Abstract
In this review we discuss the interaction between metabolic stress, mitochondrial dysfunction, and genomic instability. Unrepaired DNA damage in the nucleus resulting from excess accumulation of DNA damages and stalled replication can initiate cellular signaling responses that negatively affect metabolism and mitochondrial function. On the other hand, mitochondrial pathologies can also lead to stress in the nucleus, and cause sensitivity to DNA-damaging agents. These are examples of how hallmarks of cancer and aging are connected and influenced by each other to protect humans from disease.
1. Introduction
It has been almost two decades since Hanahan and Weinberg for the first time classified the hallmarks of cancer [1]. Ten years later, they updated that list and introduced genomic instability and dysregulation of cellular energetics and mitochondrial function as emerging hallmarks [2]. Recently, both genomic instability and mitochondrial dysfunction are considered as two of the key hallmarks of aging [3]. Both of these have been implicated in several pathologies, as reviewed in References [4,5,6]. There are other hallmarks that are common between cancer and aging, such as epigenetic changes and altered cellular communication. Moreover, other hallmarks are in opposition to each other in cancer and aging. These include the dysregulation of apoptosis and senescence, which are stimulated in aging cells and suppressed in cancer cells [2,3]. Whether we can consider cancer as the disease of aging is a topic that is beyond the scope of this review, but age is the largest risk factor in the development of cancer [7].
One of the key questions that remain to be answered is, how are these hallmarks are connected and influenced by each other [3]? As these pathologic cellular changes occur gradually, understanding the connection between them would help to develop more effective therapeutic strategies to treat cancer or rather prevent it.
It has long been known that byproducts of cellular metabolism such as reactive oxygen and nitrogen species (ROS and RNS) can damage cellular components and macromolecules including DNA [8,9]. Damage to DNA can have severe effects on cells by blocking replication, transcription, generation of DNA double and single strand breaks, as well as chromosome rearrangements [10,11]. Activation of the DNA damage response (DDR) following DNA damage is an energy-demanding process [12], and can deplete cells of substrates such as NAD+ and ATP, which can in turn lead to additional metabolic stress and mitochondrial dysfunction (Figure 1) [13,14,15].
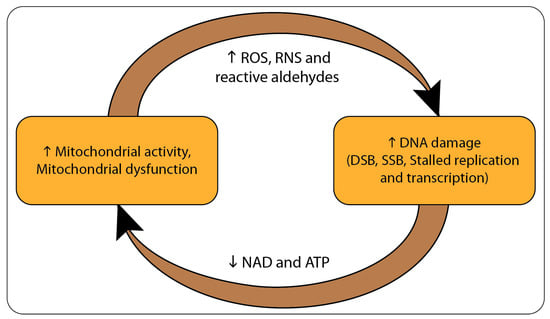
Figure 1.
Illustration of the mitochondrial-nuclear interactions in aging or cancer. Abnormal metabolism and/or metabolic defects lead to metabolic stress and mitochondrial dysfunction. This is followed by the increased generation of reactive oxygen and nitrogen species (ROS and RNS) as well as reactive aldehydes. These reactive species can react and damage macromolecules such as proteins and DNA. Damage to DNA causes genomic instability via stalled replication and transcription, and the generation of double- and single-strand breaks (DSBs and SSBs, respectively) within the genome. Increased activities of the DNA damage response (DDR) deplete cells of key cellular substrates and cofactors, mainly ATP and NAD+. This generates a positive feedback that enhances metabolic stress and mitochondrial dysfunction.
In this review, we aim to discuss the interaction between metabolic stress and mitochondrial dysfunction with genomic instability. Stress in the nucleus, such as the accumulation of DNA damage and stalled replication, negatively affects metabolism and mitochondrial function [13,14,15,16,17], while mitochondrial pathologies lead to stress in the nucleus [18] and cause sensitivity to DNA-damaging agents [19], as reviewed by Desler et al. in 2012 [20].
First, we explain how the accumulation of DNA damages and the activation of the DDR leads to mitochondrial dysfunction. Next, we explain how the dysregulation of mitochondrial function and metabolism contributes to the epigenetic changes, imbalanced dNTP pools, and genomic instability.
2. From DNA Damage to Mitochondrial Dysfunction
Activation of the main components of the DDR, including poly (ADP-ribose) polymerase (PARP) enzymes (mainly PARP1 and PARP2) as well as ataxia telangiectasia mutated (ATM) [21], DNA-dependent protein kinase (DNA-PK) [22,23] and P53 [24,25,26,27], is able to influence mitochondrial function and cellular metabolism. Chronic activation of PARP1 negatively affects cellular physiology and mitochondrial function [28]. Activation of ATM, DNA-PK, and P53 can influence mitochondrial and cellular metabolism to promote either cell survival or death (Figure 2). Here, we briefly describe how each of these enzymes are able to influence mitochondrial function.
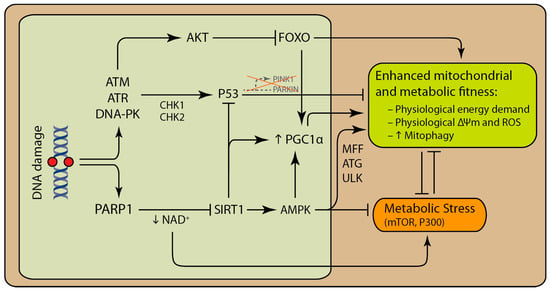
Figure 2.
Core DNA damage response proteins are able to influence mitochondrial activity and quality control via multiple pathways. Abnormal DNA structure and certain genomic lesions, such as strand breaks, activate poly (ADP-ribose) polymerase (PARP) enzymes, mainly PARP1. Chronic activation of PARP1 can deplete the cell of NAD+, which is a rate-limiting substrate for SIRT1. SIRT1 together with AMP-activated kinase (AMPK) can enhance metabolism and mitochondrial function by enhancing mitochondrial biogenesis and mitophagy. Activation of ataxia telangiectasia mutated (ATM), Rad3-related (ATR) and DNA-dependent protein kinase (DNA-PK) following DNA damage can promote the activation AKT and P53. Activation of AKT rewires cellular metabolism by inhibiting Forkhead box (FOXO) enzymes. Deacetylation of P53 by SIRT1 targets P53 for degradation. Decrease in SIRT1 activity stabilizes P53. P53 decreases mitophagy via inhibition of PTEN-induced kinase 1 (PINK1) and PARKIN transcription. SIRT1 promotes AMPK activity indirectly. Decrease in SIRT1 activity is followed by the decrease in activated AMPK.
3. PARP Modulates Mitochondrial Function and Cellular Metabolism
PARPs are a group of enzymes (16 in mice and 17 in humans) that are the main constituent of the cellular stress response [29]. PARPs cleave NAD+ to nicotinamide (NAM) and ADP-ribose (ADPR), and the ADPR is subsequently transferred to certain amino acids within the target protein. The attachment of poly ADP-ribose (PAR) to the target proteins is referred to as PARylation, and it can affect protein–protein and protein–DNA interaction as well as protein localization [30]. PAR has a short half-life and is degraded almost directly after its formation by the activity of the PAR-degrading enzyme, poly(ADP-ribose) glycohydrolase (PARG) [31]. PARylation modulates several key cellular processes such as chromatin structure, transcription, translation, cell cycle, DNA repair, mitochondrial homeostasis, apoptosis, and metabolism [29,32]. PARP1 is activated by several mechanisms, including mono(ADP-ribosyl)ation, phosphorylation, and acetylation [29]. PARP1 possesses a DNA-binding domain that recognizes abnormal DNA structures such as gapped DNA, single- and double strand breaks, cruciform structures, and nucleosome linker DNA [33,34]. In the initial steps of the repair, PARP1 PARylates histones and facilitates the chromatin relaxation that provides more space for the recruitment of DNA repair proteins. Subsequent PAR generation recruits the DNA repair proteins via their PAR-binding domains [35,36]. Under mild genotoxic stress, PARP activation results in repair and survival. However, in response to DNA damage, PARP hyper-activation results in decrease in NAD+ and ATP levels, mitochondrial dysfunction, and eventually cell death [32,35,37].
4. DNA Damage can Activate Both Pro-Survival and Pro-Death Pathways That Involve the Mitochondria
DNA damage and DDR can activate pathways that promote cell survival or death depending on the extent and type of DNA damages [38]. In addition to PARP1, other immediate sensors of DDR are enzymes belonging to the superfamily of phosphatidylinositol 3-kinase-related kinases (PIKKs), including ATM, ataxia telangiectasia and Rad3-related (ATR), and DNA-PK. Activated ATR and ATM, in turn, activate P53 through CHK1 and CHK2, respectively. Apart from preventing cell cycle progression and the recruitment of DNA repair proteins, these enzymes can modulate mitochondrial function and survival [23,39]. ATM, ATR, and DNA-PK are able to promote survival via the direct phosphorylation of AKT (also known as protein kinase B, PKB) independently of growth factor signaling [22,40,41,42,43]. However, the mechanism of this interaction is not fully understood [44]. This is particularly interesting, as in many cancer cells, the AKT is activated independently of growth factors [45]. Activated AKT stimulates glucose uptake and ATP production through glycolysis, one of the main hallmarks of cancer, also known as the Warburg effect [46]. In addition, activated AKT inhibits Forkhead box (FOXO) transcription factors [47,48]. FOXO proteins regulate the expression of key genes that are involved in mitochondrial biogenesis and homeostasis, such as peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC1a) and PTEN-induced kinase 1 (PINK1). Decrease in FOXO activity results in the decrease of mitochondrial biogenesis, mitophagy, autophagy, and lipolysis [49].
5. Mito-Nuclear Signaling in Aging and Cancer
For a long time, it was believed that mitochondria are regulated from the nucleus by the nuclear genome, and changes in mitochondria follow changes in the nucleus [50,51]. However, during recent years, accumulative evidence suggest that mitochondria and mitochondrial metabolites can influence nuclear processes and gene expression in response to various stimuli and environmental cues [52,53,54]. Mitochondrially generated ROS and intermediate metabolites are essential for several processes, including proliferation, epigenetic modifications, and post-translational modifications [54,55]. In addition, mitochondria contribute to genomic stability by replenishing dNTP pools for replication and repair of the genome (Figure 3) [56,57]. In this section we will discuss nuclear processes that are dependent on mitochondrial function and intermediate metabolites.
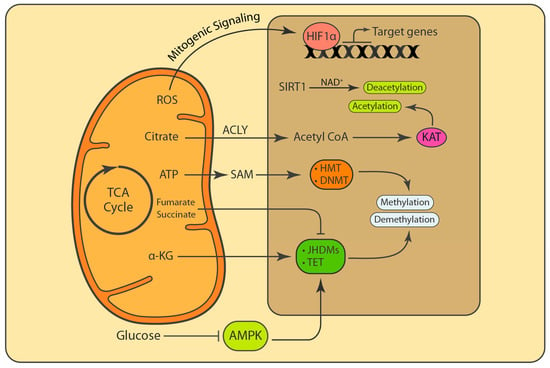
Figure 3.
Mitochondrial function and metabolites influence nuclear processes. Mitochondrial ROS and secondary metabolites act as signaling molecules and cofactors that regulate fundamental nuclear processes. Mitochondrial ROS that are released into the cytosol stabilize hypoxia-inducible factor 1- α (HIF1α) and activate the transcription of genes that are involved in proliferation. Citrate generated through the TCA cycle is released into the cytosol and in the nucleus. It is further converted into acetyl-coenzyme A (acetyl CoA) that is used for the acetylation of target proteins. Through a reverse reaction, SIRT1 uses NAD+ to deacetylate the target proteins. S-adenosylmethionine (SAM) is a methyl donor that is generated from methionine and ATP in the cytosol. Demethylases such as Jumonji C (JMJC) family members and the ten-eleven translocation (TET) methylcytosine hydroxylases use α-ketoglutarate (α-KG) as cofactor to remove methyl groups from proteins and DNA. AMPK stimulates the activity of TET enzymes, and thus the inhibition of AMPK by glucose impairs the function TET enzymes. The accumulation of fumarate and succinate due to impaired fumarate hydratase (FH) and succinate dehydrogenase (SDH) can inhibit α-KG-dependent demethylases and even cause defects in homologous recombination (HR) DNA repair.
6. Mitochondrial ROS Are Involved in Signaling and Determine Cell Fate
The mitochondrial electron transport chain (ETC) generates reactive oxygen species (ROS) as the byproduct of oxidative phosphorylation (OXPHOS) from different complexes, though mainly complexes I, II, and III. While complexes I and II exclusively create O2∙ in the mitochondrial matrix, complex III produces O2∙ in both the matrix and intermembrane space [58]. However, the ROS that are released outside of the matrix are converted into H2O2 by cytosolic superoxide dismutase 1 (SOD1) and participate in mitochondrial signaling through reversible cysteine oxidation [59]. Mitochondrially produced ROS can serve as second-messenger molecules. Mitochondrial ROS (mtROS) are required for the stabilization of HIFα (hypoxia-inducible factor 1-α) and the activation of downstream pathways that promote proliferation [60].
It is possible that the type of cell and energy demand determine the effect of ROS and mtDNA mutation over the cell. Cells that mostly rely on glycolysis will probably not be affected by mutation in mtDNA under physiological conditions. During the exposure to stress and stimuli, however, these cells might not be able to trigger an adaptive response to an increase in demand for ATP and NAD+ [61]. During stress and increased energy demand, cells respond by boosting cellular respiration and mitochondrial activity to provide the cells with ATP and NAD+ [12,62]. This is accompanied by increased mitochondrial membrane potential and ROS above the physiological level [61,63]. Increased ROS cause damage to macromolecules such as DNA, proteins, and lipids, which can cause cell death or malignancy, as reviewed by Sies et al. in 2017 [64]. The inability to enhance ATP production in response to stress and increased energy demand probably contributes to cellular deterioration during aging [65].
In contrast to increased ROS generation, a decrease in ROS in metabolically active or proliferative tissues interferes with cellular metabolism or proliferation that can promote senescence, as reviewed by Diebold and Chandel in 2016 [66].
Hematopoietic stem cells (HSCs) represent an excellent example for both situations [67]. HSCs are mainly quiescent, and they rely on glycolysis for ATP production [68]. While low levels of ROS prevent the proliferation of HSCs and maintain their quiescent state, stress and increased energy demand promote a shift toward ATP production by mitochondria and OXPHOS. This is accompanied by increased ROS and proliferation [67]. Chronic stress followed by enhanced mitochondrial activity and ROS generation leads to the depletion of HSCs, which is one of the hallmarks of aging [69,70,71].
7. Mitochondria Influence Post-Translational Modifications (PTMs) and Epigenetic Marks
Reversible acetylation and methylation are two frequently employed post-translational modifications that regulate a variety of protein functions, protein stability, gene expression [72], as well as DNA repair [73]. Epigenetic changes are also one of the main hallmarks of both aging and cancer [2,3]. The modifications include alterations in DNA methylation, modifications of histones, and chromatin remodeling. While DNA methylations show similar patterns in aging and cancer, the histone modifications show distinct patterns, as reviewed by Zane et al. in 2014 [74].
The multiple enzymatic systems assuring the generation and maintenance of epigenetic patterns include DNA methyltransferases, histone acetylases, deacetylases, methylases, and demethylases, as well as protein complexes implicated in chromatin remodeling. Most PTMs, such as phosphorylation, acetylation, methylation, and O-linked N-acetylglucosamine modification (O-GlcNAcylation), require metabolites as substrates [75]. If not all, the majority of substrates required for PTMs are intermediate metabolites generated by mitochondria [54]. Chromatin modifiers use metabolic intermediates as cofactors or substrates, but are also regulated by their availability. These metabolites include NAD+ for deacetylation, acetyl-CoA for histone acetylation, S-adenosylmethionine (SAM) for histone as well as DNA methylation, and α-ketoglutarate (α-KG) for demethylation [54].
Lysine acetyltransferases (KATs) add acetyl groups to proteins while lysine deacetylases (KDACs) remove acetyl groups from proteins [76,77]. KATs such as GCN5, CBP/p300, and MYST use acetyl-coenzyme A (acetyl CoA) as an acetyl group donor for protein acetylation [78]. KDACs are classified into two groups with different catalytic mechanisms: Zn2+-dependent histone deacetylases (HDAC1-11) and NAD+-dependent deacetylases (SIRT1-7) [76,77]. Acetyl CoA is generated in mitochondria and converted to citrate through the TCA cycle. Citrate can then be exported from mitochondria. In the cytoplasm and nucleus, citrate is converted back into acetyl CoA via the function of ATP-citrate lyase (ACLY). Citrate is the major source of acetyl CoA in the cytoplasm and the nucleus. Depletion of mtDNA and the subsequent decrease in NAD negatively affect the TCA cycle and lead to a decrease in histone acetylation. This can be rescued by the restoration of electron flow and TCA cycle [60,79]. The degree of acetylation directly correlates with the availability of cofactors such as acetyl CoA and NAD+. While increased nuclear acetyl CoA promotes increased acetylation and the formation of euchromatin, an increase in NAD+ promotes deacetylation and the formation of heterochromatin, resulting in a decrease of gene expression [79]. MYC and AKT stimulate nutrient uptake and promote acetyl CoA production via ACLY. AKT can directly phosphorylate and activate ACLY to maintain the acetyl CoA levels, regardless of glucose concentrations [80].
Another key chromatin modification that is strongly interconnected with metabolism is methylation [54,81]. Methylation is regulated by S-adenosylmethionine (SAM) abundance, whereby SAM serves as a universal methyl donor, synthesized from methionine and ATP by methionine adenosyltransferases (MATs) [54,81]. Histone and DNA methylation is removed by demethylases such as Jumonji C (JMJC) family members and the ten-eleven translocation (TET) methylcytosine hydroxylases, which use a dioxygenation reaction that requires Fe2+, O2, and α-ketoglutarate (α-KG) as cofactors [82]. α-KG is generated in the TCA cycle via the catabolism of glucose and glutamine. The function of lysine-specific histone demethylase 1A (LSD1; KDM1A) and LSD2 (KDM1B), which catalyze an amine oxidation reaction, is dependent on flavin adenine dinucleotide (FAD) [83,84].
The dysregulation of the TCA cycle and cellular metabolism affect PTMs, epigenetic changes, and choice of DNA repair pathway. Accumulation of succinate or fumarate, which occurs respectively in tumors deficient for succinate dehydrogenase (SDH) or fumarate hydratase (FH), similarly inhibits α-KG-dependent enzymes, leading to defects in homologous recombination (HR) DNA repair [53]. Changes in nutrient availability can directly affect chromatin modifications. The tumor suppressor ten-eleven translocation (TET) protein family of dioxygenases (TET1, TET2, and TET3) converts a 5mC DNA methylation to a hydroxymethylation, 5hmC [85]. AMP-activated kinase (AMPK) phosphorylates TET2 and stabilizes this tumor suppressor protein. However, increase in blood glucose levels impairs AMPK activity, which leads to destabilization of TET2 and the subsequent dysregulation of 5mCs and 5hmCs [86].
8. Regulation of dNTP Pools
In humans, nucleotide levels are maintained by the nucleotide salvage and/or de novo synthesis of ribo- and deoxyribonucleotide triphosphates (rNTPs and dNTPs).
It is generally accepted that the levels and especially the relative balance of the cytosolic dNTP pools have great influence on the replication, repair, and stability of the nuclear genome [87,88,89,90,91,92]. The efficiency and fidelity of most, if not all, polymerases and many repair enzymes are affected by the level of substrate nucleotides [87]. Too low a concentration of a nucleotide results in poor incorporation frequency, and a level which is too high risks the misincorporation of the high-concentration nucleotide [93,94,95]. Therefore, it is of no surprise that the process of synthesizing dNTPs in the right concentrations is governed by a generous amount of regulation, feedback loops, and redundancy [96,97].
Mitochondrial dysfunction, due to mutations in the mitochondrial genome or a decrease in the mitochondrial DNA (mtDNA) copy number, is associated with a poor prognosis of many types of cancer [98,99,100,101,102], also reviewed by Chatterjee et al. in 2006 [103]. The accumulation of mutations in mtDNA or impeded ETC have even been associated with tumor aggressiveness [99,101,104,105,106]. We have previously shown a relationship between mitochondrial respiration and the regulation of cytosolic dNTP pools, and demonstrated a co-occurring decrease of chromosomal stability [51].
The de novo synthesis of nucleotides is split up into purine and pyrimidine synthesis, which go through two distinct pathways.
Despite having separate synthesis pathways, both purine and pyrimidine-based ribonucleotides occupy a central role in cellular metabolism. In addition to being basal components of DNA and RNA, they function as phosphate donors in the transport of cellular energy and participate in enzymatic reactions as well as intracellular and extracellular signaling [107].
Both the salvage and the de novo synthesis pathways utilize an activated sugar intermediate: 5-phosphoribosyl-1-pyrophosphate (PRPP). PRPP is generated by the action of PRPP synthetase and is utilized in both purine and pyrimidine synthesis [108]. The purine nucleotides are synthesized from PRPP, through inosine 5-monophosphate (IMP), and further into AMP and GMP, through two separate but allosterically regulated pathways. Pyrimidines, on the other hand, originate from the precursor pyrimidine nucleotide, UMP, which is used to synthesize all the cellular ribosyl and deoxyribosyl pyrimidines including UTP, CTP, dUMP, and dTTP [108]. A key catalytic enzyme in this process is the dihydroorotate dehydrogenase (DHODH) [109], located in the inner mitochondrial membrane and functionally codependent with the OXPHOS [110]. The oxidation of dihydroorotate by DHODH, to form orotate, is a bottleneck reaction of the de novo synthesis of pyrimidines and is electrochemically coupled with the reduction of ubiquinone to ubiquinol [56,111,112,113]. Mitochondrial respiration can modulate the nucleotide synthesis at two separate steps. A decrease of the mitochondrial respiration is therefore linked to an inhibition of the DHODH enzyme, which in turn modulates the synthesis of pyrimidines [51,114]. Furthermore, the activity of the RNR complex is regulated by the binding of ATP to its active site and inhibited by dATP. By affecting the levels of cytosolic ATP, the mitochondria can influence the activity of RNR, and hence the levels of dNTPs. Furthermore, RNR is allosterically regulated by the relative levels of individual NTPs and dNTPs, and so the de novo synthesis of dNTPs and their relative balance is highly dependent on the mitochondrial supply of pyrimidines and ATP [114,115,116].
The cytosolic pool of dNTPs, which supplies the replication of the nuclear genome, is cell cycle-regulated. Synthesis is initiated at the beginning of the S-phase, and stopped upon reaching G2. In the de novo pathway, this phase-dependent synthesis of dNTPs is mediated through the RNR—specifically, through the cell cycle regulation of expression and degradation of the RNR subunit RNR-R2 [117]. In the salvage pathway, the phase-dependent synthesis is controlled by the translational regulation of constituents of the nucleotide salvage pathway [118]. Outside of S-phase, a dNTP hydrolase called SAMHD1 (SAM and HD domain-containing deoxynucleoside triphosphate triphosphohydrolase 1) depletes the dNTP pools by its hydrolase activity to block viral replication, amongst other things [119]. In response to DNA damage, however, dNTP levels can increase by up to 4-fold [96]. In this case, both the RNR and SAMHD1 are then recruited to the site of damage to tightly regulate the amount of dNTPs supplied to the DNA repair machinery [96,120].
Not only replication of the nuclear genome requires a balanced dNTP pool. Unlike the replication of the nuclear genome, the replication of mitochondrial DNA is not regulated by the cell cycle, but is carried out continuously in both mitotic and post-mitotic cells and tissue. Imbalance of the mitochondrial dNTP pools affects the replication of mtDNA, resulting in the accumulation of point mutations and deletions [121,122].
The dNTP pool of the mitochondrial compartment is somewhat separate from the much larger pool supplying the nuclear genome [123,124]. However, cytosolic de novo synthesis of dNTP is essential for mtDNA maintenance, even in post-mitotic cells and tissue [125,126,127].
P53R2 is a protein that substitutes RNR-R2 in post-mitotic cells in response to DNA damage. P53R2 is transcribed by P53 and results, when forming the complex, in the activation of RNR and the de novo synthesis of dNTPs intended as substrates for DNA repair mechanisms [128]. Mutations of the RRM2B gene encoding P53R2 have been shown to induce mtDNA replication and repair deficiency in post-mitotic, but not dividing, human fibroblasts [125]. In humans, mutations in the gene encoding the RRM2B subunit have been correlated with severe mtDNA depletion of muscle tissue, and RRM2b−/− mice further display a severe decrease of mtDNA content in liver, kidney, and muscle [126].
It is important to realize that balanced dNTP pools for mtDNA maintenance do not need to be of mitochondrial origin. Imbalances of the nuclear dNTP pools resulting from genetic predisposition, age, or even just diet [129] have the potential to start a vicious cycle, whereby failure to maintain mtDNA integrity results in the decreased synthesis of pyrimidines and further dNTP pool imbalance.
Mitochondrial dysfunction is therefore not only a risk to the mitotic cell, but also fully differentiated post-mitotic cells [130], and is involved in the etiology of a wide array of pathologies, including cancer and Alzheimer’s disease [131,132,133].
9. Conclusions
According to recent advancements, hallmarks of cancer include genomic instability, dysregulation of cellular energetics, and mitochondrial dysfunction, which also are common pathways important for cellular aging. Mitochondrial dysfunction is associated with a poor prognosis of many types of cancer, which could very well be linked to an imbalance of the cytosolic dNTP pools, as both of these conditions are related to one of the hallmarks of cancer—chromosomal instability. A better understanding of these pathological cellular processes would advance the development of therapeutic modalities in the prevention of cancer and at the same time help the understanding of biological aging.
Author Contributions
N.B.F., T.L.H., C.D., S.A., L.J.R. wrote the review.
Funding
This research was funded by Nordea-fonden and Olav Thon Foundation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Sharma, P.; Sampath, H. Mitochondrial DNA Integrity: Role in Health and Disease. Cells 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Xie, X.-H.; Chen, C.-H.; Peng, X.; Zhang, P.; Yang, C.; Wang, Y.-T. Molecular Regulation Mechanisms and Interactions Between Reactive Oxygen Species and Mitophagy. DNA Cell Biol. 2019, 38, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Aunan, J.R.; Cho, W.C.; Søreide, K. The Biology of Aging and Cancer: A Brief Overview of Shared and Divergent Molecular Hallmarks. Aging Dis. 2017, 8, 628. [Google Scholar] [CrossRef] [PubMed]
- Burcham, P.C. Internal hazards: Baseline DNA damage by endogenous products of normal metabolism. Mutat. Res./Genet. Toxicol. Environ. Mutagenesis 1999, 443, 11–36. [Google Scholar] [CrossRef]
- De Bont, R.; van Larebeke, N. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 2004, 19, 169–185. [Google Scholar] [CrossRef]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Vijg, J. Do DNA Double-Strand Breaks Drive Aging? Mol. Cell 2016, 63, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Fan, M.; Candas, D.; Jiang, G.; Papadopoulos, S.; Tian, L.; Woloschak, G.; Grdina, D.J.; Li, J.J. CDK1 Enhances Mitochondrial Bioenergetics for Radiation-Induced DNA Repair. Cell Rep. 2015, 13, 2056–2063. [Google Scholar] [CrossRef]
- Scheibye-Knudsen, M.; Mitchell, S.J.; Fang, E.F.; Iyama, T.; Ward, T.; Wang, J.; Dunn, C.A.; Singh, N.; Veith, S.; Hasan-Olive, M.M.; et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014, 20, 840–855. [Google Scholar] [CrossRef]
- Fang, E.F.; Kassahun, H.; Croteau, D.L.; Scheibye-Knudsen, M.; Marosi, K.; Lu, H.; Shamanna, R.A.; Kalyanasundaram, S.; Bollineni, R.C.; Wilson, M.A.; et al. NAD(+) Replenishment Improves Lifespan and Healthspan in Ataxia Telangiectasia Models via Mitophagy and DNA Repair. Cell Metab. 2016, 24, 566–581. [Google Scholar] [CrossRef]
- Fakouri, N.B.; Durhuus, J.A.; Regnell, C.E.; Angleys, M.; Desler, C.; Olive, M.H.; Martín-Pardillos, A.; Tsaalbi-Shtylik, A.; Thomsen, K.; Lauritzen, M.; et al. Rev1 contributes to proper mitochondrial function via the PARP-NAD+-SIRT1-PGC1α axis. Sci. Rep. 2017, 7, 12480. [Google Scholar] [CrossRef]
- Rivera-Torres, J.; Acín-Perez, R.; Cabezas-Sánchez, P.; Osorio, F.G.; Gonzalez-Gómez, C.; Megias, D.; Cámara, C.; López-Otín, C.; Enríquez, J.A.; Luque-García, J.L.; et al. Identification of mitochondrial dysfunction in Hutchinson–Gilford progeria syndrome through use of stable isotope labeling with amino acids in cell culture. J. Proteom. 2013, 91, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Scheibye-Knudsen, M.; Brace, L.E.; Kassahun, H.; SenGupta, T.; Nilsen, H.; Mitchell, J.R.; Croteau, D.L.; Bohr, V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 2014, 157, 882–896. [Google Scholar] [CrossRef]
- Qian, W.; Choi, S.; Gibson, G.A.; Watkins, S.C.; Bakkenist, C.J.; Van Houten, B. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J. Cell Sci. 2012, 125, 5745–5757. [Google Scholar] [CrossRef] [PubMed]
- Temelie, M.; Savu, D.I.; Moisoi, N. Intracellular and Intercellular Signalling Mechanisms following DNA Damage Are Modulated By PINK1. Oxid. Med. Cell. Longev. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Desler, C.; Hansen, T.L.; Frederiksen, J.B.; Marcker, M.L.; Singh, K.K.; Juel Rasmussen, L. Is There a Link between Mitochondrial Reserve Respiratory Capacity and Aging? J. Aging Res. 2012, 2012, 192503. [Google Scholar] [CrossRef]
- Cosentino, C.; Grieco, D.; Costanzo, V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011, 30, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Bozulic, L.; Surucu, B.; Hynx, D.; Hemmings, B.A. PKBα/Akt1 Acts Downstream of DNA-PK in the DNA Double-Strand Break Response and Promotes Survival. Mol. Cell 2008, 30, 203–213. [Google Scholar] [CrossRef]
- Park, S.-J.; Gavrilova, O.; Brown, A.L.; Soto, J.E.; Bremner, S.; Kim, J.; Xu, X.; Yang, S.; Um, J.-H.; Koch, L.G.; et al. DNA-PK Promotes the Mitochondrial, Metabolic, and Physical Decline that Occurs During Aging. Cell Metab. 2017, 25, 1135–1146.e7. [Google Scholar] [CrossRef] [PubMed]
- Matoba, S.; Kang, J.-G.; Patino, W.D.; Wragg, A.; Boehm, M.; Gavrilova, O.; Hurley, P.J.; Bunz, F.; Hwang, P.M. p53 regulates mitochondrial respiration. Science 2006, 312, 1650–1653. [Google Scholar] [CrossRef]
- Bensaad, K.; Vousden, K.H. p53: New roles in metabolism. Trends Cell Biol. 2007, 17, 286–291. [Google Scholar] [CrossRef]
- Hoshino, A.; Mita, Y.; Okawa, Y.; Ariyoshi, M.; Iwai-Kanai, E.; Ueyama, T.; Ikeda, K.; Ogata, T.; Matoba, S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat. Commun. 2013, 4, 2308. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.B.; Kinoshita, C.; Kinoshita, Y.; Morrison, R.S. p53 and mitochondrial function in neurons. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1186–1197. [Google Scholar] [CrossRef]
- Bai, P.; Nagy, L.; Fodor, T.; Liaudet, L.; Pacher, P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol. Metab. 2015, 26, 75–83. [Google Scholar] [CrossRef]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Krietsch, J.; Rouleau, M.; Pic, É.; Ethier, C.; Dawson, T.M.; Dawson, V.L.; Masson, J.-Y.; Poirier, G.G.; Gagné, J.-P. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Asp. Med. 2013, 34, 1066–1087. [Google Scholar] [CrossRef]
- Davidovic, L.; Vodenicharov, M.; Affar, E.B.; Poirier, G.G. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 2001, 268, 7–13. [Google Scholar] [CrossRef]
- Bürkle, A.; Virag, L. Poly(ADP-ribose): PARadigms and PARadoxes. Mol. Asp. Med. 2013, 34, 1046–1065. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011, 286, 10690–10701. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.A.E.; Timinszky, G.; Arribas-Bosacoma, R.; Kozlowski, M.; Hassa, P.O.; Hassler, M.; Ladurner, A.G.; Pearl, L.H.; Oliver, A.W. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012, 19, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, A.J.; Timinszky, G.; Kong, S.E.; Jin, J.; Cai, Y.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Ladurner, A.G.; Conaway, J.W.; et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA 2009, 106, 13770–13774. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Kraus, W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Scheibye-Knudsen, M.; Chua, K.F.; Mattson, M.P.; Croteau, D.L.; Bohr, V.A. Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 2016, 17, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef]
- Maryanovich, M.; Zaltsman, Y.; Ruggiero, A.; Goldman, A.; Shachnai, L.; Zaidman, S.L.; Porat, Z.; Golan, K.; Lapidot, T.; Gross, A. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat. Commun. 2015, 6, 7901. [Google Scholar] [CrossRef]
- Feng, J.; Park, J.; Cron, P.; Hess, D.; Hemmings, B.A. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004, 279, 41189–41196. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Park, J.; Feng, J.; Li, Y.; Hammarsten, O.; Brazil, D.P.; Hemmings, B.A. DNA-dependent protein kinase-mediated phosphorylation of protein kinase B requires a specific recognition sequence in the C-terminal hydrophobic motif. J. Biol. Chem. 2009, 284, 6169–6174. [Google Scholar] [CrossRef]
- Toulany, M.; Maier, J.; Iida, M.; Rebholz, S.; Holler, M.; Grottke, A.; Jüker, M.; Wheeler, D.L.; Rothbauer, U.; Rodemann, H.P. Akt1 and Akt3 but not Akt2 through interaction with DNA-PKcs stimulate proliferation and post-irradiation cell survival of K-RAS-mutated cancer cells. Cell Death Discov. 2017, 3, 17072. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Turner, K.M.; Yung, W.K.A.; Chen, K.; Zhang, W. Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro-Oncology 2014, 16, 1313–1323. [Google Scholar] [CrossRef]
- Stronach, E.A.; Chen, M.; Maginn, E.N.; Agarwal, R.; Mills, G.B.; Wasan, H.; Gabra, H. DNA-PK mediates AKT activation and apoptosis inhibition in clinically acquired platinum resistance. Neoplasia 2011, 13, 1069–1080. [Google Scholar] [CrossRef]
- Robey, R.B.; Hay, N. Is Akt the “Warburg kinase”-Akt-energy metabolism interactions and oncogenesis. Semin. Cancer Biol. 2009, 19, 25–31. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Kops, G.J.P.L.; de Ruiter, N.D.; De Vries-Smits, A.M.M.; Powell, D.R.; Bos, J.L.; Burgering, B.M.T. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 1999, 398, 630–634. [Google Scholar] [CrossRef]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012, 4, a011403. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yen, K.; Cohen, P. Humanin: A harbinger of mitochondrial-derived peptides? Trends Endocrinol. Metab. 2013, 24, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Son, J.M.; Benayoun, B.A.; Lee, C. The Mitochondrial-Encoded Peptide MOTS-c Translocates to the Nucleus to Regulate Nuclear Gene Expression in Response to Metabolic Stress. Cell Metab. 2018, 28, 516–524. [Google Scholar] [CrossRef]
- Sulkowski, P.L.; Sundaram, R.K.; Oeck, S.; Corso, C.D.; Liu, Y.; Noorbakhsh, S.; Niger, M.; Boeke, M.; Ueno, D.; Kalathil, A.N.; et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat. Genet. 2018, 50, 1086–1092. [Google Scholar] [CrossRef]
- Li, X.; Egervari, G.; Wang, Y.; Berger, S.L.; Lu, Z. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol. 2018, 19, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Sivanand, S.; Rhoades, S.; Jiang, Q.; Lee, J.V.; Benci, J.; Zhang, J.; Yuan, S.; Viney, I.; Zhao, S.; Carrer, A.; et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol. Cell 2017, 67, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Desler, C.; Munch-Petersen, B.; Stevnsner, T.; Matsui, S.-I.; Kulawiec, M.; Singh, K.K.; Rasmussen, L.J. Mitochondria as determinant of nucleotide pools and chromosomal stability. Mutat. Res. 2007, 625, 112–124. [Google Scholar] [CrossRef]
- Rasmussen, A.K.; Chatterjee, A.; Rasmussen, L.J.; Singh, K.K. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 3909–3917. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Diebold, L.P.; Kong, H.; Schieber, M.; Huang, H.; Hensley, C.T.; Mehta, M.M.; Wang, T.; Santos, J.H.; Woychik, R.; et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol. Cell 2016, 61, 199–209. [Google Scholar] [CrossRef]
- Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta Bioenerget. 2012, 1817, 1833–1838. [Google Scholar] [CrossRef]
- Brace, L.E.; Vose, S.C.; Stanya, K.; Gathungu, R.M.; Marur, V.R.; Longchamp, A.; Treviño-Villarreal, H.; Mejia, P.; Vargas, D.; Inouye, K.; et al. Increased oxidative phosphorylation in response to acute and chronic DNA damage. NPJ Aging Mech. Dis. 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Trifunovic, A.; Hansson, A.; Wredenberg, A.; Rovio, A.T.; Dufour, E.; Khvorostov, I.; Spelbrink, J.N.; Wibom, R.; Jacobs, H.T.; Larsson, N.-G. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl. Acad. Sci. USA 2005, 102, 17993–17998. [Google Scholar] [CrossRef]
- Diebold, L.; Chandel, N.S. Mitochondrial ROS regulation of proliferating cells. Free Radic. Biol. Med. 2016, 100, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Maryanovich, M.; Gross, A. A ROS rheostat for cell fate regulation. Trends Cell Biol. 2013, 23, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.L.; Dzeja, P.P.; Nelson, T.J.; Terzic, A. Metabolic Plasticity in Stem Cell Homeostasis and Differentiation. Cell Stem Cell 2012, 11, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Nijnik, A.; Woodbine, L.; Marchetti, C.; Dawson, S.; Lambe, T.; Liu, C.; Rodrigues, N.P.; Crockford, T.L.; Cabuy, E.; Vindigni, A.; et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature 2007, 447, 686–690. [Google Scholar] [CrossRef]
- Niedernhofer, L.J. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA Repair 2008, 7, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Flach, J.; Bakker, S.T.; Mohrin, M.; Conroy, P.C.; Pietras, E.M.; Reynaud, D.; Alvarez, S.; Diolaiti, M.E.; Ugarte, F.; Forsberg, E.C.; et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 2014, 512, 198–202. [Google Scholar] [CrossRef]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed]
- Hauer, M.H.; Gasser, S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017, 31, 2204–2221. [Google Scholar] [CrossRef]
- Zane, L.; Sharma, V.; Misteli, T. Common features of chromatin in aging and cancer: Cause or coincidence? Trends Cell Biol. 2014, 24, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wellen, K.E.; Rabinowitz, J.D. Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 2016, 30, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef]
- Ali, I.; Conrad, R.J.; Verdin, E.; Ott, M. Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics. Chem. Rev. 2018, 118, 1216–1252. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, D.C.; Sorum, A.W.; Guasch, L.; Nicklaus, M.C.; Meier, J.L. Metabolic Regulation of Histone Acetyltransferases by Endogenous Acyl-CoA Cofactors. Chem. Biol. 2015, 22, 1030–1039. [Google Scholar] [CrossRef]
- Katada, S.; Imhof, A.; Sassone-Corsi, P. Connecting Threads: Epigenetics and Metabolism. Cell 2012, 148, 24–28. [Google Scholar] [CrossRef]
- Lee, J.V.; Carrer, A.; Shah, S.; Snyder, N.W.; Wei, S.; Venneti, S.; Worth, A.J.; Yuan, Z.-F.; Lim, H.-W.; Liu, S.; et al. Akt-Dependent Metabolic Reprogramming Regulates Tumor Cell Histone Acetylation. Cell Metab. 2014, 20, 306–319. [Google Scholar] [CrossRef]
- Kinnaird, A.; Zhao, S.; Wellen, K.E.; Michelakis, E.D. Metabolic control of epigenetics in cancer. Nat. Rev. Cancer 2016, 16, 694–707. [Google Scholar] [CrossRef]
- Metzger, E.; Schüle, R. The expanding world of histone lysine demethylases. Nat. Struct. Mol. Biol. 2007, 14, 252–254. [Google Scholar] [CrossRef] [PubMed]
- Forneris, F.; Binda, C.; Vanoni, M.A.; Mattevi, A.; Battaglioli, E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005, 579, 2203–2207. [Google Scholar] [CrossRef]
- Metzger, E.; Imhof, A.; Patel, D.; Kahl, P.; Hoffmeyer, K.; Friedrichs, N.; Müller, J.M.; Greschik, H.; Kirfel, J.; Ji, S.; et al. Phosphorylation of histone H3T6 by PKCβI controls demethylation at histone H3K4. Nature 2010, 464, 792–796. [Google Scholar] [CrossRef]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hu, D.; Chen, H.; Shi, G.; Fetahu, I.S.; Wu, F.; Rabidou, K.; Fang, R.; Tan, L.; Xu, S.; et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018, 559, 637–641. [Google Scholar] [CrossRef]
- Pai, C.-C.; Kearsey, S.E. A Critical Balance: dNTPs and the Maintenance of Genome Stability. Genes 2017, 8, 57. [Google Scholar] [CrossRef]
- Kunz, B.A. Mutagenesis and deoxyribonucleotide pool imbalance. Mutat. Res. 1988, 200, 133–147. [Google Scholar] [CrossRef]
- Kunz, B.A.; Kohalmi, S.E.; Kunkel, T.A.; Mathews, C.K.; Mclntosh, E.M.; Reidy, J.A. Deoxyribonucleoside triphosphate levels: A critical factor in the maintenance of genetic stability. Mutat. Res. Toxicol. 1994, 318, 1–64. [Google Scholar] [CrossRef]
- Mathews, C.K. DNA precursor metabolism and genomic stability. FASEB J. 2006, 20, 1300–1314. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Abdulovic, A.L.; Viberg, J.; Nilsson, A.K.; Kunkel, T.A.; Chabes, A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011, 39, 1360–1371. [Google Scholar] [CrossRef] [PubMed]
- Reichard, P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988, 57, 349–374. [Google Scholar] [CrossRef]
- Meuth, M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell Res. 1989, 181, 305–316. [Google Scholar] [CrossRef]
- Meuth, M. The genetic consequences of nucleotide precursor pool imbalance in mammalian cells. Mutat. Res. Mol. Mech. Mutagen. 1984, 126, 107–112. [Google Scholar] [CrossRef]
- Haracska, L.; Prakash, S.; Prakash, L. Replication pastO6-Methylguanine by Yeast and Human DNA Polymerase η. Mol. Cell. Biol. 2000, 20, 8001–8007. [Google Scholar] [CrossRef]
- Niida, H.; Shimada, M.; Murakami, H.; Nakanishi, M. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci. 2010, 101, 2505–2509. [Google Scholar] [CrossRef] [PubMed]
- Niida, H.; Katsuno, Y.; Sengoku, M.; Shimada, M.; Yukawa, M.; Ikura, M.; Ikura, T.; Kohno, K.; Shima, H.; Suzuki, H.; et al. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev. 2010, 24, 333–338. [Google Scholar] [CrossRef]
- Tseng, L.-M.; Yin, P.-H.; Chi, C.-W.; Hsu, C.-Y.; Wu, C.-W.; Lee, L.-M.; Wei, Y.-H.; Lee, H.-C. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 2006, 45, 629–638. [Google Scholar] [CrossRef]
- Yu, M.; Zhou, Y.; Shi, Y.; Ning, L.; Yang, Y.; Wei, X.; Zhang, N.; Hao, X.; Niu, R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 2007, 59, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Nomoto, S.; Fujii, T.; Kaneko, T.; Takeda, S.; Inoue, S.; Kanazumi, N.; Nakao, A. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur. J. Surg. Oncol. 2006, 32, 303–307. [Google Scholar] [CrossRef]
- Matsuyama, W.; Nakagawa, M.; Wakimoto, J.; Hirotsu, Y.; Kawabata, M.; Osame, M. Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Hum. Mutat. 2003, 21, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Lièvre, A.; Chapusot, C.; Bouvier, A.-M.; Zinzindohoué, F.; Piard, F.; Roignot, P.; Arnould, L.; Beaune, P.; Faivre, J.; Laurent-Puig, P. Clinical value of mitochondrial mutations in colorectal cancer. J. Clin. Oncol. 2005, 23, 3517–3525. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Mambo, E.; Sidransky, D. Mitochondrial DNA mutations in human cancer. Oncogene 2006, 25, 4663–4674. [Google Scholar] [CrossRef] [PubMed]
- Simonnet, H. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis 2002, 23, 759–768. [Google Scholar] [CrossRef]
- Petros, J.A.; Baumann, A.K.; Ruiz-Pesini, E.; Amin, M.B.; Sun, C.Q.; Hall, J.; Lim, S.; Issa, M.M.; Flanders, W.D.; Hosseini, S.H.; et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Shidara, Y.; Yamagata, K.; Kanamori, T.; Nakano, K.; Kwong, J.Q.; Manfredi, G.; Oda, H.; Ohta, S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005, 65, 1655–1663. [Google Scholar] [CrossRef]
- Ralevic, V.; Burnstock, G. Receptors for purines and pyrimidines. Pharmacol. Rev. 1998, 50, 413–492. [Google Scholar] [PubMed]
- Smith, J.L. Enzymes of nucleotide synthesis. Curr. Opin. Struct. Biol. 1995, 5, 752–757. [Google Scholar] [CrossRef]
- Fang, J.; Uchiumi, T.; Yagi, M.; Matsumoto, S.; Amamoto, R.; Takazaki, S.; Yamaza, H.; Nonaka, K.; Kang, D. Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci. Rep. 2013, 33, e00021. [Google Scholar] [CrossRef]
- Bader, B.; Knecht, W.; Fries, M.; Löffler, M. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr. Purif. 1998, 13, 414–422. [Google Scholar] [CrossRef]
- Löffler, M.; Jöckel, J.; Schuster, G.; Becker, C. Dihydroorotat-ubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol. Cell. Biochem. 1997, 174, 125–129. [Google Scholar] [CrossRef]
- Beuneu, C.; Auger, R.; Löffler, M.; Guissani, A.; Lemaire, G.; Lepoivre, M. Indirect inhibition of mitochondrial dihydroorotate dehydrogenase activity by nitric oxide. Free Radic. Biol. Med. 2000, 28, 1206–1213. [Google Scholar] [CrossRef]
- Rawls, J.; Knecht, W.; Diekert, K.; Lill, R.; Löffler, M. Requirements for the mitochondrial import and localization of dihydroorotate dehydrogenase. Eur. J. Biochem. 2000, 267, 2079–2087. [Google Scholar] [CrossRef]
- Uhlin, U.; Eklund, H. Structure of ribonucleotide reductase protein R1. Nature 1994, 370, 533–539. [Google Scholar] [CrossRef]
- Jordan, A.; Reichard, P. Ribonucleotide reductases. Annu. Rev. Biochem. 1998, 67, 71–98. [Google Scholar] [CrossRef] [PubMed]
- Kashlan, O.B.; Scott, C.P.; Lear, J.D.; Cooperman, B.S. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry 2002, 41, 462–474. [Google Scholar] [CrossRef]
- Chabes, A.L.; Pfleger, C.M.; Kirschner, M.W.; Thelander, L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA 2003, 100, 3925–3929. [Google Scholar] [CrossRef] [PubMed]
- Munch-Petersen, B.; Cloos, L.; Jensen, H.K.; Tyrsted, G. Human thymidine kinase 1. Regulation in normal and malignant cells. Adv. Enzyme Regul. 1995, 35, 69–89. [Google Scholar] [CrossRef]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.T.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- Clifford, R.; Louis, T.; Robbe, P.; Ackroyd, S.; Burns, A.; Timbs, A.T.; Wright Colopy, G.; Dreau, H.; Sigaux, F.; Judde, J.G.; et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 2014, 123, 1021–1031. [Google Scholar] [CrossRef]
- Song, S.; Wheeler, L.J.; Mathews, C.K. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J. Biol. Chem. 2003, 278, 43893–43896. [Google Scholar] [CrossRef]
- López, L.C.; Akman, H.O.; García-Cazorla, A.; Dorado, B.; Martí, R.; Nishino, I.; Tadesse, S.; Pizzorno, G.; Shungu, D.; Bonilla, E.; et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum. Mol. Genet. 2009, 18, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Pontarin, G.; Gallinaro, L.; Ferraro, P.; Reichard, P.; Bianchi, V. Origins of mitochondrial thymidine triphosphate: Dynamic relations to cytosolic pools. Proc. Natl. Acad. Sci. USA 2003, 100, 12159–12164. [Google Scholar] [CrossRef] [PubMed]
- Desler, C.; Munch-Petersen, B.; Rasmussen, L.J. The Role of Mitochondrial dNTP Levels in Cells with Reduced TK2 Activity. Nucleosides Nucleotides Nucleic Acids 2006, 25, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Pontarin, G.; Ferraro, P.; Bee, L.; Reichard, P.; Bianchi, V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. USA 2012, 109, 13302–13307. [Google Scholar] [CrossRef]
- Bourdon, A.; Minai, L.; Serre, V.; Jais, J.-P.; Sarzi, E.; Aubert, S.; Chrétien, D.; de Lonlay, P.; Paquis-Flucklinger, V.; Arakawa, H.; et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007, 39, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Pontarin, G.; Ferraro, P.; Rampazzo, C.; Kollberg, G.; Holme, E.; Reichard, P.; Bianchi, V. Deoxyribonucleotide metabolism in cycling and resting human fibroblasts with a missense mutation in p53R2, a subunit of ribonucleotide reductase. J. Biol. Chem. 2011, 286, 11132–11140. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Arakawa, H.; Yamaguchi, T.; Shiraishi, K.; Fukuda, S.; Matsui, K.; Takei, Y.; Nakamura, Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 2000, 404, 42–49. [Google Scholar] [CrossRef]
- James, S.J.; Miller, B.J.; McGarrity, L.J.; Morris, S.M. The effect of folic acid and/or methionine deficiency on deoxyribonucleotide pools and cell cycle distribution in mitogen-stimulated rat lymphocytes. Cell Prolif. 1994, 27, 395–406. [Google Scholar] [CrossRef]
- Micheli, V.; Camici, M.; G Tozzi, M.; L Ipata, P.; Sestini, S.; Bertelli, M.; Pompucci, G. Neurological Disorders of Purine and Pyrimidine Metabolism. Curr. Top. Med. Chem. 2011, 11, 923–947. [Google Scholar] [CrossRef]
- Desler, C.; Marcker, M.L.; Singh, K.K.; Rasmussen, L.J. The importance of mitochondrial DNA in aging and cancer. J. Aging Res. 2011, 2011, 407536. [Google Scholar] [CrossRef] [PubMed]
- Desler, C.; Frederiksen, J.H.; Angleys, M.; Maynard, S.; Keijzers, G.; Fagerlund, B.; Mortensen, E.L.; Osler, M.; Lauritzen, M.; Bohr, V.A.; et al. Increased deoxythymidine triphosphate levels is a feature of relative cognitive decline. Mitochondrion 2015, 25, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Maynard, S.; Hejl, A.-M.; Dinh, T.-S.T.; Keijzers, G.; Hansen, Å.M.; Desler, C.; Moreno-Villanueva, M.; Bürkle, A.; Rasmussen, L.J.; Waldemar, G.; et al. Defective mitochondrial respiration, altered dNTP pools and reduced AP endonuclease 1 activity in peripheral blood mononuclear cells of Alzheimer’s disease patients. Aging 2015, 7, 793–815. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).