Genome-Wide Association Study Identifies Loci Associated with Sensitive Skin
Abstract
1. Introduction
2. Methods
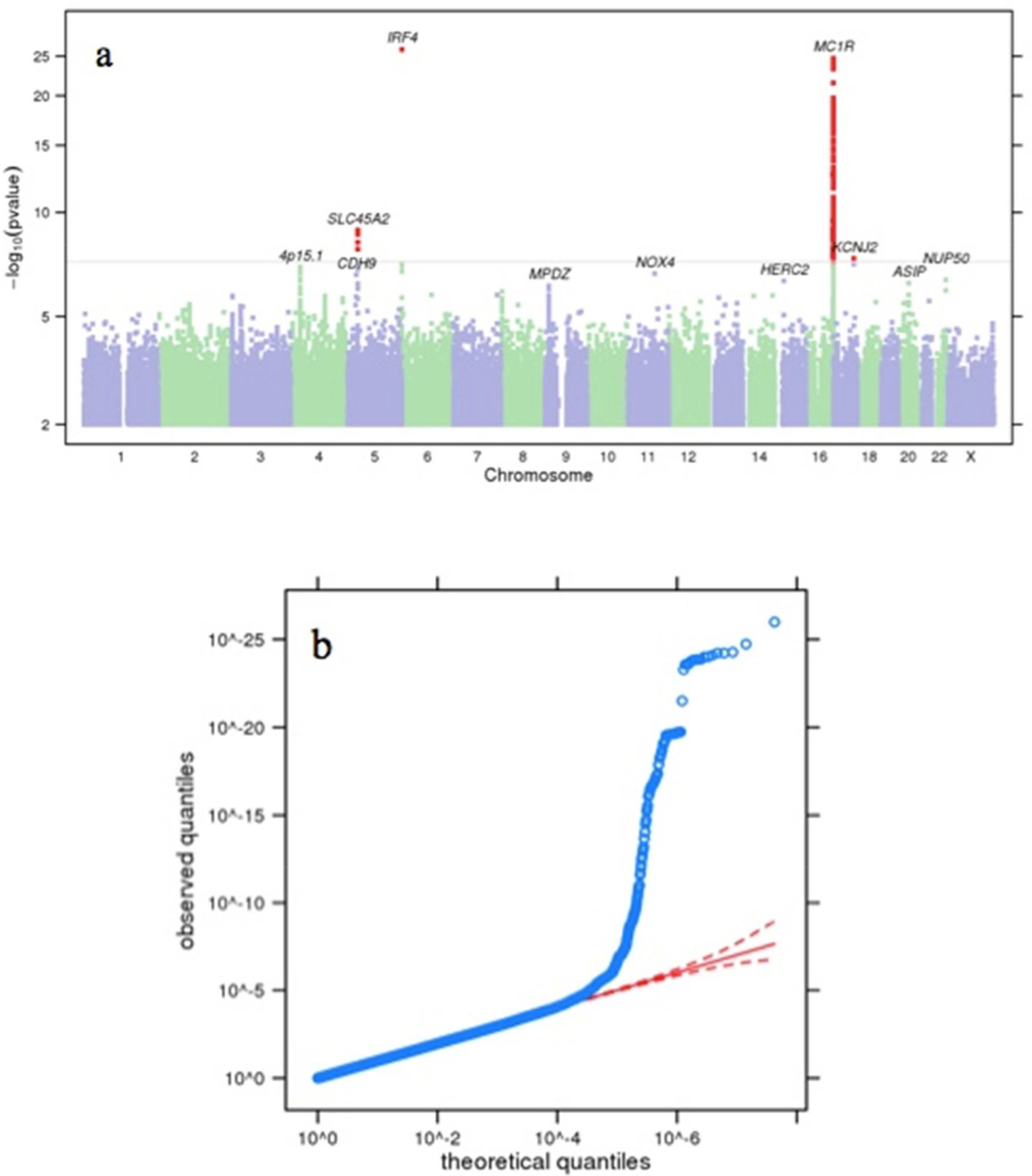
3. Results and Discussion
Supplementary Materials
Data Availability
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Misery, L.; Ständer, S.; Szepietowski, J.C.; Reich, A.; Wallengren, J.; Evers, A.W.; Takamori, K.; Brenaut, E.; Le Gall-Ianotto, C.; Fluhr, J.; et al. Definition of sensitive skin: An expert position paper from the special interest group on sensitive skin of the international forum for the study of itch. Acta. Derm. Venereol. 2017, 97, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Misery, L.; Loser, K.; Ständer, S. Sensitive Skin. J. Eur. Acad. Dermatol. Venereol. 2016, 1 (Suppl. 30), 2–8. [Google Scholar]
- Misery, L. Sensitive skin. Exper. Rev. Dermatol. 2013, 8, 631–637. [Google Scholar] [CrossRef]
- Farage, M.A. The prevalence of sensitive skin. Front. Med. 2019, 6, 98. [Google Scholar] [CrossRef] [PubMed]
- Quillen, E.E.; Norton, H.L.; Parra, E.J.; Lona-Durazo, F.; Ang, K.C.; Illiescu, F.M.; Pearson, L.N.; Shriver, M.D.; Lasisi, T.; Gokcumen, O. Shades of complexity. New perspectives on the evolution and genetic architecture of human skin. Am. J. Phys. Anthropol. 2019, 67 (Suppl. 168), 4–26. [Google Scholar]
- Aponte, J.L.; Chiano, M.N.; Yerges-Armstrong, L.M.; Hinds, D.A.; Tian, C.; Gupta, A.; Guo, C.; Fraser, D.J. Assessment of Rosacea symptom severity by genome-wide association study and expression analysis highlights immuno-inflammatory and skin pigmentation genes. Hum. Mol. Genet. 2018, 27, 2762–2772. [Google Scholar] [CrossRef] [PubMed]
- Petridis, C.; Navarini, A.A.; Dand, N.; Saklatvala, J.; Baudry, D.; Duckworth, M.; Allen, M.H.; Curtis, C.J.; Lee, S.H.; Burden, A.D. Genome-wide meta-analysis implicates mediators of hair follicle development and morphogenesis in risk for severe acne. Nat. Commun. 2018, 9, 5075. [Google Scholar] [CrossRef] [PubMed]
- Paternoster, L.; Standl, M.; Waage, J.; Baurecht, H.; Hotze, M.; Strachan, D.P.; Curtin, J.A.; Bonnelykke, K.; Tian, C.; Takahashi, A.; et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet. 2015, 47, 1449–1456. [Google Scholar] [PubMed]
- Tsoi, L.C.; Stuart, P.E.; Tian, C.; Gudjonsson, J.E.; Das, S.; Zawistowski, M.; Ellinghaus, E.; Barker, J.N.; Chandran, V.; Dand, N.; et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat. Commun. 2017, 8, 15382. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.L.S.; Raber, I.; Xu, J.; Li, R.; Spitale, R.; Chen, J.; Kiefer, A.K.; Tian, C.; Eriksson, N.K. Assessment of the genetic basis of Rosacea by genome-wide association study. J. Invest. Dermatol. 2015, 135, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Open Targets Genetics. Available online: https://genetics.opentargets.org/ (accessed on 16 June 2020).
- Chen, W.; Dai, R.; Li, L. The prevalence of self-declared sensitive skin: A systematic review and meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2019. [Google Scholar] [CrossRef]
- Tian, C.; Hromatka, B.S.; Kiefer, A.K.; Eriksson, N.; Noble, S.M.; Tung, J.Y.; Hinds, D.A. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017, 8, 599. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information NLOM: Database of Single Nucleotide Polymorphisms (dbSNP). Available online: https://www.ncbi.nlm.nih.gov/snp (accessed on 18 February 2019).
- National Library of Medicine: Genetics Home Reference: Genes. Available online: https://ghr.nlm.nih.gov/gene (accessed on 9 April 2019).
- Praetorius, C.; Grill, C.; Stacey, S.N.; Metcalf, A.M.; Gorkin, D.U.; Robinson, K.C.; Van Otterloo, E.; Kim, R.S.; Bergsteinsdottir, K. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell 2013, 155, 1033. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Achuthan, P.; Akanni, V.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic. Acids. Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- NHGRI-EBI: GWAS catalog: The NHGRI-EBI Catalog of published genome-wide association studies. Available online: https://www.ebi.ac.uk/gwas/downloads (accessed on 13 November 2019).
| Skin Conditions and Diseases: | Proportion |
|---|---|
| Contact dermatitis | 42.3% |
| Acne | 41.3% |
| Freckles | 33.8% |
| Atopic dermatitis (eczema) | 30.5% |
| Seborrheic dermatitis (dandruff) | 25.0% |
| Cold sores | 24.9% |
| Rosacea | 19.9% |
| Nail fungus | 15.1% |
| Pseudofolliculitis barbae | 14.0% |
| Actinic keratosis | 9.7% |
| Keloids | 9.5% |
| Melasma | 9.4% |
| Psoriasis | 9.2% |
| Skin cancer | 9.2% |
| Severe bruising | 8.1% |
| Chronic itch | 8.0% |
| Seborrheic keratosis | 5.7% |
| Dermatosis papulosis nigra | 3.4% |
| Top Variant | Location | Alleles | MAF | OR [95% CI] | p-Value | p-Value after Skin Color Correction | Lead Variant | Gene(s) | GWAS Traits |
|---|---|---|---|---|---|---|---|---|---|
| rs12203592 | chr6:396321 | C/T | 0.1718 | 1.36 [1.28–1.44] | 1.0 × 10−26 | 5.3 × 10−17 | rs12203592 | IRF4 | Pigmentation, skin cancer, mole, hair loss |
| rs1805007 | chr16:89986117 | C/T | 0.0746 | 1.54 [1.40–1.64] | 1.9 × 10−25 | 1.5 × 10−17 | rs1805007 (Arg151Cys) | MC1R | Pigmentation, skin cancer, mole, acne, rosacea |
| rs35407 | chr5:33946571 | G/A | 0.0351 | 0.69 [0.61–0.77] | 1.2 × 10−9 | 6.5 × 10−3 | rs35407 | SLC45A2 | Pigmentation, skin cancer, mole, acne, rosacea |
| rs61795905 | chr4:32588461 | C/T | 0.0884 | 0.82 [0.76–0.84] | 8.9 × 10−8 | 7.0 × 10−8 | - | - | None identified |
| rs199956012 | ch11:89058277 | TTAA/- | 0.1703 | 1.18 [1.10–1.25] | 1.9 × 10−7 | 2.6 × 10−7 | rs1126809 (Arg402Gln) | TYR | Pigmentation, skin cancer, mole, acne, rosacea |
| rs764649965 | chr5:27631799 | C/T | 0.0003 | 165.37 [7.61–Inf] | 2.0 × 10−7 | 1.4 × 10−3 | − | − | |
| rs10599246 | chr22:45508123 | A/- | 0.4968 | 1.12 [1.07–1.16] | 3.4 × 10−7 | 7.9 × 10−7 | rs5766565 | KIAA0930/NUP50 | Pigmentation |
| rs1667392 | chr15:28533565 | C/G | 0.3051 | 0.84 [0.78–0.90] | 3.9 × 10−7 | 5.8 × 10−3 | rs62007494 | HERC2 | Pigmentation, skin cancer, mole |
| rs540655099 | chr20:32819222 | -/G | 0.0753 | 1.22 [1.13–1.32] | 4.9 × 10-7 | 3.8 × 10-4 | rs540655099 | ASIP | Pigmentation, skin cancer, mole |
| rs77013500 | chr9:12978473 | G/C | 0.0083 | 0.51 [0.39–0.67] | 6.2 × 10−7 | 2.7 × 10−6 | rs12340775 | MPDZ | Hair loss |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farage, M.A.; Jiang, Y.; Tiesman, J.P.; Fontanillas, P.; Osborne, R. Genome-Wide Association Study Identifies Loci Associated with Sensitive Skin. Cosmetics 2020, 7, 49. https://doi.org/10.3390/cosmetics7020049
Farage MA, Jiang Y, Tiesman JP, Fontanillas P, Osborne R. Genome-Wide Association Study Identifies Loci Associated with Sensitive Skin. Cosmetics. 2020; 7(2):49. https://doi.org/10.3390/cosmetics7020049
Chicago/Turabian StyleFarage, Miranda A., Yunxuan Jiang, Jay P. Tiesman, Pierre Fontanillas, and Rosemarie Osborne. 2020. "Genome-Wide Association Study Identifies Loci Associated with Sensitive Skin" Cosmetics 7, no. 2: 49. https://doi.org/10.3390/cosmetics7020049
APA StyleFarage, M. A., Jiang, Y., Tiesman, J. P., Fontanillas, P., & Osborne, R. (2020). Genome-Wide Association Study Identifies Loci Associated with Sensitive Skin. Cosmetics, 7(2), 49. https://doi.org/10.3390/cosmetics7020049






