Abstract
Plant disease control has long been a critical issue in agricultural production and relies heavily on the identification of plant diseases, but traditional disease identification requires extensive experience. Most of the existing deep learning-based plant disease classification methods run on high-performance devices to meet the requirements for classification accuracy. However, agricultural applications have strict cost control and cannot be widely promoted. This paper presents a novel method for plant disease classification using a binary neural network with dual attention (DABNN), which can save computational resources and accelerate by using binary neural networks, and introduces a dual-attention mechanism to improve the accuracy of classification. To evaluate the effectiveness of our proposed approach, we conduct experiments on the PlantVillage dataset, which includes a range of diseases. The and of our method reach 99.39% and 99.4%, respectively. Meanwhile, compared to AlexNet and VGG16, the of our method is reduced by 72.3% and 98.7%, respectively. The of our algorithm is 5.4% of AlexNet and 2.3% of VGG16. The experimental results show that DABNN can identify various diseases effectively and accurately.
1. Introduction
Plant disease can lead to a reduction in crop yields. Different control measures are needed for different plant diseases. Plant disease classification is helpful to identify the category of plant disease. The external manifestations of plant diseases are physical changes in leaf contours and veins. Earlier, the manual plant disease classification method relied on agricultural experts and could not timely accomplish large-scale disease identification tasks. In addition, there are problems, such as high subjective influence and the low efficiency of identification. Therefore, it is extremely urgent to elevate the accuracy and efficiency of plant disease classification.
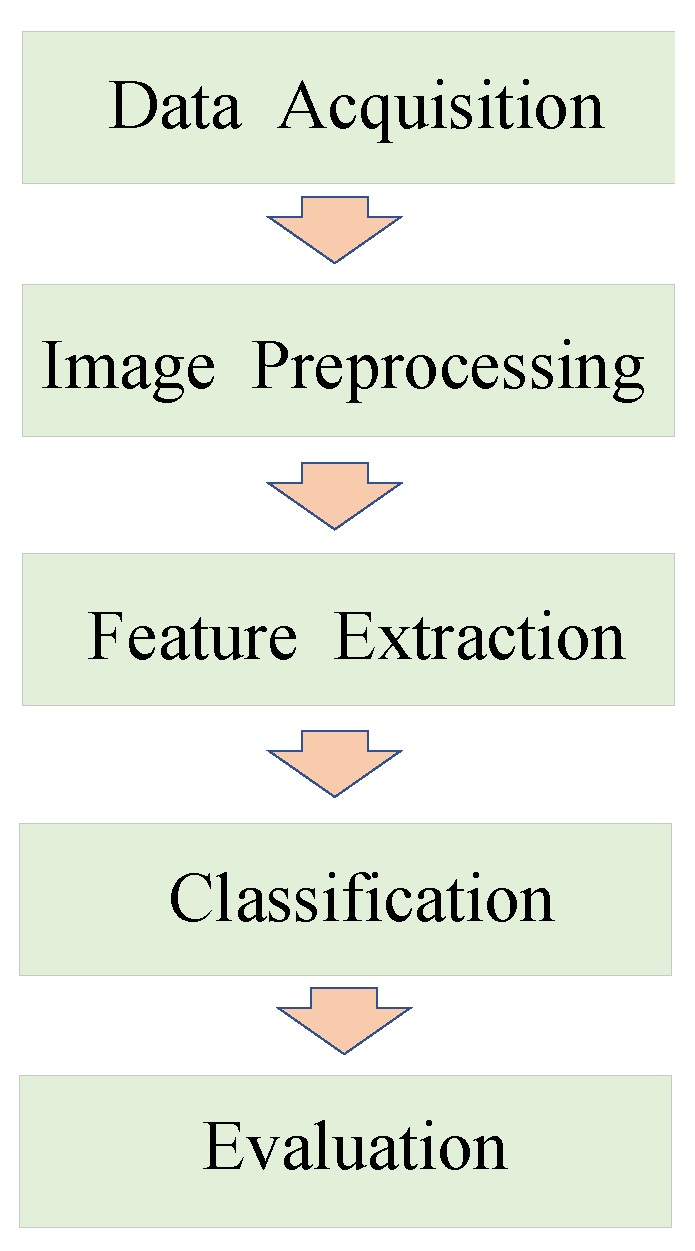
Currently, researchers tend to favor computer vision approaches for plant disease classification. The general process of disease classification using computer vision approaches is shown in Figure 1, consisting of five parts: data acquisition, image preprocessing, feature extraction, classification, and evaluation. Previous visual detection algorithms need to manually extract features such as the texture and shape of disease spots to establish feature vectors and then use machine learning (ML) algorithms [1] such as the support vector machine (SVM) [2], K-means clustering algorithm [3], and random forest algorithm [4] to classify. These algorithms rely heavily on disease data processing in the early phases. With the increase in computational performance, deep learning techniques [5] began to develop rapidly. AlexNet [6] claimed the ImageNet Image Classification Championship in 2012. Subsequently, many image classifiers including GoogLeNet [7], VGGNet [8], and ResNet [9] have been developed for extracting higher-level semantic information through a smaller convolution kernel and deeper network structures, thus improving the accuracy of image classification. With the automatic extraction and easy processing of feature information and high-density data, the convolutional neural network (CNN) [10] is enabled to generate feature representations with higher accuracy and robustness than traditional methods. Building appropriate CNN models has emerged as a hot topic in plant disease classification recently.
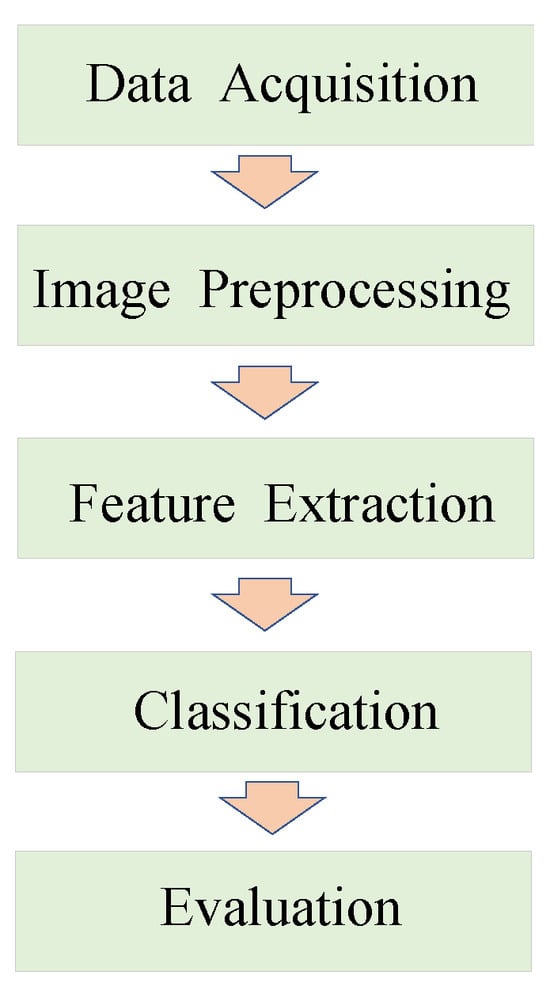
Figure 1.
The general process of disease classification using computer vision approaches.
Applying CNN-based computer vision technology to identify agricultural diseases would be far less influenced by subjective factors and achieve large-scale disease classification. To enhance identification efficiency, some methods are proposed to reduce model parameters. For instance, adjusting the combination of a network hyperparameter and pooling to identify corn diseases [11]; locating the disease region of buckwheat faster in combination with maximally stable extremal regions (MSER) [12]; using separable convolutions instead of standard convolutions in the network to compress tomato disease recognition models [13]; and optimizing the region recommendation network in the Faster CNN network [14] to improve the generalization performance of grape disease models [15]. However, reducing model parameters cannot change the complex and bulky nature of a CNN, which requires high Graphic Processing Unit (GPU) memory and is time-consuming.
Agricultural applications require strict cost control, and high-performance computing equipment cannot be used in agriculture on a large scale, but low-performance computing equipment cannot satisfy the demand for computing resources for existing agricultural disease identification models. In response to the above situation, researchers have proposed methods to accelerate network operation by compressing models, such as matrix decomposition, weight sparsification [16], network pruning [17,18], knowledge distillation [19], low-rank decomposition [20], lightweight networks [21,22,23], Neural Architecture Search [24,25], and low-bit quantization (quantifying data with high-bit width to data with low-bit width) [26]. A low-bit quantization approach to network computing is more adaptable to low computational performance limitations, because it saves more computing resources and usage time. Among them, binarization is the most promising method among the low-bit quantization methods. Applying binary networks to agricultural disease identification reduces the cost and computational complexity of the task. It provides the ability to perform the task more quickly and efficiently in real time.
High precision and minimal computing resource demands are essential for practical agricultural applications. The existing plant disease classification models have high complexity and a large number of parameters, so they can only work on the equipment with high computational performance. This is contrary to the need for low cost in agricultural applications, so it is necessary to investigate plant disease classification models suitable for working on devices with low computational performance. Although some existing lightweight networks can break through the limitation of computing resources, they cannot achieve satisfactory disease classification results. We propose a novel method for plant disease classification using a binary neural network with dual attention (DABNN). The main contributions of this paper are as follows:
- (1)
- We introduce the binary neural network into the plant disease classification task, which can achieve a good balance between speed and accuracy under the limitation of computational resources.
- (2)
- A dual-attention mechanism is introduced to reweight the feature map to obtain more efficient channel and spatial features, thus achieving better performance.
- (3)
- We conduct experiments on the PlantVillage dataset [27] and achieve promising results, indicating that our proposed approach has great potential for plant disease classification.
The remainder of this paper is organized as follows. Closely related works are presented in Section 2. The details of our method are provided in Section 3. The experiments are conducted on the PlantVillage dataset, and the results are shown in Section 4. We present the discussions in Section 5. Finally, Section 6 concludes this work.
2. Related Work
In this section, we introduce two aspects of the work related to our study: (i) a binary neural network and (ii) an attention mechanism.
2.1. Binary Neural Network
Binary neural networks [28,29,30,31,32] limit the network weight to +1 and −1 through a binarization function. A 32-bit single-precision floating-point parameter is quantified to a 1-bit fixed-point parameter. If the parameter’s value is greater than 0, the result will be converted to +1; otherwise, the result will be −1. The binary function is computed as:
In forward propagation, the floating-point convolution operation after binarization is rephrased as:
where ∗ denotes the floating-point convolution operation. ⊛ represents the convolution operation implemented by the Bitcount operation and the bitwise XNOR operation. w and a represent the floating-point weight and activation, respectively.
By adopting binary neural networks, the memory demand of full-precision neural networks can be compressed up to 32 times, saving storage resources for model deployment. By using Bitcount operation and XNOR operation instead of floating-point calculations, the computational process of neural networks can be improved by up to 58 times.
In the existing work, classification tasks based on binary neural networks are performed for infrared images on GPUs and are at least four times faster and have three orders of magnitude less energy consumption [33]. The binary weight and binary input data network (BWBDN) [34] achieved 62 times the acceleration and saved 32 times the storage space. Compared to full-precision networks, the binary model is able to represent the weights of the network at extremely low precision. It saves a lot of storage space and forward propagation time. However, quantization of the network would cause the loss of its detailed information and it would have problems with accuracy degradation.
2.2. Attention Mechanism
The attention mechanism draws on the principles of human vision to select more critical information for the task and ignore irrelevant information. This attention mechanism can quickly and effectively focus on useful information in complex scenes. With limited computational resources, the use of attention mechanisms can enhance model accuracy. In 2015, the attention mechanism was first introduced to Natural Language Processing (NLP) and achieved good results in machine translation tasks [35]. Then, the attention mechanism was also applied to computer vision, such as image classification [36,37], object detection [38,39,40], and semantic segmentation [41,42]. In plant disease classification, the attention modules highlight important features in the output feature map of the CNN architecture [43]. The lightweight convolutional neural networks with different attention modules [44,45] were designed for tomato and grape diseases. By combining the attention mechanism with multi-task learning, the grape leaf disease detection network (GLDDN) [46] improved the accuracy of grape disease classification.
Channel attention [36] is trained to obtain weight on each channel, which highlights useful information and suppresses useless information. Firstly, global pooling and average pooling are performed on the input feature maps in the spatial dimension to compress the spatial size and facilitate the subsequent learning of channel features. Then, the results of the global and average pooling are fed to the multilayer perceptron (MLP) to learn the features of the channel dimensions and the importance of each channel. The interdependence between channels is modeled and the content that needs to be focused on each layer’s feature maps is determined. Finally, the output of two MLP branches is subjected to an addition operation and then mapped through the non-linear function to obtain the final output. The channel attention is computed as:
where F is the input feature map and is the output feature map of the channel attention module.
Spatial attention [47] is concerned with where the features are more important to our task. Firstly, global pooling and average pooling are performed on the input feature map in the channel dimension, which can compress the channel size and facilitate the subsequent learning of spatial features. Then, the convolution and non-linear operation are performed on the results of the pooling to extract the spatial location information. Finally, the spatial attention map is used on the corresponding feature maps to mark the locations that need to be focused on. The spatial attention is computed as:
where denotes the convolution operation and is the output feature map of the spatial attention module.
At present, the attention mechanism is mainly applied to ordinary neural networks. We introduce the channel attention and spatial attention mechanisms into the binary neural network to reinforce the feature representation of binary neural networks.
3. Method
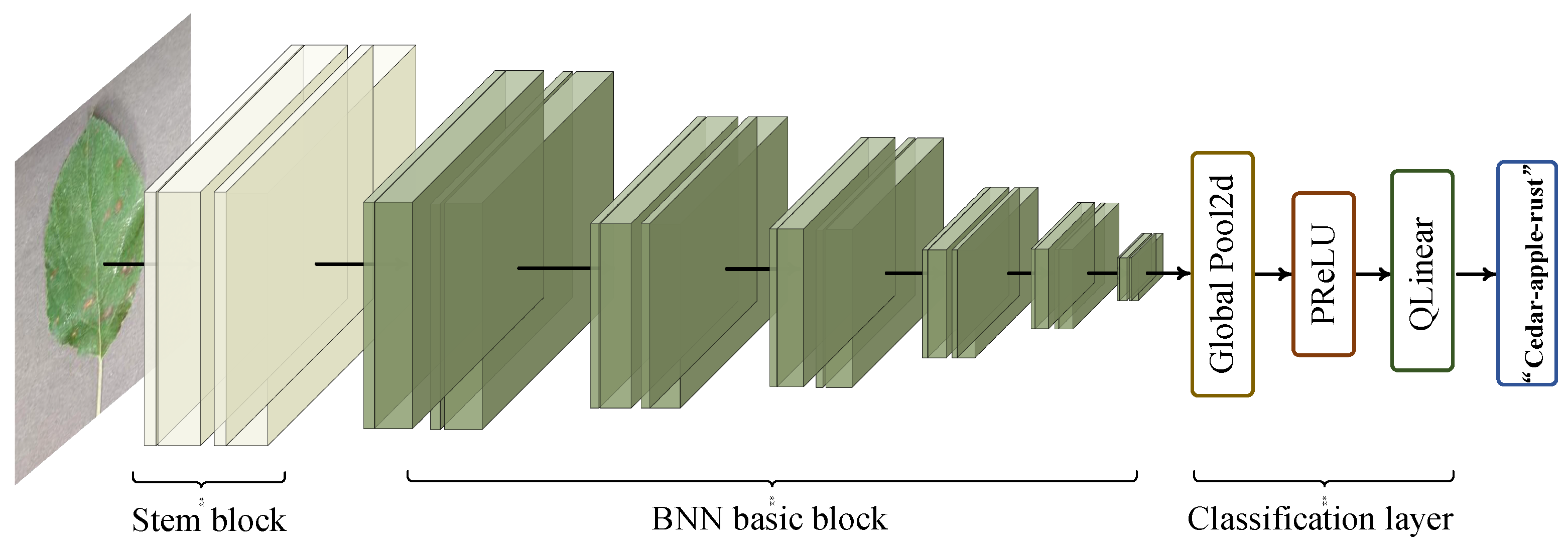
In this paper, a binary neural network with dual attention is proposed for plant disease classification. Our overall network is shown in Figure 2. The network has three stages, organized as the stem block, feature extractor, and classification layer. The details of each stage are revealed in Table 1. In the first stage, the stem block contains quantized convolution, batch normalization (BN), 3 × 3 binary convolution with dual attention (3 × 3 DABconv), and 1 × 1 pointwise binary convolution with dual attention (1 × 1 DABconv), which implements the downsampling operation to diminish the parameters and adjust the number of channels. In the second stage, the feature extractor contains six BNN basic blocks used to extract valid features, which will be explained in detail later. In the third stage, the feature maps from the previous stage are input to the adaptive averaging pooling layer, which is adjusted by PReLU, and then the results are input to the quantized linear classifier to obtain the final label.
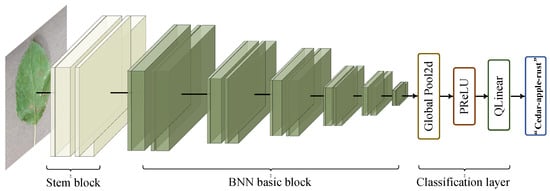
Figure 2.
The overall structure of our network consists of three parts, including stem block, BNN basic block, and classification layer.

Table 1.
Overview of DABNN architecture.
3.1. DABConv Module
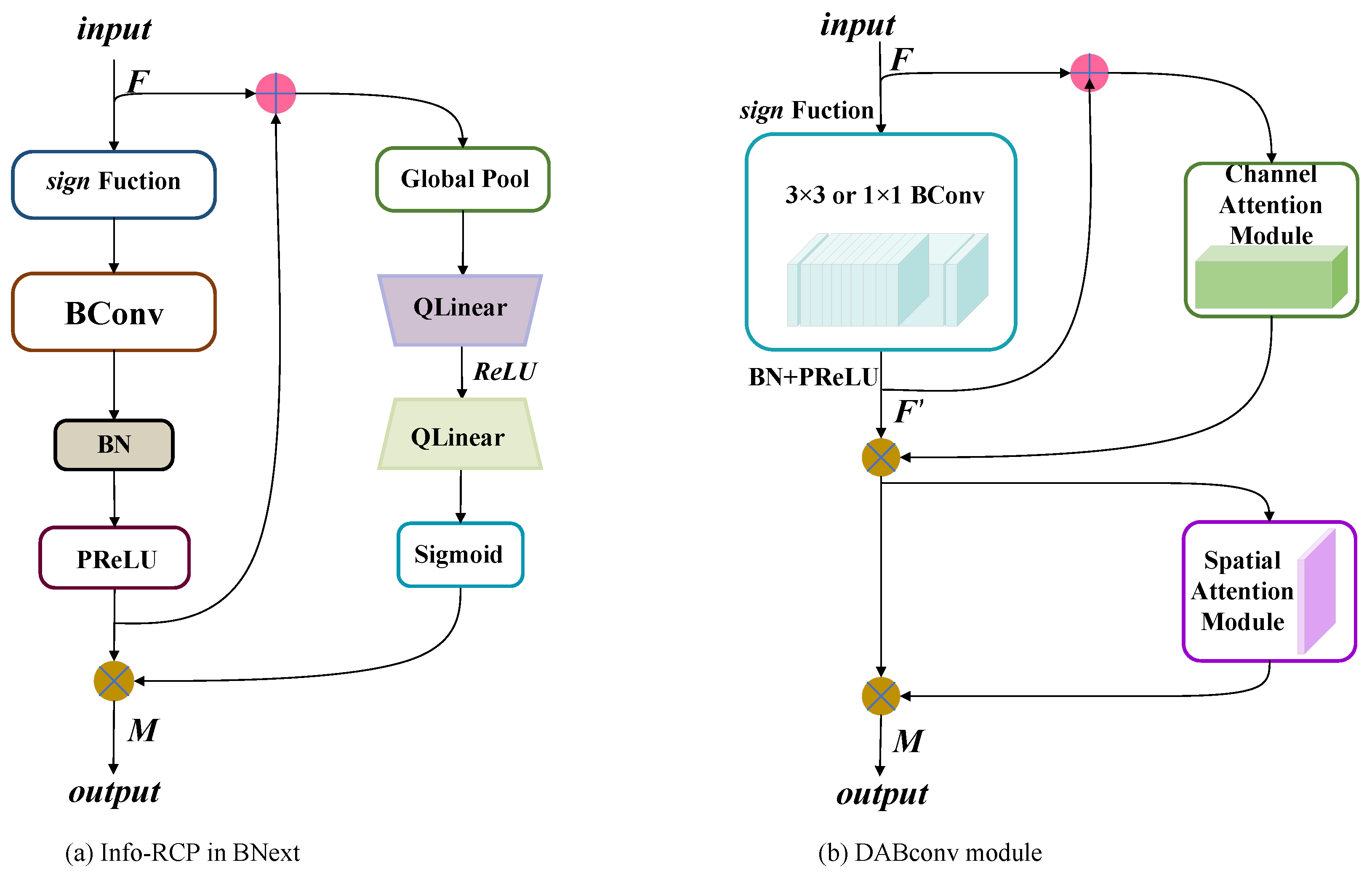
To improve the precision of the binary convolutional neural network for disease identification, a dual-attention module is designed based on the binary convolution. The entire BNN basic block consists of the binary convolution module (Bconv) and dual-attention module. This module can be described as:
where ⊗ denotes the element-wise multiplication. is the output of the BConv module. M is the output of the DABconv module. and represent the binarized channel attention module and the binarized spatial attention module, respectively.
In the DABconv module, BN and PReLU are adopted to adjust the output distribution of the BConv module during the forward propagation of the information flow. To make the information flow spread more smoothly, the input and output of the BConv module are fused by addition. The fused data are input into a channel attention module, which could scale the input space size and perceive the changes in the information flow. The feature channels are weighted to protrude effective features and weaken the interfering features, and in addition, the output distribution can be adaptively adjusted by learning. Subsequently, the final result is imported into the spatial module. Spatial attention is used to scale the size of the input channel, perceive key locations of spatial feature information, and strengthen the representation of feature regions.
3.2. The Network of BNN Basic Block
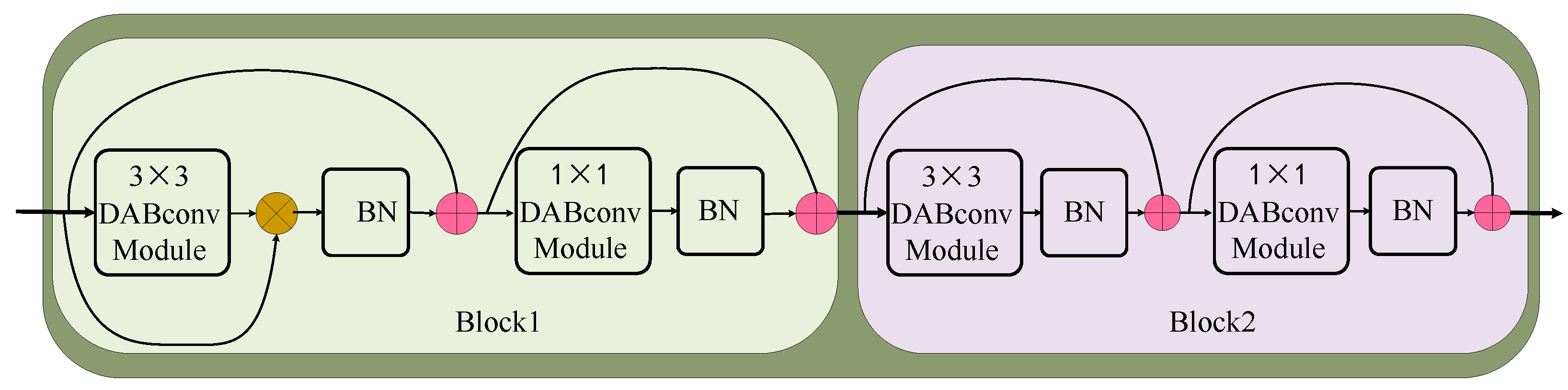
The BNN basic block is the basic component of our network, and the structure of the BNN basic block is shown in Figure 3. The BNN basic block consists of multiple DABconv blocks, as shown in Figure 4. The 3 × 3 DABconv module is used to extract the disease features of each input channel. Then, the 1 × 1 DABconv module is employed to adjust the feature dimension. Lastly, the inputs and outputs are connected via a residual structure. The output of each convolutional module is normalized by using BN to adjust the module output distribution. To alleviate the information bottleneck in the forward propagation of the information flow, a continuous residual structure is used to encapsulate each convolution and BN combination. This structure consistently maintains larger values during back propagation and does not decrease its gradient during the expansion process of network layers.
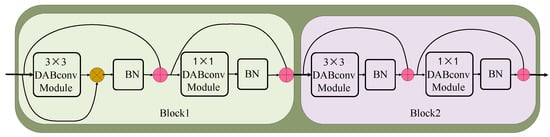
Figure 3.
BNN basic block consists of 2 blocks, both composed of 3 × 3 DABconv module, 1 × 1 DABconv module, and batch normalization. Their difference is that the shortcut is different.
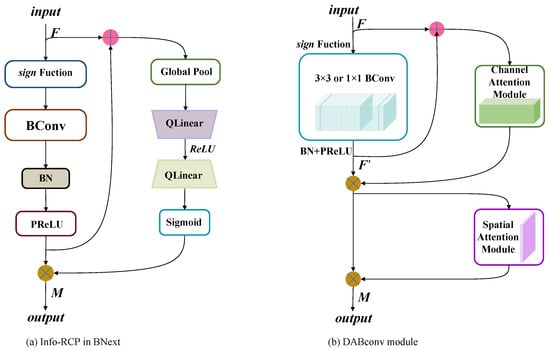
Figure 4.
Contrast Info-RCP module and DABconv module; DABconv module is an improvement on Info-RCP module.
4. Experiments
The details of the experimental design and results are presented in this section. First, we briefly describe the implementation details and image preprocessing in Section 4.1 and Section 4.2. Section 4.3 presents the performance metrics. Then, the experimental results are presented in Section 4.4. Finally, the computational complexity analysis is shown in Section 4.5.
4.1. Implementation Details
To evaluate the effectiveness of DABNN, the authors of this paper conduct comparative experiments on the PlantVillage dataset. Our experiments are conducted on a computer with an AMD Ryzen 5900X CPU and NVIDIA RTX 3090 GPU. All the models are trained and tested on this computer. The batch size and the initial learning rate are set to 32 and 0.01. The optimizer we use is Adam; the weight decay is set to 0.0001.
The loss function serves to measure the difference between the true value and the predicted value of the model. The label and prediction are fed into the loss function to obtain the loss. Then, the parameters of the model are optimized by reducing the loss by using gradient descent and back propagation. Cross-entropy is used to describe the gap between two distributions and can help the model estimate the closest predictive probability objective function to the target distribution from the training samples. The smaller the cross-entropy, the closer the hypothesized distribution is to the true distribution and the better the model. The cross-entropy loss function can be described as:
where L is the loss function. y and are the prediction and its label. o is the output of the model. If the prediction is correct, y is closer to , and then L is smaller.
4.2. Image Preprocessing
To evaluate our algorithm, we use the PlantVillage dataset, which contains a dataset of 55,448 images of 13 different plant leaves. This dataset consists of 38 different classes, and all the image samples are defined as healthy or infected plants using labels. There is also a class with 1143 background images that do not contain plant leaves.
Image transformations are used to increase the number of images in the dataset and to reduce overfitting by adding some distorted images to the training data. Gamma correction, image flipping, PCA color augmentation, noise injection, rotation, and scaling transforms are used to create augmented images of the training data.
Finally, the dataset containing the augmented images has a total of 61,486 images. Some images of the dataset are shown in Figure 5. The entire dataset is divided into training and validation sets with an 8:2 ratio. Both our algorithm and the algorithms compared in this paper are trained and evaluated on the dataset.

Figure 5.
Example images of PlantVillage dataset.
4.3. Performance Metrics
In this paper, , , , and the are used to evaluate these models. refers to the ratio of the number of samples correctly predicted by the model to the total number of samples. is the ratio of the number of positive samples correctly predicted by the model to the number of samples predicted as positive by the model. is concerned with how accurately the model predicts positive samples and whether it can misclassify negative samples as positive samples. is the ratio of the number of positive samples correctly predicted by the model to the number of true positive samples. is concerned with the model’s ability to find all true positive samples and whether the model misses positive samples. The is a reconciled average of the and that provides a more comprehensive assessment of the model’s performance. These metrics can be calculated by (7)–(10):
where , , , and are true positive, true positive, false positive, and false negative, respectively. In plant disease classification tasks, these evaluation metrics can be used to measure the accuracy and reliability of the model, helping to improve the classification results. Higher values of these metrics indicate better performance of the model. The computational complexity and the size of the model are also important performance metrics.
4.4. Results
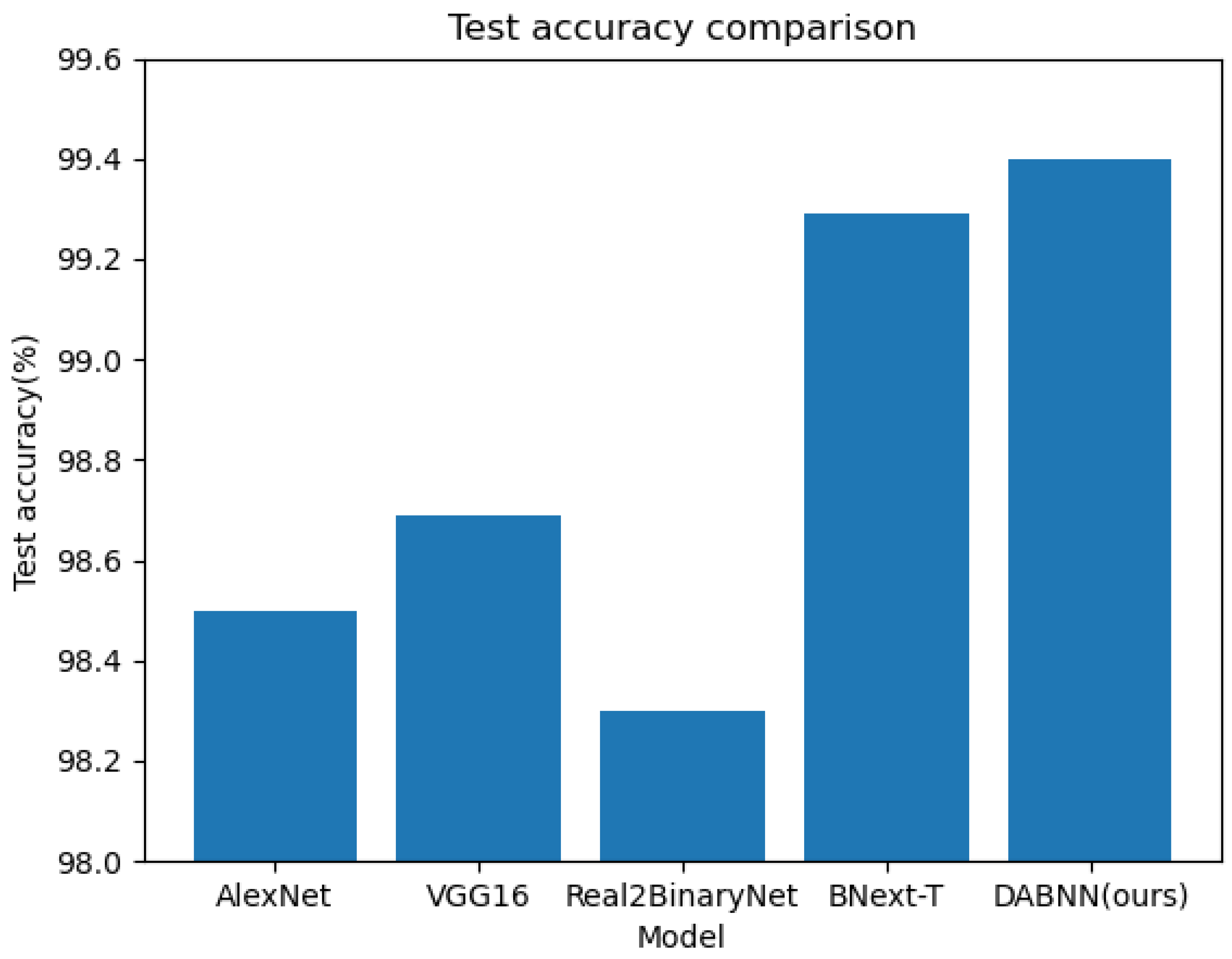
The results are shown in Table 2 and reveal that the of our algorithm improves 0.91% over AlexNet, 0.72% over VGG16, 1.12% over Real2BinaryNet, and 0.11% over our baseline algorithm BNext-T [48]. The comparison of the with the other algorithms is shown in Figure 6, which shows that our algorithm has a certain improvement in accuracy. In addition, the and of our algorithm reveal some improvement. Theoretically, our method is able to weight the feature information in the channel dimension and spatial dimension to enhance the effective information and suppress the interfering information. In summary, DABNN focuses more on the representation learning of plant disease features, which improves the expressive ability and classification performance of the model. Additionally, DABNN employs a residual structure to encapsulate the binary convolution module, which alleviates the information bottleneck in the forward propagation of the information flow. This structure allows the gradient to flow throughout the network, always maintaining a large value, which can alleviate the gradient vanishing during training. Overall, the experiments show that our algorithm has good performance.

Table 2.
Quantitative results.
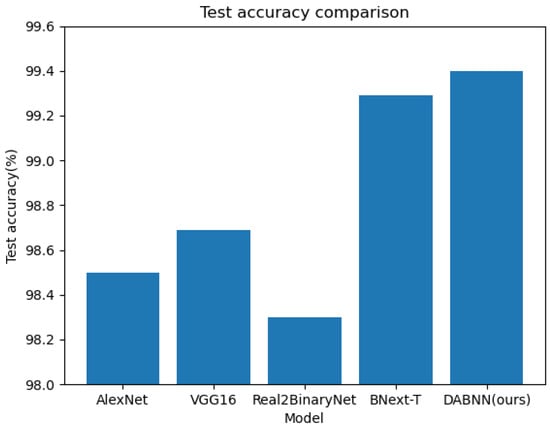
Figure 6.
Accuracy comparison of different network structures.
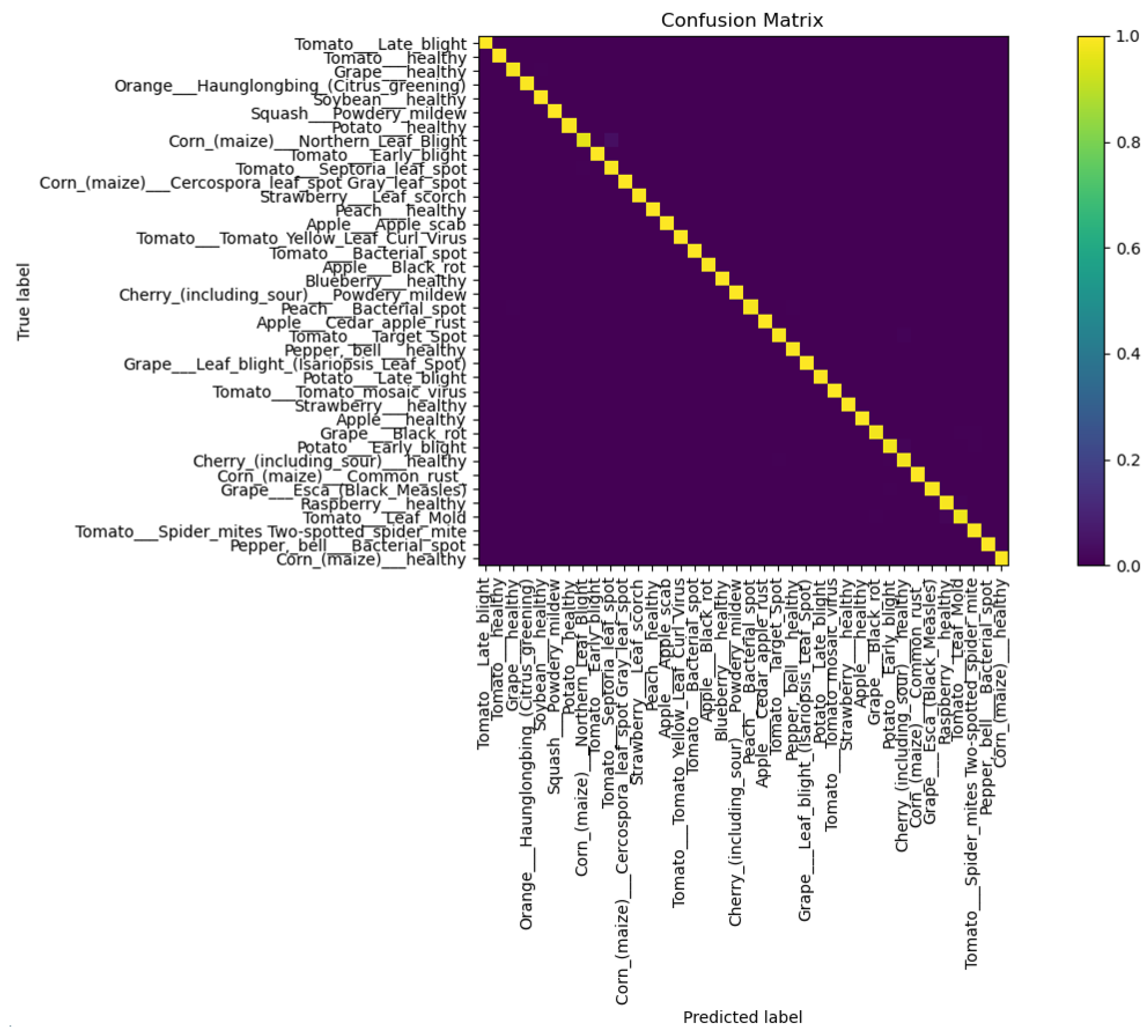
To visually display the classification error of our algorithm for various diseases, we drew a confusion matrix, as shown in Figure 7. According to the results reflected in the confusion matrix, we can see that the proposed model has a high classification accuracy and a low probability of misclassification.
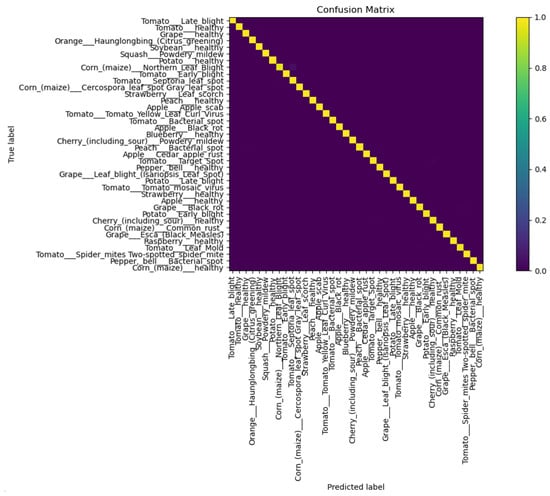
Figure 7.
Confusion matrix of the proposed model.
From the experimental results, the DABNN model shows superior performance in the plant disease classification task. Compared with other models, the model achieved excellent results in the evaluation metrics such as the , , , and , indicating that the DABNN model has high accuracy and reliability for classifying plant disease images.
4.5. Computational Complexity Analysis
The and metrics are very important to plant disease classification models. reflects the computational resources and time required for the model to perform a single forward propagation or back propagation. For plant disease classification tasks, models with high computational complexity typically require a longer training time and more computational resources. The reflects the size of the model.
Our method utilizes binarized convolution instead of ordinary convolution, where the original convolution operation is implemented by the bitwise XNOR operation and Bitcount operation, and the parameters of the model are quantized as 1-bit fixed-point parameters. To verify the lightweight of the DABNN model, we conducted comparison experiments with four other models, as shown in Table 3. It indicates that the of our method is decreased by 72.3% compared to AlexNet and by 98.7% compared to VGG16. Because our model is based on BNext-T and incorporates the dual-attention module, its computational complexity is slightly increased compared to BNext-T and Real2BinaryNet. The of our algorithm is 5.4% of AlexNet and 2.3% of VGG16. In terms of the , our model is much smaller than AlexNet and VGG16 but slightly larger than BNext-T and Real2BinaryNet. Based on the analysis above, our model is relatively lightweight. In summary, by evaluating the and of the model, it is demonstrated that DABNN effectively reduces the model complexity and size while maintaining a high performance.

Table 3.
Comparison of computational complexity results between different models.
When selecting a model, these indicators need to be considered in combination with the actual hardware platform and application scenarios in order to obtain the best plant disease classification effect and computational resource utilization efficiency, so as to improve the model’s classification accuracy and generalization ability. For plant disease classification application scenarios with limited computational resources, DABNN with a lower and can be the best.
5. Discussion
The implementation of a dual-attention mechanism can effectively improve the accuracy of binary convolution, and binary neural networks are well suited for low-performance computing equipment. DABNN can reduce the demand for computational performance while expanding the application scope of plant disease classification. DABNN is compared quantitatively and qualitatively with other full-precision networks, such as AlexNet, VGG16, and so on. When these algorithms are trained from scratch, DABNN has advantages in accelerating computations and saving storage space over full-precision convolutional neural networks. DABNN can support general-purpose hardware with limited computational resources (including FPGA, CPU, etc.). However, binary neural networks may lose some accuracy as a trade-off. The dual-attention mechanism can increase the weight of effective features and ignore the interference of irrelevant information. Meanwhile, with the increase in convolutional layers, the residual structure can effectively alleviate the information bottleneck in forward propagation and avoid the problem of gradient vanishing in optimization. Therefore, using DABNN can effectively elevate the accuracy and efficiency of plant disease classification.
However, there are some limitations to our approach. Due to the relatively simple environment for collecting plant disease datasets, the classification algorithm needs to adapt based on crops, environments, and scenes. In real scenes, DABNN may lack some generalization ability and robustness. Additionally, background noise, overshot images, motion blur, or occlusion can make it difficult for DABNN to efficiently handle these types of disease recognition tasks and obtain the desired results. This is a common problem in the current algorithms. In subsequent research, the generalization performance of the model should be further improved by increasing the types of scenes in the collection of datasets and simultaneously enhancing its robustness in dealing with various unexpected situations.
In the future, developing application-specific integrated circuits (ASICs) for binary neural networks can fully leverage the advantages of DABNN to achieve low-cost and high-performance agricultural applications. And the algorithm can be transplanted to various portable devices, making it convenient for users to timely identify disease categories and apply the correct pesticides to improve crop yield.
6. Conclusions
In this paper, we propose an improved binary neural network model for plant disease classification, called DABNN. Firstly, a binary neural network is introduced into disease classification, which requires low GPU memory and saves computational resources. Secondly, a dual-attention mechanism is introduced and combined with the binary neural network to enhance the feature expression. Finally, experiments are conducted on the PlantVillage dataset, and the results show that our algorithm achieves 99.4% accuracy. Meanwhile, we also carry out the computational complexity analysis. Compared with other models, DABNN has lower computational complexity while maintaining high accuracy. DABNN strikes a good balance between computational complexity and precision. In addition, due to its lightweight structure, DABNN can be run on devices with low computational performance, which is more suitable for application in the agricultural field.
Author Contributions
Conceptualization, P.M. and J.Z.; methodology, P.M.; software, P.M. and G.Z.; validation, P.M. and J.Z.; formal analysis, P.M.; investigation, G.Z.; resources, P.M.; data curation, P.M.; writing—original draft preparation, P.M. and G.Z.; writing—review and editing, P.M. and J.Z.; funding acquisition, P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Development Plan Project of Jilin Province under Grant 20200201294JC.
Data Availability Statement
The PlantVillage dataset referred to in this study is openly available in “Data for: Identification of Plant Leaf Diseases Using a 9-layer Deep Convolutional Neural Network” at https://data.mendeley.com/datasets/tywbtsjrjv/1, accessed on 10 April 2023.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Flach, P. Machine Learning: The Art and Science of Algorithms That Make Sense of Data; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Oakland, CA, USA, 1 January 1967; Volume 1, pp. 281–297. [Google Scholar]
- Breiman, L. Random forest. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, UK, 2016. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Lecun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional Networks and Applications in Vision. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems, Paris, France, 30 May–2 June 2010. [Google Scholar]
- Sm, A.; Rs, A.; Dr, A. Deep Convolutional Neural Network based Detection System for Real-time Corn Plant Disease Recognition. Procedia Comput. Sci. 2020, 167, 2003–2010. [Google Scholar]
- Matas, J.; Chum, O.; Urban, M.; Pajdla, T. Robust wide-baseline stereo from maximally stable extremal regions. Image Vis. Comput. 2004, 22, 761–767. [Google Scholar] [CrossRef]
- Li, M.; Zhou, G.; Chen, A.; Li, L.; Hu, Y. Identification of tomato leaf diseases based on LMBRNet. Eng. Appl. Artif. Intell. 2023, 123, 106195. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Xie, X.; Ma, Y.; Liu, B.; He, J.; Li, S.; Wang, H. A Deep-Learning-Based Real-Time Detector for Grape Leaf Diseases Using Improved Convolutional Neural Networks. Front. Plant Sci. 2020, 11, 751. [Google Scholar] [CrossRef]
- Wen, W.; Wu, C.; Wang, Y.; Chen, Y.; Li, H. Learning Structured Sparsity in Deep Neural Networks. arXiv 2016, arXiv:1608.03665. [Google Scholar]
- Srinivas, S.; Babu, R.V. Data-free parameter pruning for Deep Neural Networks. arXiv 2015, arXiv:1507.06149. [Google Scholar]
- Han, S.; Pool, J.; Tran, J.; Dally, W.J. Learning both Weights and Connections for Efficient Neural Networks. In Proceedings of the 29th Annual Conference on Neural Information Processing Systems (NIPS), Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the Knowledge in a Neural Network. arXiv 2015, arXiv:1503.02531. [Google Scholar]
- Sainath, T.N.; Kingsbury, B.; Sindhwani, V.; Arisoy, E.; Ramabhadran, B. Low-rank matrix factorization for Deep Neural Network training with high-dimensional output targets. In Proceedings of the IEEE International Conference on Acoustics, Vancouver, BC, Canada, 26–31 May 2013. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Zoph, B.; Le, Q.V. Neural Architecture Search with Reinforcement Learning. arXiv 2017, arXiv:1611.01578. [Google Scholar]
- Liu, H.; Simonyan, K.; Yang, Y. DARTS: Differentiable Architecture Search. arXiv 2019, arXiv:1806.09055. [Google Scholar]
- Jacob, B.; Kligys, S.; Chen, B.; Zhu, M.; Tang, M.; Howard, A.; Adam, H.; Kalenichenko, D. Quantization and Training of Neural Networks for Efficient Integer-Arithmetic-Only Inference. In Proceedings of the 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 2704–2713. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathe, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2016, arXiv:1511.08060. [Google Scholar]
- Courbariaux, M.; Bengio, Y. BinaryNet: Training Deep Neural Networks with Weights and Activations Constrained to +1 or -1. arXiv 2016, arXiv:1602.02830. [Google Scholar]
- Liu, Z.; Shen, Z.; Savvides, M.; Cheng, K.T. ReActNet: Towards Precise Binary Neural Network with Generalized Activation Functions. In Proceedings of the European Conference on Computer Vision. (ECCV), Glasgow, UK, 23–28 August 2020; pp. 143–159. [Google Scholar]
- Liu, Z.; Wu, B.; Luo, W.; Yang, X.; Liu, W.; Cheng, K. Bi-Real Net: Enhancing the Performance of 1-bit CNNs With Improved Representational Capability and Advanced Training Algorithm. arXiv 2018, arXiv:1808.00278. [Google Scholar]
- Martínez, B.; Yang, J.; Bulat, A.; Tzimiropoulos, G. Training Binary Neural Networks with Real-to-Binary Convolutions. arXiv 2020, arXiv:2003.11535. [Google Scholar]
- Rastegari, M.; Ordonez, V.; Redmon, J.; Farhadi, A. XNOR-Net: ImageNet Classification Using Binary Convolutional Neural Networks. In Proceedings of the 14th European Conference on Computer Vision (ECCV), Amsterdam, The Netherlands, 8–16 October 2016; Volume 9908, pp. 525–542. [Google Scholar] [CrossRef]
- Kung, J.; Zhang, D.; Gooitzen, V.; Chai, S.; Mukhopadhyay, S. Efficient Object Detection Using Embedded Binarized Neural Networks. J. Signal Process. Syst. 2018, 90, 877–890. [Google Scholar] [CrossRef]
- Wang, X.; Sun, S.; Yin, Y.; Xu, D.; Wu, W.; Gu, Q. Fast Object Detection Based on Binary Deep Convolution Neural Networks. Caai Trans. Intell. Technol. 2018, 3, 191–197. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural Machine Translation by Jointly Learning to Align and Translate. arXiv 2016, arXiv:1409.0473. [Google Scholar]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; Volume 11211, pp. 3–19. [Google Scholar] [CrossRef]
- Dai, J.; Qi, H.; Xiong, Y.; Li, Y.; Zhang, G.; Hu, H.; Wei, Y. Deformable Convolutional Networks. In Proceedings of the 16th IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 764–773. [Google Scholar] [CrossRef]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. arXiv 2020, arXiv:2005.12872. [Google Scholar]
- Xie, D.; Chen, L.; Liu, L.; Chen, L.; Wang, H. Actuators and Sensors for Application in Agricultural Robots: A Review. Machines 2022, 10, 913. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, J. OCNet: Object Context Network for Scene Parsing. arXiv 2018, arXiv:1809.00916. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Fang, Z.; Lu, H. Dual Attention Network for Scene Segmentation. arXiv 2018, arXiv:1809.02983. [Google Scholar]
- Alirezazadeh, P.; Schirrmann, M.; Stolzenburg, F. Improving Deep Learning-based Plant Disease Classification with Attention Mechanism. Gesunde Pflanz. 2023, 75, 49–59. [Google Scholar] [CrossRef]
- Bhujel, A.; Kim, N.E.; Arulmozhi, E.; Basak, J.K.; Kim, H.T. A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification. Agriculture 2022, 12, 228. [Google Scholar] [CrossRef]
- Guo, W.; Feng, Q.; Li, X.; Yang, S.; Yang, J. Grape leaf disease detection based on attention mechanisms. Int. J. Agric. Biol. Eng. 2022, 15, 205–212. [Google Scholar] [CrossRef]
- Dwivedi, R.; Dey, S.; Chakraborty, C.; Tiwari, S. Grape Disease Detection Network Based on Multi-Task Learning and Attention Features. IEEE Sens. J. 2021, 21, 17573–17580. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, H.; Xiao, J.; Nie, L.; Shao, J.; Liu, W.; Chua, T.S. SCA-CNN: Spatial and Channel-Wise Attention in Convolutional Networks for Image Captioning. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 6298–6306. [Google Scholar] [CrossRef]
- Guo, N.; Bethge, J.; Meinel, C.; Yang, H. Join the high accuracy club on ImageNet with a binary neural network ticket. arXiv 2022, arXiv:2211.12933. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).