionalis strain, W1297. This allele was called S57-j (j for Jpn2).
S57 is located in the 430.6 kb chromosomal region encompassed by the two DNA markers KGC1_1.24 and KGC1_.67. We are undertaking fine-mapping of the S57 gene to identify the causal DNA sequence diversifying the function of the S57 gene. According to Rice Genome Annotation Project (Rice Genome Annotation Project. Available online: http://rice.uga.edu/index.shtml accessed on 15 January 2022) [35], 69 genes with the gene name LOC_Os01g0**** are located on that region in O. sativa cultivar Nipponbare genome IRGSP 1.0 pseudomolecule [35]. We are undertaking fine-mapping of the S57 gene to identify the causal DNA sequence diversifying the function of the S57 gene. In the case of the S1 gene, the corresponding DNA sequence of killer allele in O. glaberrima is not present only in O. sativa genome [15]. After delimiting the candidate chromosomal region, we will search for the candidate gene in both O. sativa and O. meridionalis genomes. The project of sequencing the Jpn2 genome is underway to uncover S57, SDV1, SDV2 and long grain gene(s) from Jpn2 [36].
In a review by Calvo-Baltanás et al. [37] on hybrid incompatibility, genetic and molecular studies of hybrid incompatibility in numerous plant species revealed that such self-destructing symptoms in most cases are attributed to autoimmunity: plant immune responses are inadvertently activated in the absence of pathogenic invasion. Most of the hybrid incompatibility, such as hybrid weakness and hybrid necrosis, is explained by a conflict involving a member of the major plant immune receptor family, the nucleotide-binding domain and leucine-rich repeat-containing protein (NLR; formerly known as NBS-LRR). NLR genes are associated with disease resistance traits. According to [37], hybrid sterility can be explained in the same context. The S5 gene encodes a disease resistance-related aspartic protease (AP) [11]. Although the AP family has not yet been investigated in line with NLR activity, members of this family are involved in defense responses with activated salicylic acid in addition to pollen and ovule development in multiple plant species. Furthermore, the S5 gene is actually a gene complex of three open reading frames, ORF3, ORF4 and ORF5 [11]. ORF5 encodes AP. AP from indica S5 allele behaves as a killer. Because products from indica ORF3 behave as the protector from killer AP, eggs carrying indica S5 alleles are viable. A pollen-killer gene qHMS7 is actually composed of two tightly linked genes, ORF2 and ORF3 [28]. ORF2 encodes a ribosome-inactivating protein (RIP) domain-containing protein, which behaves as a toxic genetic element that aborts pollen in a sporophytic manner, whereas ORF3 is predicted to encode homologous, grass family-specific proteins with a mitochondrial targeting signal at the N terminus, which behaves as an antidote that protects pollen in a gametophytic manner. RIPs are toxic RNA N-glycosidases that affect translation processes and have been implicated in apoptotic pathways in mammalian cells and antiviral, antifungal and insecticidal activities in plants [38]. The above two abortion models involve proteins related to plant immunity. Therefore, the S57 egg-killer system might evolve from genes conferring plant immunity and be composed of tightly linked plural genes. Fine mapping of this gene will test the universality of killer-protector systems reported earlier.
According to [27], the S53 gene [23], a pollen-killer gene found in the cross between O. sativa and O. meridiolis, is located on the same location as other pollen-killer genes, S22A, S22B [39] and S29 [40] on the short arm of chromosome 2. In the cross between O. sativa and O. meridiolis, O. sativa allele behaves as pollen-killer and O. meridionalis allele as abortive allele at S53 locus. In the cross between O. sativa and O. glumaepatula, the O. sativa allele behaves as pollen-killer and O. glumaepatula allele as abortive allele at S22A and S22B loci. In the cross between O. sativa and O. glaberrima, the O. glaberrima allele behaves as pollen-killer and O. sativa allele as abortive allele at S29 locus. If S53, S29 and S22A or S22B are located on the same locus, the hierarchy of tentative allelic interaction would be that O. glaberrima carries the strongest egg-killer allele, followed by O. sativa, and O. meridionalis and O. glumaepatula carry abortive alleles. These kinds of allelic hierarchy could be detected by testcross: The progeny of homozygotes of S57-j of BC5F6 will be the tester lines for the allele of S57. Introgression lines of AA genome wild rice chromosomal segments under T65 genetic background were developed [41,42]. The testcross with homozygotes of S57-j and intercrosses among introgression lines carrying the introgressed chromosomal segments of the distal end of chromosome 1 would clarify the hierarchy of alleles at the S57 locus.
Lam et al. [22] reported that the hybrid between Jpn2 and W1297, that between Jpn2 and another O. meridionalis strain W1299, showed very low seed fertility and that the hybrid between W1297 and W1299 showed seed fertility comparable to parental lines. It suggests that W1299 also does not carry the egg-killer allele at S57. Additionally, W1297, W1299 and Jpn2 have originated from different places in Australia: W1297and W1299 are from the Northern Territory, and Jpn2 is from Queensland. According to Juliano et al. [43], most crosses between Northern Territory and Queensland accessions produced sterile hybrids, and DNA marker-based analyzes showed O. meridionalis genetic differentiation corresponding to geographic origin. In Oryza sativa, the carriers of the egg-killer allele and those of the abortive allele at S5 corresponded to indica and japonica [8]. The distribution of alleles of S57 in the wild populations of O. meridiolnais will test the role of the egg-killer gene in population differentiation in nature.
4. Materials and Methods
4.1. Plant Material
Three wild rice strains, Jpn1, Jpn2 and W1297, and one cultivated rice cultivar, Taichung 65 (T65) were used in this study. Detailed information on the three strains and the procedure of backcrossing has been already described in [24]. The number of plants in BC2F1 and BC3F1 generation in Figure 4 was larger than [24] because spare plants in each generation were planted and checked for seed fertility as T65 was used as a pollen donor. The year of cultivation, sowing date and transplanting date are shown in Table 7. Plant cultivation conditions followed Toyomoto et al. [24]: Germinated seeds were sown in nursery beds in a greenhouse. About two weeks after sowing, seedlings were transferred out of the greenhouse. Then, seedlings were planted in a paddy field at the Experimental Farm of Kagoshima University, Kagoshima, Japan.

Table 7.
Year of cultivation, sowing date and transplanting date of the materials in this study.
4.2. Trait Evaluation
The recording of the heading date and evaluation of seed fertility and pollen fertility followed Toyomoto et al. [24].
4.3. DNA Analysis
DNA from leaves was extracted according to Toyomoto et al. [24]. PCR mixture, cycle, electrophoresis, DNA staining and gel image documentation also followed Ichitani et al. [44]. The linkage map was constructed using Antmap [45]. Kosambi function was adopted to calculate map distance.
4.4. DNA Markers
For the rough mapping of the causal gene of hybrid sterility, 76 PCR-based DNA markers covering the whole chromosomes in K.I.’s lab were examined (data not shown).
For the linkage analysis of the S57 gene, new PCR-based DNA markers were designed because no available polymorphic DNA markers were closely located to S13374. Our strategy for designing co-dominant DNA markers was that using Nipponbare genome (GCF_001433945.1) [35] as a reference, insertion/deletion (indel) ranging from 5 to 100 base pairs shared by the two O. meridionalis accessions, W2112 (sequence information ID: SAMN02870169) [19] and IRGC105298 (SAMN03263073) [18], and not shared by the following AA genome species sequences was selected: O. barthii accession IRGC105608 (GCA_000182155.4) [19], O. glumaepatula accession GEN1233_2 ((SAMN02941241) [19], O. nivara accession IRGC100897 (SAMN02941243) [18], O. rufipogon accession IRGC100897 (SAMN02941243) [19], O. rufipogon accession W1943 (SAMEA1523624) [18], O. longistaminata accession W1413 (SAMD00003589) [46], O. longistaminata accession W1508 (SAMD00003590) [46], O. sativa cultivar Taicung 65 (SAMN03463201) [47] with the aid of TASUKE+ (https://ricegenome.dna.affrc.go.jp/ accessed online 15 December 2020) [48]. The selected indels were screened based on sequence similarity surrounding indels between genome assemblies of Nipponbare (Os-Nipponbare-Reference-IRGSP-1.0) and the O. meridionalis W2112 accession (GCA_000338895.3). The primer design followed Busung et al. [49]. The mismatch in primer sequences and genome sequences were checked using Oryzabase BLAST (https://shigen.nig.ac.jp/rice/oryzabase/locale/change?lang=en accessed online 15 December 2020) [31] and MBKBASE BLAST (http://mbkbase.org/rice/blastOL accessed online 15 December 2020) [50]. One base mismatch inside the primer sequence could be permitted, and we obtained enough PCR product.
5. Conclusions
A new egg-killer gene S57 was detected in the cross between O. sativa and O. meridionalis. This gene was located on the distal end of the short arm of chromosome 1. An O. meridionalis strain Jpn2 carried an egg-killer allele, and O. sativa cultivar Taichung 65 carried an abortive allele. Fine mapping of this gene will test the universality of killer-protector systems reported before. Not all O. meridionalis strains carry the egg-killer allele. Analysis of the distribution of alleles of S57 in the wild populations of O. meridiolnais will test the role of the egg-killer gene in population differentiation in nature.
Author Contributions
Conceptualization, K.I.; methodology, K.I.; validation, M.I., K.M., D.T. and M.U.; formal analysis, M.I., K.M., D.T., M.U. and K.I.; investigation, M.I., K.M., D.T. and M.U.; resources, S.T., T.S., R.H., R.I. and K.I.; data curation, M.I., K.M., D.T., M.U. and K.I.; writing—original draft preparation, K.I.; writing—review and editing, T.S., R.H., R.I. and K.I.; visualization, M.I., K.M., D.T., M.U. and K.I.; supervision, K.I.; project administration, R.I.; funding acquisition, R.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI Grant Number JP16H05777 from the Japan Society for the Promotion of Science.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Acknowledgments
We are grateful to the National Institute of Genetics for their kind provision of W1297 seeds. We thank Masaaki Ikenoue, Noboru Takasaki, Rieko Nomura, Yoko Nakashima, Asako Kobai, and all the members of the Plant Breeding Laboratory, Kagoshima University, Japan, for their technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ichitani, K.; Namigoshi, K.; Sato, M.; Taura, S.; Aoki, M.; Matsumoto, Y.; Saitou, T.; Marubashi, W.; Kuboyama, T. Fine mapping and allelic dosage effect of Hwc1, a complementary hybrid weakness gene in rice. Theor. Appl. Genet. 2007, 114, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Kuboyama, T.; Saito, T.; Matsumoto, T.; Wu, J.; Kanamori, H.; Taura, S.; Sato, M.; Marubashi, W.; Ichitani, K. Fine mapping of HWC2, a complementary hybrid weakness gene, and haplotype analysis around the locus in rice. Rice 2009, 2, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Ichitani, K.; Taura, S.; Tezuka, T.; Okiyama, Y.; Kuboyama, T. Chromosomal location of HWA1 and HWA2, complementary hybrid weakness genes in rice. Rice 2011, 4, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Ichitani, K.; Takemoto, Y.; Iiyama, K.; Taura, S.; Sato, M. Chromosomal location of HCA1 and HCA2, hybrid chlorosis genes in rice. Int. J. Plant Genom. 2012, 2012, 649081. [Google Scholar] [CrossRef]
- Nadir, S.; Khan, S.; Zhu, Q.; Henry, D.; Wei, L.; Lee, D.S.; Chen, L. An overview on reproductive isolation in Oryza sativa Complex. AoB Plants 2018, 10, ply060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stebbins, G.L. The inviability, weakness, and sterility of interspecic hybrids. Adv. Genet. 1958, 9, 147–215. [Google Scholar]
- Orr, H.A. Dobzhansky, Bateson, and the genetics of speciation. Genetics 1996, 144, 1331–1335. [Google Scholar] [CrossRef]
- Ouyang, Y.; Liu, Y.-G.; Zhang, Q. Hybrid sterility in plant: Stories from rice. Curr. Opin. Plant Biol. 2010, 13, 186–192. [Google Scholar] [CrossRef]
- Ouyang, Y.; Zhang, Q. Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol. 2013, 64, 111–135. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G. Prospects of Utilization of inter-subspecific heterosis between indica and japonica rice. J. Integr. Agric. 2020, 19, 1–10. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, X.; Cheng, K.; Du, H.; Ouyang, Y.; Chen, J.; Qiu, S.; Huang, J.; Jiang, Y.; Jiang, L.; et al. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 2012, 337, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhao, Z.; Shi, Y.; Tian, H.; Liu, L.; Bian, X.; Xu, Y.; Zheng, X.; Gan, L.; Shen, Y.; et al. Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein. Genetics 2016, 203, 1439–1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Yamaguchi, Y.; Kato, H.; Ikehashi, H. Two new loci for hybrid sterility in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 1996, 92, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Koide, Y.; Ogino, A.; Yoshikawa, T.; Kitashima, Y.; Saito, N.; Kanaoka, Y.; Onishi, K.; Yoshitake, Y.; Tsukiyama, T.; Saito, H.; et al. Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice. Proc. Natl. Acad. Sci. USA 2018, 115, E1955–E1962. [Google Scholar] [CrossRef] [Green Version]
- Koide, Y.; Onishi, K.; Kanazawa, A.; Sano, Y. Genetics of speciation in rice. In Rice Biology in the Genomics Era; Hirano, H.-Y., Sano, Y., Hirai, A., Sasaki, T., Eds.; Biotechnology in Agriculture and Forestry; Springer: Berlin/Heidelberg, Germany, 2008; pp. 247–259. ISBN 978-3-540-74250-0. [Google Scholar]
- Zin Mar, M.; Koide, Y.; Ogata, M.; Kuniyoshi, D.; Tokuyama, Y.; Hikichi, K.; Obara, M.; Kishima, Y. Genetic mapping of the gamete eliminator locus, S2, causing hybrid sterility and transmission ratio distortion found between Oryza sativa and Oryza glaberrima cross combination. Agriculture 2021, 11, 268. [Google Scholar] [CrossRef]
- Henry, R.J.; Rice, N.; Waters, D.L.E.; Kasem, S.; Ishikawa, R.; Hao, Y.; Dillon, S.; Crayn, D.; Wing, R.; Vaughan, D. Australian Oryza: Utility and conservation. Rice 2010, 3, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-J.; Zhu, T.; Xia, E.-H.; Shi, C.; Liu, Y.-L.; Zhang, Y.; Liu, Y.; Jiang, W.-K.; Zhao, Y.-J.; Mao, S.-Y.; et al. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, E4954–E4962. [Google Scholar] [CrossRef] [Green Version]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zwickl, D.J.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, D.; Iwata, A.; Goicoechea, J.L.; et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef]
- Naredo, M.E.B.; Juliano, A.B.; Lu, B.-R.; Jackson, M.T. Hybridization of AA genome rice species from Asia and Australia I. Crosses and development of hybrids. Genet. Resour. Crop. Evol. 1997, 44, 17–23. [Google Scholar] [CrossRef]
- Naredo, M.E.B.; Juliano, A.B.; Lu, B.-R.; Jackson, M.T. Taxonomic status of Oryza glumaepatula Steud. II. Hybridization between NewWorld diploids and AA genome species from Asia and Australia. Genet. Resour. Crop. Evol. 1998, 45, 205–214. [Google Scholar] [CrossRef]
- Lam, D.T.; Ichitani, K.; Henry, R.J.; Ishikawa, R. Molecular and morphological divergence of australian wild rice. Plants 2020, 9, E224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhou, J.; Xu, P.; Deng, X.; Deng, W.; Zhang, Y.; Yang, Y.; Tao, D. Mapping five novel interspecific hybrid sterility loci between Oryza sativa and Oryza meridionalis. Breed. Sci. 2018, 68, 516–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toyomoto, D.; Uemura, M.; Taura, S.; Sato, T.; Henry, R.; Ishikawa, R.; Ichitani, K. segregation distortion observed in the progeny of crosses between Oryza sativa and O. meridionalis caused by abortion during seed development. Plants 2019, 8, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harushima, Y.; Yano, M.; Shomura, A.; Sato, M.; Shimano, T.; Kuboki, Y.; Yamamoto, T.; Lin, S.Y.; Antonio, B.A.; Parco, A.; et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 1998, 148, 479–494. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, J.; Zhang, Y.; Yang, Y.; Pu, Q.; Tao, D. New insights into the nature of interspecific hybrid sterility in rice. Front Plant Sci. 2020, 11, 555572. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, Z.; Zheng, X.; Zhou, J.; Kong, W.; Wang, P.; Bai, W.; Zheng, H.; Zhang, H.; Li, J.; et al. A selfish genetic element confers non-mendelian inheritance in rice. Science 2018, 360, 1130–1132. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Cao, C.; Ruan, Y.; Deng, Y.; Liu, Y.; Zhang, K.; Tan, L.; Zhu, Z.; Cai, H.; Liu, F.; et al. ESA1 Is involved in embryo sac abortion in interspecific hybrid progeny of rice. Plant Physiol. 2019, 180, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, Z.; Liu, L.; Kong, W.; Lin, Y.; You, S.; Bai, W.; Xiao, Y.; Zheng, H.; Jiang, L.; et al. Genetic analysis of a hybrid sterility gene that causes both pollen and embryo sac sterility in hybrids between Oryza sativa L. and Oryza longistaminata. Heredity 2017, 119, 166–173. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, Y.; Sakaniwa, S.; Tsuchiya, R.; Nonomura, K.-I.; Kurata, N. Oryzabase: An integrated information resource for rice science. Breed. Sci. 2010, 60, 544–548. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhou, J.; Li, J.; Yang, Y.; Xu, P.; Tao, D. Mapping of S56(t) responsible for interspecific hybrid sterility between Oryza sativa and Oryza glumaepatula. Breed. Sci. 2018, 68, 242–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimura, A.; Nagato, Y. Report of the committee on gene symbolizatiion, nomenclature and linkage groups. Rice Genet. Newsl. 2008, 24, 7–9. [Google Scholar]
- McCouch, S.R.; CGSNL. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Sotowa, M.; Ootsuka, K.; Kobayashi, Y.; Hao, Y.; Tanaka, K.; Ichitani, K.; Flowers, J.M.; Purugganan, M.D.; Nakamura, I.; Sato, Y.-I.; et al. Molecular relationships between Australian annual wild rice, Oryza meridionalis, and two related perennial forms. Rice 2013, 6, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo-Baltanás, V.; Wang, J.; Chae, E. Hybrid Incompatibility of the Plant Immune System: An opposite force to heterosis equilibrating hybrid performances. Front. Plant Sci. 2021, 11, 576796. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Boston, R.S. Ribosome-Inactivating Proteins: A plant perspective. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 2001, 52, 785–816. [Google Scholar] [CrossRef] [Green Version]
- Sakata, M.; Yamagata, Y.; Doi, K.; Yoshimura, A. Two linked genes on rice chromosome 2 for F1 pollen sterility in a hybrid between Oryza sativa and O. glumaepatula. Breed. Sci. 2014, 64, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Xu, P.; Deng, X.; Zhou, J.; Li, J.; Tao, D. Molecular mapping of a pollen killer gene S29(t) in Oryza glaberrima and co-linear analysis with S22 in O. glumaepatula. Euphytica 2006, 151, 273–278. [Google Scholar] [CrossRef]
- Yoshimura, A.; Nagayama, H.; Sobrizal; Kurakazu, T.; Sanchez, P.L.; Doi, K.; Yamagata, Y.; Yasui, H. Introgression lines of rice (Oryza sativa L.) carrying a donor genome from the wild species, O. glumaepatula Steud. and O. meridionalis Ng. Breed. Sci. 2010, 60, 597–603. [Google Scholar] [CrossRef] [Green Version]
- Ogami, T.; Yasui, H.; Yoshimura, A.; Yamagata, Y. Identification of anther length QTL and construction of chromosome segment substitution lines of Oryza longistaminata. Plants 2019, 8, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juliano, A.B.; Naredo, M.E.B.; Lu, B.-R.; Jackson, M.T. Genetic differentiation in Oryza meridionalis Ng based on molecular and crossability analyses. Genet. Resour. Crop. Evol. 2005, 52, 435–445. [Google Scholar] [CrossRef]
- Ichitani, K.; Yamaguchi, D.; Taura, S.; Fukutoku, Y.; Onoue, M.; Shimizu, K.; Hashimoto, F.; Sakata, Y.; Sato, M. Genetic analysis of ion-beam induced extremely late heading mutants in rice. Breed. Sci. 2014, 64, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, H.; Ninomiya, S. AntMap: Constructing genetic linkage maps using an ant colony optimization algorithm. Breed. Sci. 2006, 56, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Ohyanagi, H.; Ebata, T.; Huang, X.; Gong, H.; Fujita, M.; Mochizuki, T.; Toyoda, A.; Fujiyama, A.; Kaminuma, E.; Nakamura, Y.; et al. OryzaGenome: Genome diversity database of wild Oryza species. Plant Cell Physiol. 2016, 57, e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.-J.; Tsai, Y.-C.; Wu, H.-P.; Huang, L.-T.; Chen, Y.-C.; Chen, Y.-F.; Wu, C.-C.; Tseng, Y.-T.; Hsing, Y.-I.C. Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 2016, 242, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, M.; Nishikawa, D.; Kawahara, Y.; Wakimoto, H.; Itoh, R.; Tabei, N.; Tanaka, T.; Itoh, T. TASUKE+: A web-based platform for exploring genome-wide association studies results and large-scale resequencing data. DNA Res. 2019, 26, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busungu, C.; Taura, S.; Sakagami, J.-I.; Ichitani, K. Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 2016, 66, 636–645. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Wang, K.; Chen, Z.; Cao, Y.; Gao, Q.; Li, Y.; Li, X.; Lu, H.; Du, H.; Lu, M.; et al. MBKbase for rice: An integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Research 2019, gkz921. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).