Effect of Genetically Reduced Maternal Myostatin on Late Gestation Maternal, Fetal, and Placental Metabolomes in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Intraperitoneal Glucose Tolerance Testing (IPGTT)
2.3. Insulin and Triglyceride Assays
2.4. Metabolomics
2.5. Statistical Analysis
3. Results
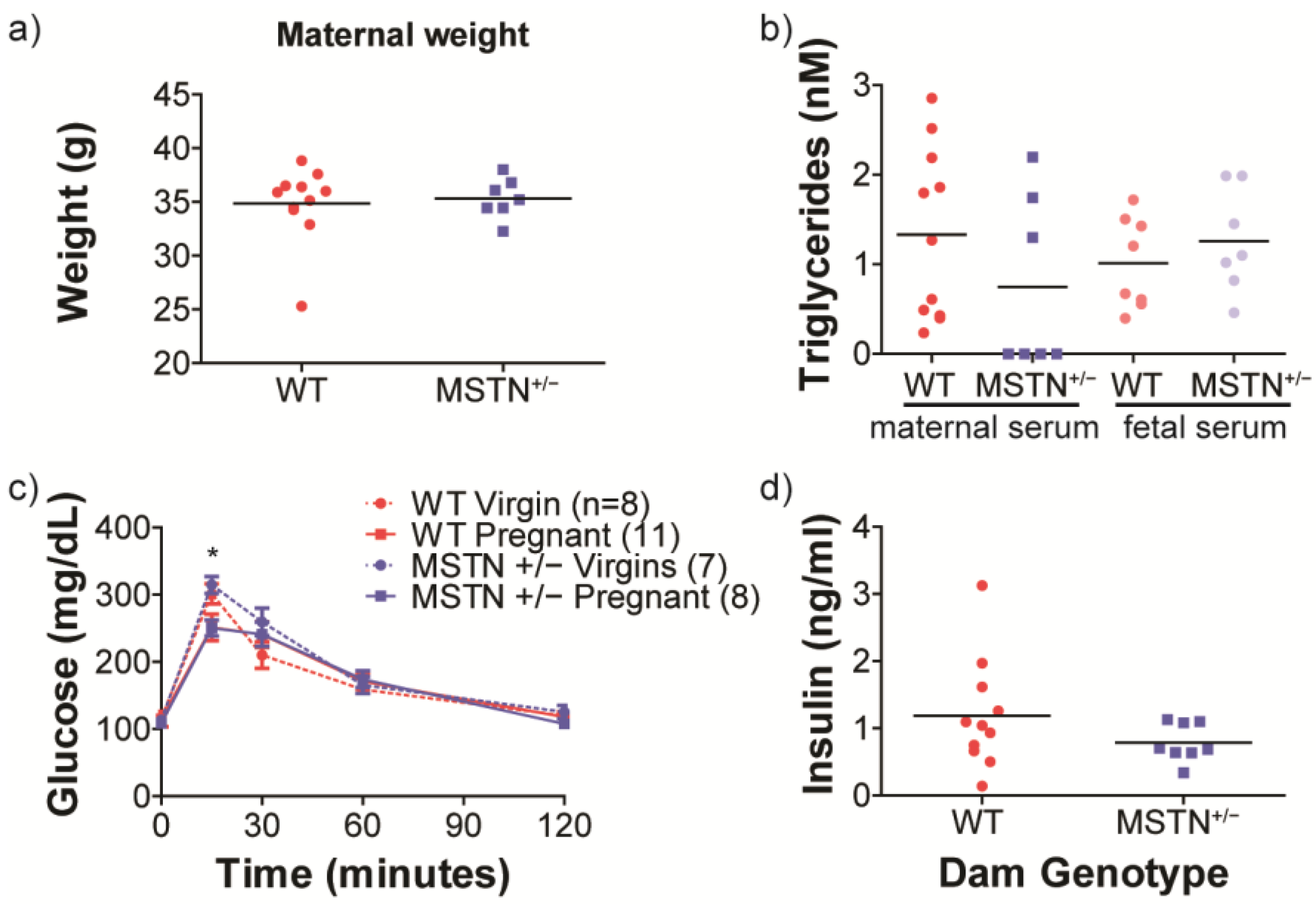
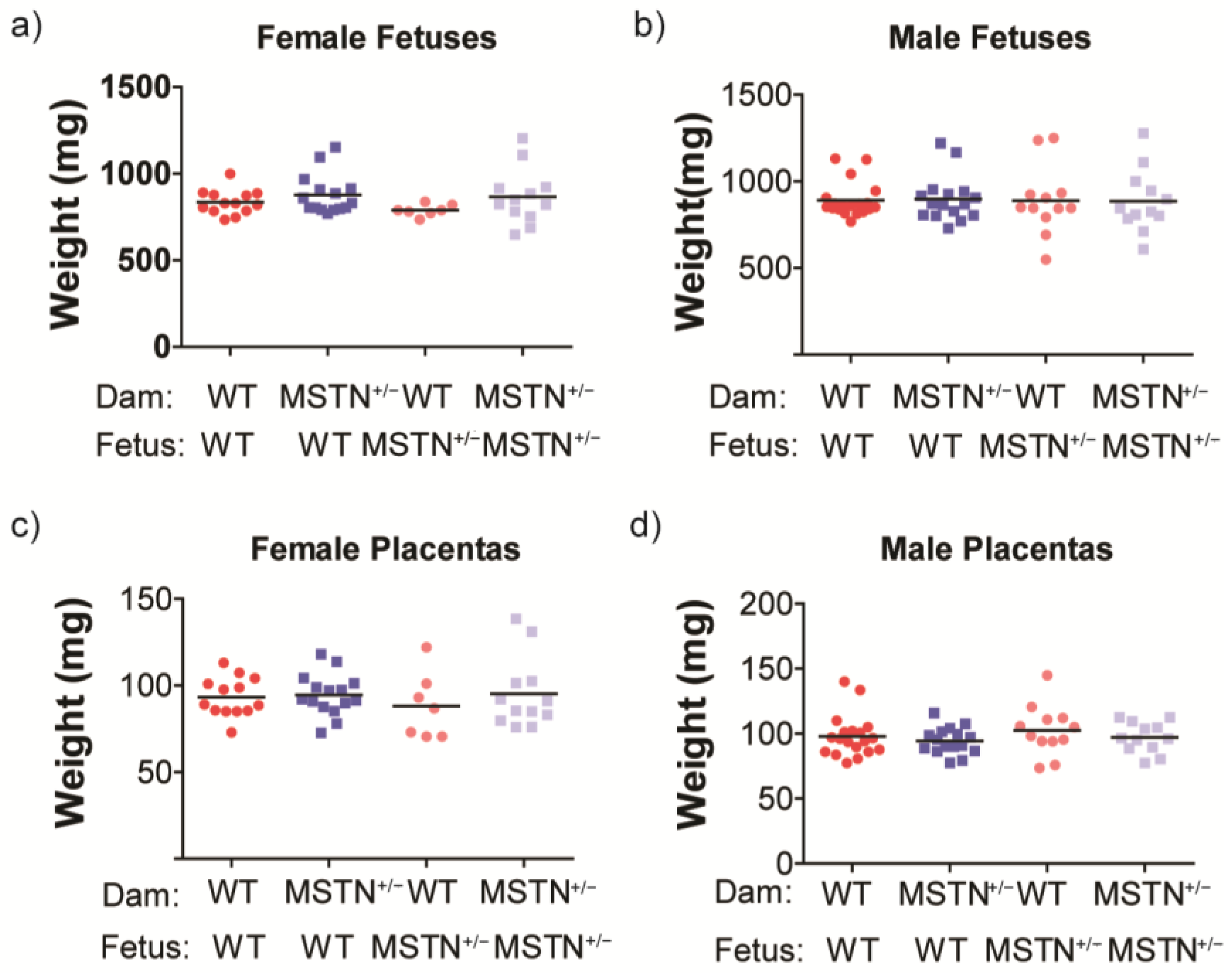
3.1. Growth and Maternal Metabolic Assessment
3.2. Global Metabolomic Assessment
3.3. Maternal Sera Metabolite Differences Based on Maternal Myostatin Genotype
3.4. Fetal Sera Metabolite Differences Based on Maternal Myostatin Genotype
3.5. Placental Metabolite Differences Based on Maternal Myostatin Genotype
3.6. Lipid Metabolism
3.7. Purine Metabolism Differences Based on Maternal Myostatin Genotype
3.8. Branched-Chain Amino Acids
3.9. Vitamins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, L.D.; Hay, W.W., Jr. Impact of placental insufficiency on fetal skeletal muscle growth. Mol. Cell. Endocrinol. 2016, 435, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Salam, R.A.; Das, J.K.; Bhutta, Z.A. Impact of intrauterine growth restriction on long-term health. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Romano, T.; Wark, J.D.; Wlodek, M.E. Developmental programming of bone deficits in growth-restricted offspring. Reprod. Fertil. Dev. 2015, 27, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Cuthbertson, D.; Nagamani, S.C.S.; Sutton, V.R.; Lee, B.H.; Krischer, J.; Krakow, D. Pregnancy in women with osteogenesis imperfecta: Pregnancy characteristics, maternal, and neonatal outcomes. Am. J. Obs. Gynecol. MFM 2021, 3, 100362. [Google Scholar] [CrossRef]
- Regnault, T.R.; Galan, H.L.; Parker, T.A.; Anthony, R.V. Placental development in normal and compromised pregnancies—A review. Placenta 2002, 23 (Suppl. SA), S119–S129. [Google Scholar] [CrossRef]
- Hay, W.W., Jr.; Brown, L.D.; Rozance, P.J.; Wesolowski, S.R.; Limesand, S.W. Challenges in nourishing the intrauterine growth-restricted foetus—Lessons learned from studies in the intrauterine growth-restricted foetal sheep. Acta Paediatr. 2016, 105, 881–889. [Google Scholar] [CrossRef]
- Jansson, T.; Powell, T.L. Role of the placenta in fetal programming: Underlying mechanisms and potential interventional approaches. Clin. Sci. 2007, 113, 1–13. [Google Scholar] [CrossRef]
- Lee, S.J.; McPherron, A.C. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA 2001, 98, 9306–9311. [Google Scholar] [CrossRef]
- Lee, S.J. Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2007, 2, e789. [Google Scholar] [CrossRef]
- Salzler, R.R.; Shah, D.; Dore, A.; Bauerlein, R.; Miloscio, L.; Latres, E.; Papadopoulos, N.J.; Olson, W.C.; MacDonald, D.; Duan, X. Myostatin deficiency but not anti-myostatin blockade induces marked proteomic changes in mouse skeletal muscle. Proteomics 2016, 16, 2019–2027. [Google Scholar] [CrossRef]
- Girgenrath, S.; Song, K.; Whittemore, L.A. Loss of myostatin expression alters fiber-type distribution and expression of myosin heavy chain isoforms in slow- and fast-type skeletal muscle. Muscle Nerve 2005, 31, 34–40. [Google Scholar] [CrossRef]
- Oestreich, A.K.; Kamp, W.M.; McCray, M.G.; Carleton, S.M.; Karasseva, N.; Lenz, K.L.; Jeong, Y.; Daghlas, S.A.; Yao, X.; Wang, Y.; et al. Decreasing maternal myostatin programs adult offspring bone strength in a mouse model of osteogenesis imperfecta. Proc. Natl. Acad. Sci. USA 2016, 113, 13522–13527. [Google Scholar] [CrossRef] [PubMed]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Gentry, B.A.; Ferreira, J.A.; Phillips, C.L.; Brown, M. Hindlimb skeletal muscle function in myostatin-deficient mice. Muscle Nerve 2011, 43, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.F.; Li, N.; Rodgers, B.D. Myostatin regulates tissue potency and cardiac calcium-handling proteins. Endocrinology 2014, 155, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- AMDCC. Intraperitoneal Glucose Tolerance Testing (IPGTT). Available online: http://www.diacomp.org/shared/showFile.aspx?doctypeid=3&docid=11 (accessed on 27 March 2023).
- Tunster, S.J. Genetic sex determination of mice by simplex PCR. Biol. Sex. Differ. 2017, 8, 31. [Google Scholar] [CrossRef]
- Guo, L.; Milburn, M.V.; Ryals, J.A.; Lonergan, S.C.; Mitchell, M.W.; Wulff, J.E.; Alexander, D.C.; Evans, A.M.; Bridgewater, B.; Miller, L.; et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc. Natl. Acad. Sci. USA 2015, 112, E4901–E4910. [Google Scholar] [CrossRef]
- DeHaven, C.D.; Evans, A.M.; Dai, H.P.; Lawton, K.A. Software Techniques for Enabling High-Throughput Analysis of Metabolomic Datasets. In Metabolomics; Intech: London, UK, 2012; pp. 167–192. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Roszkowska, A.; Klimek, J.; Kaletha, K. Expression patterns of AMP-deaminase and cytosolic 5′-nucleotidase genes in human term placenta. Mol. Cell. Biochem. 2008, 311, 249–251. [Google Scholar] [CrossRef]
- Bowers-Komro, D.M.; McCormick, D.B.; King, G.A.; Sweeny, J.G.; Iacobucci, G.A. Confirmation of 2-O-methyl ascorbic acid as the product from the enzymatic methylation of L-ascorbic acid by catechol-O-methyltransferase. Int. J. Vitam. Nutr. Res. 1982, 52, 186–193. [Google Scholar]
- Barnea, E.R.; MacLusky, N.J.; DeCherney, A.H.; Naftolin, F. Catechol-o-methyl transferase activity in the human term placenta. Am. J. Perinatol. 1988, 5, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, S.; Bupp, C.P.; Leimanis-Laurens, M.; Shukla, A.; Russell, C.; Junewick, J.; Gleason, E.; VanSickle, E.A.; Edgerly, Y.; Wittmann, B.M.; et al. Repurposing eflornithine to treat a patient with a rare ODC1 gain-of-function variant disease. eLife 2021, 10, e67097. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Wang, X.; Yin, Y.; Li, X.; Gao, H.; Bazer, F.W.; Wu, G. Putrescine stimulates the mTOR signaling pathway and protein synthesis in porcine trophectoderm cells. Biol. Reprod. 2014, 91, 106. [Google Scholar] [CrossRef] [PubMed]
- Pendeville, H.; Carpino, N.; Marine, J.C.; Takahashi, Y.; Muller, M.; Martial, J.A.; Cleveland, J.L. The ornithine decarboxylase gene is essential for cell survival during early murine development. Mol. Cell. Biol. 2001, 21, 6549–6558. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Chi, Y.J.; Yu, Y.S.; Liu, J.L.; Su, R.W.; Ma, X.H.; Shan, C.H.; Yang, Z.M. Polyamines are essential in embryo implantation: Expression and function of polyamine-related genes in mouse uterus during peri-implantation period. Endocrinology 2008, 149, 2325–2332. [Google Scholar] [CrossRef]
- Fozard, J.R.; Part, M.L.; Prakash, N.J.; Grove, J. Inhibition of murine embryonic development by alpha-difluoromethylornithine, an irreversible inhibitor of ornithine decarboxylase. Eur. J. Pharm. 1980, 65, 379–391. [Google Scholar] [CrossRef]
- Fenelon, J.C.; Murphy, B.D. Inhibition of polyamine synthesis causes entry of the mouse blastocyst into embryonic diapause. Biol. Reprod. 2017, 97, 119–132. [Google Scholar] [CrossRef]
- Fenelon, J.C.; Banerjee, A.; Lefevre, P.; Gratian, F.; Murphy, B.D. Polyamine-Mediated Effects of Prolactin Dictate Emergence from Mink Obligate Embryonic Diapause. Biol. Reprod. 2016, 95, 6. [Google Scholar] [CrossRef]
- Hosoi, T.; Yakabe, M.; Sasakawa, H.; Sasako, T.; Ueki, K.; Kato, S.; Tokuoka, S.M.; Oda, Y.; Abe, M.; Matsumoto, T.; et al. Sarcopenia phenotype and impaired muscle function in male mice with fast-twitch muscle-specific knockout of the androgen receptor. Proc. Natl. Acad. Sci. USA 2023, 120, e2218032120. [Google Scholar] [CrossRef]
- Lee, N.K.; Skinner, J.P.; Zajac, J.D.; MacLean, H.E. Ornithine decarboxylase is upregulated by the androgen receptor in skeletal muscle and regulates myoblast proliferation. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E172–E179. [Google Scholar] [CrossRef]
- McCann, M.R.; George De la Rosa, M.V.; Rosania, G.R.; Stringer, K.A. L-Carnitine and Acylcarnitines: Mitochondrial Biomarkers for Precision Medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef]
- Shekhawat, P.; Bennett, M.J.; Sadovsky, Y.; Nelson, D.M.; Rakheja, D.; Strauss, A.W. Human placenta metabolizes fatty acids: Implications for fetal fatty acid oxidation disorders and maternal liver diseases. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E1098–E1105. [Google Scholar] [CrossRef]
- Ramanathan, R.; Ibdah, J.A. Mitochondrial Dysfunction and Acute Fatty Liver of Pregnancy. Int. J. Mol. Sci. 2022, 23, 3595. [Google Scholar] [CrossRef]
- Xin, X.B.; Yang, S.P.; Li, X.; Liu, X.F.; Zhang, L.L.; Ding, X.B.; Zhang, S.; Li, G.P.; Guo, H. Proteomics insights into the effects of MSTN on muscle glucose and lipid metabolism in genetically edited cattle. Gen. Comp. Endocrinol. 2020, 291, 113237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; McFarlane, C.; Lokireddy, S.; Masuda, S.; Ge, X.; Gluckman, P.D.; Sharma, M.; Kambadur, R. Inhibition of myostatin protects against diet-induced obesity by enhancing fatty acid oxidation and promoting a brown adipose phenotype in mice. Diabetologia 2012, 55, 183–193. [Google Scholar] [CrossRef]
- Tan, S.T.; Ramesh, T.; Toh, X.R.; Nguyen, L.N. Emerging roles of lysophospholipids in health and disease. Prog. Lipid Res. 2020, 80, 101068. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Nanayakkara, G.; Cheng, J.; Cueto, R.; Yang, W.Y.; Park, J.Y.; Wang, H.; Yang, X. Lysophospholipids and Their Receptors Serve as Conditional DAMPs and DAMP Receptors in Tissue Oxidative and Inflammatory Injury. Antioxid. Redox Signal. 2018, 28, 973–986. [Google Scholar] [CrossRef]
- Wu, X.; Ma, Y.; Su, N.; Shen, J.; Zhang, H.; Wang, H. Lysophosphatidic acid: Its role in bone cell biology and potential for use in bone regeneration. Prostaglandins Other Lipid Mediat. 2019, 143, 106335. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Zheng, Y.; Hu, M.; Xu, P.; Lin, L.; Liu, X.; Wu, Y.; Huang, B.; Ye, X.; Li, S.; et al. Impaired Sphingosine-1-Phosphate Synthesis Induces Preeclampsia by Deactivating Trophoblastic YAP (Yes-Associated Protein) Through S1PR2 (Sphingosine-1-Phosphate Receptor-2)-Induced Actin Polymerizations. Hypertension 2022, 79, 399–412. [Google Scholar] [CrossRef]
- Nagamatsu, T.; Iwasawa-Kawai, Y.; Ichikawa, M.; Kawana, K.; Yamashita, T.; Osuga, Y.; Fujii, T.; Schust, D.J. Emerging roles for lysophospholipid mediators in pregnancy. Am. J. Reprod. Immunol. 2014, 72, 182–191. [Google Scholar] [CrossRef]
- Gil-Sanchez, A.; Demmelmair, H.; Parrilla, J.J.; Koletzko, B.; Larque, E. Mechanisms involved in the selective transfer of long chain polyunsaturated Fatty acids to the fetus. Front. Genet. 2011, 2, 57. [Google Scholar] [CrossRef]
- Blackburn, M.R.; Wakamiya, M.; Caskey, C.T.; Kellems, R.E. Tissue-specific rescue suggests that placental adenosine deaminase is important for fetal development in mice. J. Biol. Chem. 1995, 270, 23891–23894. [Google Scholar] [CrossRef]
- Blackburn, M.R.; Knudsen, T.B.; Kellems, R.E. Genetically engineered mice demonstrate that adenosine deaminase is essential for early postimplantation development. Development 1997, 124, 3089–3097. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, J.; Espinoza, A.F.; Power, G.G. High fetal plasma adenosine concentration: A role for the fetus in preeclampsia? Am. J. Obs. Gynecol. 2011, 205, 485.e24–485.e27. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.I.; Wesolowski, S.R.; Gilje, E.A.; Baker, P.R., 2nd; Reisz, J.A.; D’Alessandro, A.; Hay, W.W., Jr.; Rozance, P.J.; Brown, L.D. Skeletal muscle amino acid uptake is lower and alanine production is greater in late gestation intrauterine growth restricted fetal sheep hindlimb. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 317, R615–R629. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.H.; Wu, C.P.; Chen, S.F. Differential Changes in Akt and AMPK Phosphorylation Regulating mTOR Activity in the Placentas of Pregnancies Complicated by Fetal Growth Restriction and Gestational Diabetes Mellitus with Large-For-Gestational Age Infants. Front. Med. 2021, 8, 788969. [Google Scholar] [CrossRef] [PubMed]
- Lorca, R.A.; Houck, J.A.; Laurent, L.C.; Matarazzo, C.J.; Baker, K.; Horii, M.; Nelson, K.K.; Bales, E.S.; Euser, A.G.; Parast, M.M.; et al. High altitude regulates the expression of AMPK pathways in human placenta. Placenta 2021, 104, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Fukami, T.; Sun, X.; Li, T.; Desai, M.; Ross, M.G. Mechanism of programmed obesity in intrauterine fetal growth restricted offspring: Paradoxically enhanced appetite stimulation in fed and fasting states. Reprod. Sci. 2012, 19, 423–430. [Google Scholar] [CrossRef]
- Gu, M.; Wei, Z.; Wang, X.; Gao, Y.; Wang, D.; Liu, X.; Bai, C.; Su, G.; Yang, L.; Li, G. Myostatin Knockout Affects Mitochondrial Function by Inhibiting the AMPK/SIRT1/PGC1alpha Pathway in Skeletal Muscle. Int. J. Mol. Sci. 2022, 23, 13703. [Google Scholar] [CrossRef]
- Coats, L.E.; Bakrania, B.A.; Bamrick-Fernandez, D.R.; Ariatti, A.M.; Rawls, A.Z.; Ojeda, N.B.; Alexander, B.T. Soluble guanylate cyclase stimulation in late gestation does not mitigate asymmetric intrauterine growth restriction or cardiovascular risk induced by placental ischemia in the rat. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H1923–H1934. [Google Scholar] [CrossRef]
- Beaumann, M.; Delhaes, F.; Menetrey, S.; Joye, S.; Vial, Y.; Baud, D.; Magaly, J.G.; Tolsa, J.F.; Peyter, A.C. Intrauterine growth restriction is associated with sex-specific alterations in the nitric oxide/cyclic GMP relaxing pathway in the human umbilical vein. Placenta 2020, 93, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Gould, B.S.; Woessner, J.F. Biosynthesis of collagen; the influence of ascorbic acid on the proline, hydroxyproline, glycine, and collagen content of regenerating guinea pig skin. J. Biol. Chem. 1957, 226, 289–300. [Google Scholar] [CrossRef]
- Conaway, H.H.; Henning, P.; Lerner, U.H. Vitamin a metabolism, action, and role in skeletal homeostasis. Endocr. Rev. 2013, 34, 766–797. [Google Scholar] [CrossRef] [PubMed]
- Balasuriya, C.N.D.; Larose, T.L.; Mosti, M.P.; Evensen, K.A.I.; Jacobsen, G.W.; Thorsby, P.M.; Stunes, A.K.; Syversen, U. Maternal serum retinol, 25(OH)D and 1,25(OH)2D concentrations during pregnancy and peak bone mass and trabecular bone score in adult offspring at 26-year follow-up. PLoS ONE 2019, 14, e0222712. [Google Scholar] [CrossRef] [PubMed]
- Jishage, K.; Tachibe, T.; Ito, T.; Shibata, N.; Suzuki, S.; Mori, T.; Hani, T.; Arai, H.; Suzuki, H. Vitamin E is essential for mouse placentation but not for embryonic development itself. Biol. Reprod. 2005, 73, 983–987. [Google Scholar] [CrossRef]
- Oestreich, A.K.; Carleton, S.M.; Yao, X.; Gentry, B.A.; Raw, C.E.; Brown, M.; Pfeiffer, F.M.; Wang, Y.; Phillips, C.L. Myostatin deficiency partially rescues the bone phenotype of osteogenesis imperfecta model mice. Osteoporos. Int. 2016, 27, 161–170. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Suppression of body fat accumulation in myostatin-deficient mice. J. Clin. Investig. 2002, 109, 595–601. [Google Scholar] [CrossRef]

| ANOVA Contrast | WT Fetal WT Maternal | MSTN+/− Fetal MSTN+/− Maternal | MSTN+/− Maternal WT Maternal | MSTN+/− Fetal WT Fetal | MSTN+/− Placenta WT Placenta |
|---|---|---|---|---|---|
| Total biochemicals p < 0.05 | 704 | 682 | 33 | 50 | 27 |
| Biochemicals ↑|↓ | 393|311 | 412|270 | 17|16 | 40|10 | 4|23 |
| MSTN+/− vs. WT | |||
|---|---|---|---|
| Biochemical Name | Ratio | p-Value | q-Value |
| cysteine sulfinic acid | 6.26 | 0.0367 | 0.9906 |
| glu-gly-asn-val ** | 2.28 | 0.0456 | 0.9906 |
| Erythronate * | 2.16 | 0.0422 | 0.9906 |
| 3-bromo-5-chloro-2,6-dihydroxybenzoic acid * | 2.09 | 0.0068 | 0.9906 |
| 2,3-dihydroxyisovalerate | 1.90 | 0.0469 | 0.9906 |
| heptanoate (7:0) | 1.82 | 0.0283 | 0.9906 |
| N,N,N-trimethyl-alanylproline betaine (TMAP) | 1.68 | 0.0175 | 0.9906 |
| lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) | 1.62 | 0.0216 | 0.9906 |
| dodecanedioate (C12-DC) | 1.50 | 0.0464 | 0.9906 |
| 3-acetylphenol sulfate | 1.47 | 0.0104 | 0.9906 |
| quinolinate | 1.45 | 0.0103 | 0.9906 |
| N-acetylkynurenine (2) | 1.35 | 0.0355 | 0.9906 |
| N-acetylthreonine | 1.28 | 0.0225 | 0.9906 |
| 3,4-dihydroxybutyrate | 1.28 | 0.0332 | 0.9906 |
| 2-O-methylascorbic acid | 1.26 | 0.0113 | 0.9906 |
| arabitol/xylitol | 1.26 | 0.0479 | 0.9906 |
| 2-ketogulonate | 1.24 | 0.0494 | 0.9906 |
| N-acetylarginine | 0.78 | 0.0294 | 0.9906 |
| 1-stearoyl-2-linoleoyl-GPE (18:0/18:2) * | 0.71 | 0.0173 | 0.9906 |
| alpha-tocopherol | 0.62 | 0.0376 | 0.9906 |
| 1-myristoyl-2-arachidonoyl-GPC (14:0/20:4) * | 0.61 | 0.0421 | 0.9906 |
| 1-(1-enyl-stearoyl)-2-oleoyl-GPE (P-18:0/18:1) | 0.60 | 0.0058 | 0.9906 |
| 4-vinylcatechol sulfate | 0.59 | 0.0334 | 0.9906 |
| 1-stearoyl-2-oleoyl-GPE (18:0/18:1) | 0.57 | 0.0317 | 0.9906 |
| N-acetylproline | 0.53 | 0.0011 | 0.9853 |
| N-(2-furoyl)glycine | 0.50 | 0.0110 | 0.9906 |
| pyrraline | 0.48 | 0.0131 | 0.9906 |
| N-acetylpyrraline | 0.40 | 0.0155 | 0.9906 |
| glutathione, oxidized (GSSG) | 0.37 | 0.0091 | 0.9906 |
| thromboxane B2 | 0.37 | 0.0313 | 0.9906 |
| cysteine-glutathione disulfide | 0.35 | 0.0074 | 0.9906 |
| ribose 1-phosphate | 0.33 | 0.0452 | 0.9906 |
| cysteinylglycine disulfide * | 0.15 | 0.0140 | 0.9906 |
| MSTN+/− vs. WT | |||
|---|---|---|---|
| Biochemical Name | Ratio | p-Value | q-Value |
| glycylleucine | 3.85 | 0.0275 | 0.7158 |
| 1-arachidonoyl-GPA (20:4) | 3.76 | 0.0479 | 0.8504 |
| palmitoyl-arachidonoyl-glycerol (16:0/20:4) [1] * | 3.15 | 0.0219 | 0.7158 |
| oleoylcholine | 2.71 | 0.0432 | 0.8504 |
| N-acetylputrescine | 2.63 | 1.9 × 10−6 | 0.0017 |
| nicotinate ribonucleoside | 2.29 | 0.0029 | 0.7158 |
| 1-arachidonylglycerol (20:4) | 2.25 | 0.0003 | 0.1153 |
| biopterin | 2.20 | 0.0161 | 0.7158 |
| caproate (6:0) | 2.13 | 0.0050 | 0.7158 |
| N-acetyl-3-methylhistidine * | 1.97 | 0.0175 | 0.7158 |
| 2-docosahexaenoylglycerol (22:6) * | 1.92 | 0.0163 | 0.7158 |
| dihomo-linolenoylcarnitine (C20:3n3 or 6) * | 1.78 | 0.0262 | 0.7158 |
| isovalerate (i5:0) | 1.73 | 0.0463 | 0.8504 |
| cis-3,4-methyleneheptanoylcarnitine | 1.67 | 0.0112 | 0.7158 |
| N-acetyltyrosine | 1.66 | 0.0236 | 0.7158 |
| linolenoylcarnitine (C18:3) * | 1.64 | 0.0147 | 0.7158 |
| butyrate/isobutyrate (4:0) | 1.64 | 0.0195 | 0.7158 |
| N-acetylphenylalanine | 1.61 | 0.0079 | 0.7158 |
| methylsuccinate | 1.59 | 0.0230 | 0.7158 |
| S-(3-hydroxypropyl)mercapturic acid (HPMA) | 1.57 | 0.0188 | 0.7158 |
| N2,N5-diacetylornithine | 1.56 | 0.0075 | 0.7158 |
| 3-acetylphenol sulfate | 1.53 | 0.0418 | 0.8503 |
| N-acetyltryptophan | 1.51 | 0.0400 | 0.8503 |
| mevalonate | 1.48 | 0.0199 | 0.7158 |
| succinoyltaurine | 1.48 | 0.0255 | 0.7158 |
| 1-methyl-5-imidazolelactate | 1.47 | 0.0349 | 0.7994 |
| 1-linoleoyl-GPC (18:2) | 1.42 | 0.0259 | 0.7158 |
| 1-arachidonoyl-GPI (20:4) * | 1.42 | 0.0263 | 0.7158 |
| myristoyl dihydrosphingomyelin (d18:0/14:0) * | 1.42 | 0.0410 | 0.8503 |
| 1-oleoyl-GPC (18:1) | 1.41 | 0.0091 | 0.7158 |
| N-acetylkynurenine (2) | 1.41 | 0.0146 | 0.7158 |
| 1-stearoyl-GPC (18:0) | 1.34 | 0.0281 | 0.7158 |
| N2-acetyllysine | 1.34 | 0.0315 | 0.7409 |
| N-myristoyltaurine * | 1.33 | 0.0146 | 0.7158 |
| 2-palmitoyl-GPC (16:0) * | 1.32 | 0.0215 | 0.7158 |
| N-acetylisoleucine | 1.32 | 0.0299 | 0.7202 |
| 1-palmitoyl-GPC (16:0) | 1.31 | 0.0105 | 0.7158 |
| 1-palmitoleoyl-GPC (16:1) * | 1.29 | 0.0457 | 0.8504 |
| 3-methylhistidine | 1.28 | 0.0297 | 0.7202 |
| N6-acetyllysine | 1.19 | 0.0147 | 0.7158 |
| N1-methyladenosine | 0.77 | 0.0276 | 0.7158 |
| guanine | 0.66 | 0.0139 | 0.7158 |
| cytidine 5′-diphosphocholine | 0.64 | 0.0388 | 0.8503 |
| glycerol 3-phosphate | 0.64 | 0.0406 | 0.8503 |
| dehydroascorbate | 0.58 | 0.0209 | 0.7158 |
| gamma-glutamyl-2-aminobutyrate | 0.47 | 0.0069 | 0.7158 |
| guanosine 5′-monophosphate (5′-GMP) | 0.33 | 0.0144 | 0.7158 |
| adenosine | 0.29 | 0.0127 | 0.7158 |
| adenosine 5′-monophosphate (AMP) | 0.12 | 0.0187 | 0.7158 |
| ascorbate (vitamin C) | 0.07 | 0.0482 | 0.8504 |
| MSTN+/− vs. WT | |||
|---|---|---|---|
| Biochemical Name | Ratio | p-Value | q-Value |
| 4-acetamidobutanoate | 1.53 | 0.0112 | 0.7972 |
| 2′-deoxyinosine | 1.50 | 0.0011 | 0.7753 |
| dihydroxyacetone phosphate (DHAP) | 1.46 | 0.0153 | 0.7972 |
| N-acetyl-cadaverine | 1.42 | 0.0112 | 0.7972 |
| 1-palmitoyl-2-linoleoyl-GPC (16:0/18:2) | 0.93 | 0.0434 | 0.7972 |
| N2-acetyllysine | 0.92 | 0.0472 | 0.7972 |
| sphingomyelin (d18:1/20:1, d18:2/20:0) * | 0.91 | 0.0468 | 0.7972 |
| isoleucine | 0.90 | 0.0300 | 0.7972 |
| 1-palmitoleoyl-2-linoleoyl-GPC (16:1/18:2) * | 0.90 | 0.0331 | 0.7972 |
| alanine | 0.90 | 0.0419 | 0.7972 |
| myo-inositol | 0.89 | 0.0267 | 0.7972 |
| fumarate | 0.89 | 0.0349 | 0.7972 |
| 1-palmitoyl-2-palmitoleoyl-GPC (16:0/16:1) * | 0.88 | 0.0464 | 0.7972 |
| S-adenosylhomocysteine (SAH) | 0.87 | 0.0267 | 0.7972 |
| alpha-ketoglutarate | 0.86 | 0.0194 | 0.7972 |
| retinol (vitamin A) | 0.85 | 0.0271 | 0.7972 |
| glycerophosphorylcholine (GPC) | 0.83 | 0.0490 | 0.7972 |
| 3-methylcytidine | 0.80 | 0.0170 | 0.7972 |
| pyridoxamine phosphate (vitamin B6) | 0.79 | 0.0048 | 0.7972 |
| 3-aminoisobutyrate | 0.77 | 0.0197 | 0.7972 |
| 1-palmitoyl-2-oleoyl-GPS (16:0/18:1) | 0.74 | 0.0110 | 0.7972 |
| alpha-tocopherol | 0.73 | 0.0021 | 0.7866 |
| gluconate | 0.71 | 0.0443 | 0.7972 |
| pyrraline | 0.69 | 0.0048 | 0.7972 |
| phytosphingosine | 0.66 | 0.0477 | 0.7972 |
| genistein | 0.64 | 0.0314 | 0.7972 |
| taurocholate | 0.61 | 0.0437 | 0.7972 |
| Pathway | Biochemical Name | Ratio | ||||
|---|---|---|---|---|---|---|
| WT Fetal WT Maternal | MSTN+/− Fetal MSTN+/− Maternal | MSTN+/− Maternal WT Maternal | MSTN+/− Fetal WT Fetal | MSTN+/− Placenta WT Placenta | ||
| Acylcarnitine Medium Chain | cis-3,4-methylene heptanoylcarnitine | 0.39 * | 0.66 * | 0.99 | 1.67 * | 1.10 |
| Acylcarnitine Monounsaturated | cis-4-decenoyl carnitine (C10:1) | 0.21 * | 0.26 * | 1.07 | 1.36 † | 1.02 |
| Myristoleoyl carnitine (C14:1) * | 0.30 * | 0.35 * | 1.14 | 1.33 † | 1.09 | |
| Acylcarnitine Polyunsaturated | Linolenoyl carnitine (C18:3) * | 0.43 * | 0.65 * | 1.08 | 1.64 * | 1.05 |
| dihomo-linoleoyl carnitine (C20:2) * | 1.31 | 1.76 * | 1.18 | 1.58 † | 1.0 | |
| Arachidonoyl carnitine (C20:4) | 0.79 | 1.06 | 1.02 | 1.38 † | 0.98 | |
| dihomo-linolenoyl carnitine (C20:3n3 or 6) * | 1.00 | 1.76 * | 1.02 | 1.78 * | 1.07 | |
| docosapentaenoylcarnitine (C22:5n3) * | 1.94 * | 2.90 * | 1.09 | 1.62 † | 1.00 | |
| docosahexaenoylcarnitine (C22:6) * | 0.63 * | 0.93 | 1.02 | 1.51 † | 1.00 | |
| Carnitine | Carnitine | 1.02 | 1.12 | 0.97 | 1.06 | 1.01 |
| Pathway | Biochemical Name | Ratio | ||||
|---|---|---|---|---|---|---|
| WT Fetal WT Maternal | MSTN+/− Fetal MSTN+/− Maternal | MSTN+/− Maternal WT Maternal | MSTN+/− Fetal WT Fetal | MSTN+/− Placenta WT Placenta | ||
| Lysophospholipids | 1-linoleoyl-GPA (18:2) * | 0.28 * | 0.59 | 1.47 | 3.11 † | ND |
| 1-arachidonoyl-GPA (20:4) | 0.57 | 1.19 | 1.80 | 3.76 * | ND | |
| 1-palmitoyl-GPC (16:0) | 0.39 * | 0.5 * | 1.02 | 1.31 * | 0.98 | |
| 2-palmitoyl-GPC (16:0) * | 0.24 * | 0.33 * | 0.98 | 1.32 * | 0.94 | |
| 1-palmitoleoyl-GPC (16:1) * | 1.74 * | 2.31 * | 0.97 | 1.29 * | 0.88 | |
| 1-stearoyl-GPC (18:0) | 0.27 * | 0.38 * | 0.98 | 1.34 * | 1.01 | |
| 1-oleoyl-GPC (18:1) | 1.11 | 1.51 * | 1.04 | 1.41 * | 0.95 | |
| 1-linoleoyl-GPC (18:2) | 0.38 * | 0.54 * | 0.98 | 1.42 * | 0.92 | |
| 1-arachidonoyl-GPC (20:4n6) * | 0.83 | 1.08 | 1.01 | 1.33 † | 0.83 | |
| 2-stearoyl-GPE (18:0) * | 0.28 * | 0.38 * | 0.79 † | 1.04 | 0.84 | |
| 1-oleoyl-GPE (18:1) | 0.94 | 1.63 * | 0.64 | 1.11 | 0.98 | |
| 1-linoleoyl-GPE (18:2) * | 0.34 * | 0.5 * | 0.85 | 1.27 | 0.82 † | |
| 1-arachidonoyl-GPE (20:4n6) * | 0.65 * | 0.75 | 0.99 | 1.16 | 0.86 † | |
| 1-oleoyl-GPS (18:1) | ND | ND | ND | ND | 0.76 † | |
| 1-linoleoyl-GPS (18:2) * | ND | ND | ND | ND | 0.78 † | |
| 1-oleoyl-GPG (18:1) * | 12.93 * | 9.59 * | 1.61 | 1.19 | 0.82 † | |
| 1-linoleoyl-GPG (18:2) * | 0.98 | 1.04 | 1.23 | 1.3 † | 0.84 | |
| 1-arachidonoyl-GPI (20:4) * | 0.22 * | 0.26 * | 1.19 | 1.42 * | 0.88 | |
| Pathway | Biochemical Name | Ratio | ||||
|---|---|---|---|---|---|---|
| WT Fetal WT Maternal | MSTN+/− Fetal MSTN+/− Maternal | MSTN+/− Maternal WT Maternal | MSTN+/− Fetal WT Fetal | MSTN+/− Placenta WT Placenta | ||
| Purine Metabolism, (Hypo)Xanthine/Inosine containing | 2′-deoxyinosine | ND | ND | ND | ND | 1.5 * |
| inosine 5′-monophosphate (IMP) | 15.16 * | 2.00 | 1.00 | 0.13 † | ND | |
| N1-methylinosine | 8.01 * | 7.15 * | 1.26 † | 1.12 | 1.10 | |
| allantoic acid | 0.41 * | 0.59 * | 1.11 | 1.61 † | ND | |
| Purine Metabolism, Adenine containing | adenosine 5′-monophosphate (AMP) | 68.59 * | 8.83 * | 0.93 | 0.12 * | 1.47 |
| adenosine 3′-monophosphate (3′-AMP) | 6.95 † | 5.8 * | 0.78 | 0.66 | 1.47 † | |
| adenosine 2′-monophosphate (2′-AMP) | 8.27 | 18.52 * | 0.22 | 0.50 | 1.85 † | |
| adenosine | 11.27 * | 4.38 * | 0.75 | 0.29 * | 0.84 | |
| N1-methyladenosine | 3.04 * | 2.32 * | 1.00 | 0.77 * | 0.78 † | |
| N6-succinyladenosine | 30.35 | 16.6 * | 1.22 | 0.67 † | 0.92 | |
| Purine Metabolism, Guanine containing | guanosine 5′-monophosphate (5′-GMP) | 57.36 * | 36.38 * | 0.52 | 0.33 * | 0.96 |
| guanosine | 53.73 * | 132.89 * | 0.38 † | 0.95 | 0.95 | |
| guanine | 22.04 * | 14.46 * | 1.00 | 0.66 * | 0.89 | |
| Pathway | Biochemical Name | Ratio | ||||
|---|---|---|---|---|---|---|
| WT Fetal WT Maternal | MSTN+/− Fetal MSTN+/− Maternal | MSTN+/− Maternal WT Maternal | MSTN+/− Fetal WT Fetal | MSTN+/− Placenta WT Placenta | ||
| Leucine, isoleucine, and valine metabolisms | N-acetylleucine | 0.63 * | 0.89 | 0.91 | 1.28 | 1.08 |
| isovalerate (i5:0) | 0.83 | 1.01 | 1.42 | 1.73 * | ND | |
| isovalerylcarnitine (C5) | 2.58 * | 3.96 * | 0.85 | 1.30 | 0.91 † | |
| isoleucine | 1.17 | 1.15 | 0.99 | 0.97 | 0.9 * | |
| N-acetylisoleucine | 0.78 * | 1.11 | 0.92 | 1.32 * | 0.96 | |
| 2-methylbutyrylglycine | 0.29 * | 0.27 * | 1.57 | 1.46 | 1.68 † | |
| methylsuccinate | 0.69 * | 0.81 | 1.36 | 1.59 * | 1.08 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opoku, R.; DeCata, J.; Phillips, C.L.; Schulz, L.C. Effect of Genetically Reduced Maternal Myostatin on Late Gestation Maternal, Fetal, and Placental Metabolomes in Mice. Metabolites 2023, 13, 719. https://doi.org/10.3390/metabo13060719
Opoku R, DeCata J, Phillips CL, Schulz LC. Effect of Genetically Reduced Maternal Myostatin on Late Gestation Maternal, Fetal, and Placental Metabolomes in Mice. Metabolites. 2023; 13(6):719. https://doi.org/10.3390/metabo13060719
Chicago/Turabian StyleOpoku, Ruth, Jenna DeCata, Charlotte L. Phillips, and Laura C. Schulz. 2023. "Effect of Genetically Reduced Maternal Myostatin on Late Gestation Maternal, Fetal, and Placental Metabolomes in Mice" Metabolites 13, no. 6: 719. https://doi.org/10.3390/metabo13060719
APA StyleOpoku, R., DeCata, J., Phillips, C. L., & Schulz, L. C. (2023). Effect of Genetically Reduced Maternal Myostatin on Late Gestation Maternal, Fetal, and Placental Metabolomes in Mice. Metabolites, 13(6), 719. https://doi.org/10.3390/metabo13060719







