Simple Summary
The Colorectal cancer Study of Austria (CORSA), an ongoing multicenter prospective case‒control study, was initiated to discover prognostic as well as diagnostic biomarkers for colorectal cancer (CRC) risk prediction mainly based on OMICS research, such as genomics, metabolomics, and metagenomics centered at the Institute of Cancer Research at the Medical University of Vienna; recruitment for CORSA started in 2003. Until now, we have generated genomics data, untargeted and targeted metabolomics data, folate-dependent one-carbon metabolism data, and leukocyte telomere length data using the CORSA biobank. The generated data, the collection of biological samples (genomic DNA, plasma, fecal samples) and the comprehensive CORSA database represents a valuable resource for ongoing and future national and international cooperation projects on CRC research.
Abstract
The Colorectal cancer Study of Austria (CORSA) is comprised more than 13,500 newly diagnosed colorectal cancer (CRC) patients, patients with high- and low-risk adenomas as well as population-based controls. The recruitment for the CORSA biobank is performed in close cooperation with the invited two-stage CRC screening project “Burgenland PREvention trial of colorectal Disease with ImmunologiCal Testing” (B-PREDICT). Annually, more than 150,000 inhabitants of the Austrian federal state Burgenland aged between 40 and 80 are invited to participate using FIT-tests as an initial screening. FIT-positive tested participants are offered a diagnostic colonoscopy and are asked to take part in CORSA, sign a written informed consent, complete questionnaires concerning dietary and lifestyle habits and provide an ethylenediaminetetraacetic acid (EDTA) blood sample as well as a stool sample. Additional CRC cases have been recruited at four hospitals in Vienna and a hospital in lower Austria. A major strength of CORSA is the population-based controls who are FIT-positive and colonoscopy-confirmed to be free of polyps and/or CRC.
1. Introduction
Colorectal cancer (CRC) is the fourth most common cancer and the third highest cause of cancer-related deaths in Europe, thereby representing a severe public health problem [1]. In Austria, the CRC incidence rate is observed in the lower third within the European Union with about 49.2 per 100,000 inhabitants (Statistics Austria, 2017). CRC is a complex disease with both genetic and lifestyle factors contributing to individual risk of CRC [2]. Early detection of CRC is an important issue since the stage at diagnosis remains the most important prognostic factor for CRC. The 5-year survival rate is about 90% for those patients diagnosed at an early stage but decreases to 10% for advanced or metastasized cancer [3]. The majority of sporadic CRCs develop from normal epithelium through sequentially worsening degrees of adenomatous dysplasia, known as the adenoma-carcinoma sequence [4]. Due to the slow progression of CRC, there is a window of opportunity of about ten years for prevention and intervention.
Screening programs have the potential to detect early precancerous lesions and perform endoscopic removal of adenomas, thereby contributing to the reduction of CRC incidence and mortality [5]. Therefore, population-wide screening programs are recommended in several European countries. Many CRC screening programs utilize a two-stage process wherein patients testing positive on a fecal occult test are followed up by colonoscopy. Nowadays, the preferred approach in testing for occult blood in feces used for CRC screening programs is the fecal immunochemical test (FIT), despite its relatively low specificity and sensitivity. Moreover, FIT tests can detect only a small percentage of non-advanced colorectal adenomas [6].
In Austria, the organization of screening programs is not regulated nationwide; it is a matter of the federal states. To date, only two invited screening programs are performed in Austria, in the federal states Vorarlberg and Burgenland. In Vorarlberg, a colonoscopy-based screening program is offered to the insured population older than 50 years since 2007. In Burgenland, the screening program “Burgenland Prevention Trial of Colorectal Cancer Disease with Immunological Testing” (BPREDICT), initiated in 2003, is a two-stage invited screening program for individuals aged between 40 and 80 using FIT as the initial screening. Participants with a positive FIT were invited for further diagnostic work-up such as a diagnostic colonoscopy. These participants were asked to take part in the “Colorectal Cancer Study of Austria” (CORSA), sign a written informed consent, complete questionnaires, and provide an ethylenediaminetetraacetic acid (EDTA) blood sample as well as a stool sample for the CORSA biobank. In cooperation with B-PREDICT, several hospitals in Vienna and the Hospital Wiener Neustadt, we have established the CORSA biobank, which currently comprises more than 13,500 participants. From each participant, genomic DNA and plasma samples are stored at −80 °C. Two years ago, we also started recruitment of stool samples within the CRC screening program B-PREDICT.
The recruitment of participants for the ongoing CORSA started over 18 years ago at the Medical University of Vienna, Austria. The initial motivation of CORSA was to identify genetic risk variants that modify individual predisposition to CRC. At the beginning, the aim was the identification of single nucleotide polymorphisms (SNPs) as well as tandem repeat minisatellites and their association with CRC risk in a case‒control study design [7]. With the progress in molecular biologically methods, we have focused on high-throughput OMICS-based projects, in particular, genomics, metabolomics and metagenomics. We conducted a genome-wide association study (GWAS) for sporadic CRC and untargeted as well as targeted metabolomics profiling. The most recent CORSA project aims at the identification of diagnostic signatures along the colorectal adenoma-carcinoma sequence analyzing fecal samples via gut metagenomics. Altogether, CORSA aims to discover and validate prognostic as well as diagnostic biomarkers for CRC risk prediction nowadays based mainly on OMICS research.
The aim of CORSA is to identify new robust diagnostic biomarkers for advanced adenomas and early-stage CRC by using state-of-the-art OMICS technologies such as metabolomics, proteomics, and metagenomics in order to improve the identification of patients at higher CRC risk.
2. Materials and Methods
2.1. Participant Recruitment
CORSA is an ongoing population case‒control study of women and men recruiting newly diagnosed CRC patients (colon, rectum or rectosigmoid cancer (ICD-10 C18-20), histopathologically confirmed invasive cancer of any stage), high-/low-risk adenomas and population-based colonoscopy-negative controls, with an age range between 30 and 90 years. Blood and fecal samples, dietary/lifestyle questionnaires, and anthropometrics/demographics were obtained from each participant.
Since 2003, more than 13,500 participants have been recruited across nine sites in Austria. The multicenter recruitment within CORSA follows standardized protocols, resulting in consistent data from all recruitment sites. These sites include the Medical University of Vienna (Department of Surgery), three further hospitals in Vienna (Sozialmedizinisches Zentrum Süd, Hospital Rudolfstiftung and Hospital Elisabeth), and the Hospital Wiener Neustadt in Lower Austria. Furthermore, the recruitment for CORSA was performed in four hospitals in Burgenland (Hospital Kittsee, Hospital Oberpullendof, Hospital Oberwart, Hospital Güssing) within the population-based screening program “Burgenland PREvention Trial of colorectal cancer DIsease with ImmunologiCal Testing” (B-PREDICT) (Figure 1).
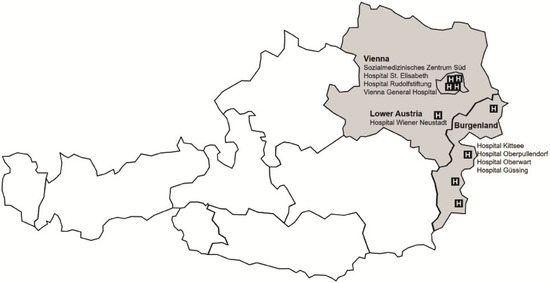
Figure 1.
Study sites of CORSA recruitment in Vienna, Burgenland, and lower Austria. Since 2003, more than 13,500 participants have been recruited across nine sites in Austria. Source of illustration: CORSA study team, 2020.
B-PREDICT is an invited two-stage screening project initiated by gastroenterologist Karl Mach at the Hospital Oberpullendorf, Burgenland. Annually, more than 150,000 inhabitants of Burgenland aged between 40 and 80 are invited to participate in this program using a fecal immunochemical test (FIT) as an initial screening. Nowadays, FIT is the preferred approach in testing for occult blood in feces used for CRC screening programs [6]. FIT-positive (≥10 µg hemoglobin/g feces) tested individuals are offered a complete colonoscopy and are asked to participate in CORSA and provide an EDTA blood sample, questionnaire, and written informed consent for the CORSA biobank. Within B-PREDICT, we have recruited CRC patients, patients with high- and low-risk adenomas as well as population-based controls. These controls are triggered by a positive FIT result, and because all of them underwent a colonoscopy, they were known to be free of polyps and CRC. The high-risk adenoma group included patients with adenomatous tubular polyps >1 cm, adenomatous tubulo-villous polyps, adenomatous villous polyps, sessile serrated polyps (SSA) and traditional serrated polyps (TSA). Adenomatous tubular polyps <1 cm were considered as low-risk polyps. Baseline characteristics of recruited CORSA participants are given in Table 1.

Table 1.
Demographic and clinical characteristics of CORSA participants (as of May 2020).
FIT-positive participants from the B-PREDICT screening are recruited when undergoing colonoscopy and comprise CRC patients, high- and low-risk polyps and colonoscopy-negative controls. CORSA participants from other centers are recruited before surgery or at the time of their clinical follow-up and are mainly CRC cases. All subjects gave written informed consent, and the study was approved by the institutional review boards.
2.2. Biospecimen Collection
Genomic DNA, plasma (divided into aliquots in barcoded tubes) and stool samples are collected at each CORSA site using harmonized protocols and stored at −80 °C. Paraffin-embedded (FFPE) tissue from CRC cases is stored at room temperature.
Within CORSA, no regular sequential follow-up recruitment is performed.
Blood draws are performed at least at baseline (first participation in CORSA). In addition, 2263 CORSA participants (17%) have been repeatedly recruited. On average, these participants were recruited about 2.3 times. The mean time period between single participations amounts to 2.9 years. Some participants have a follow-up of about 15 years.
Survival data, representing a comprehensive output file comprising confirmed or unconfirmed date of death, date of last contact, and information on missing or incorrect data input, are provided through a biennial clinical data abstraction from the IT database of the Medical University of Vienna, “Allgemeines Krankenhaus Information Management” (AKIM) in cooperation with Statistics Austria. In addition, we abstract survival data in cooperation with the Main Association of Austrian Social Insurance Institutions (“Hauptverband der österreichischen Sozialversicherungsträger”). Therefore, the social insurance number, first and last name, date of birth, and sex of all participants were processed through a database pipeline. For a subset of the data, the cause of death in terms of ICD-10 category codes of the World Health Organization can be obtained.
2.3. Clinical Data
Clinical data were abstracted from medical records and processed in a structured database following standardized documentation guidelines and according to the General Data Protection Regulation (GDPR). The CORSA databank comprises structured information on diagnosis, treatment, histology, progression (recurrence and metastasis), and survival data (Table 2). Follow-up on clinical data is regularly performed. Some CORSA participants have been followed up for about 15 years.

Table 2.
CORSA data resources: questionnaire data and clinical databank.
2.4. Questionnaires
CORSA participants provided at least a basic CORSA questionnaire assessing data on body mass index (BMI), smoking history, alcohol consumption, education level, family status, profession, basic dietary habits, and diabetes (Table 2). Patient information and interviewing was performed by trained study personnel.
Additionally, CORSA participants were asked to complete a slightly modified version of the VITamins And Lifestyle cohort study (VITAL) questionnaire and the European Prospective Investigation into Cancer and Nutrition Food Frequency Questionnaire (EPIC-Potsdam FFQ2). The VITAL questionnaire assesses the use of nutritional supplements, vitamins, medication, physical activity, and social environment [8]. The EPIC-Potsdam FFQ2 questionnaire was designed to capture detailed dietary habits and is used to receive habitual dietary food or nutrient intake [9].
3. Result
3.1. Generated Data
So far, we have generated genomics data, metabolomics data, folate-dependent one-carbon metabolism data, and leukocyte telomere length data from the CORSA biobank (Table 3). GWAS data are available from 2677 CORSA participants using the Axiom Array Human Genome-Wide CEU1 array (Affymetrix, Santa Clara, CA, USA) comprising 1060 CRC, 689 high-risk polyps, and 928 colonoscopy-negative controls. Metabolomics data, targeted as well as untargeted, are available from 88 CRC, 200 high-risk polyps, 200 low-risk polyps, and 400 controls. Untargeted metabolomics was conducted using ultrahigh-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-qTOF-MS, Agilent Technologies, Santa Clara, CA, USA). Samples for targeted metabolomics were analyzed with the AbsoluteIDQTM p180 kit (Biocrates Life Sciences, Innsbruck, Austria). Both analyses were performed at the biomarker lab headed by Augustin Scalbert (International Agency for Research on Cancer, Lyon, France).

Table 3.
Available CORSA data derived from GWAS, metabolomics, folate biomarkers, and leukocyte telomere length analyses.
Furthermore, data on folate and biomarkers related to one-carbon metabolism are available from 245 CRC patients at three follow-up time points: baseline (n = 218), 6 months (n = 10), and 12 months (n = 17). The analytical measurements were performed at Bevital AS, Bergen, Norway. Conducting liquid chromatography tandem mass spectrometry (LC-MS-MS) circulating folate, folate derivatives, and folic acid were detected. Gas chromatography tandem mass spectrometry (GC-MS-MS) was performed to analyze other amino acids. C-reactive protein, cystatin C, and its variants were measured by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometric analysis.
Telomere length data from 2011 CORSA participants (384 CRC, 544 high-risk polyps, 537 low-risk polyps and 546 controls) were determined using monochrome multiplex quantitative PCR (MMQPCR).
3.2. Key Findings and Publications
CORSA key publications are based on genomics and metabolomics: “Bayesian and frequentist analysis of an Austrian genome-wide association study of colorectal cancer and advanced adenomas”, “Plasma metabolites associated with colorectal cancer: a discovery replication study” and “Plasma metabolites associated with colorectal cancer stage: findings from an international consortium” [10,11,12]. All publications related to CORSA are listed in Supplementary Table S1 [13,14,15,16,17,18,19,20,21,22].
We have performed a GWAS of CORSA participants comprising 1060 CRC patients, 689 patients with advanced colorectal adenomas, 928 colonoscopy-negative controls, and additionally, 3439 controls from the “Cooperative Health Research in the Region of Augsburg” (KORA) using Axiom Arrays CEU 1 (Affymetrix, Santa Clara, CA, USA). We pursued a dual approach to investigate genome-wide associations with disease risk applying both single marker analysis as well as model selection based on the modified Bayesian information criterion, mBIC2, implemented in the software package MOSGWA. The advantage of the model selection approach is its larger power to detect candidate SNPs compared to single marker tests. Furthermore, 56 SNPs that are already known to influence CRC susceptibility from previous studies were tested in a hypothesis-driven approach and some of them were also found to be relevant in CORSA. Furthermore, we found some so-far unreported SNPs [10].
This GWAS was the basis for many international cooperation projects: the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) at the Fred Hutchinson Cancer Center, Seattle, Washington, two European Cooperation in Science and Technology (COST) Actions “Cooperation studies on inherited susceptibility to colorectal cancer” (BMI1206) and “Identifying biomarkers through translation research and prevention and stratification of colorectal cancer” (CA17118).
Within the “Metabolomic profiles throughout the continuum of colorectal carcinogenesis” (MetaboCCC) consortium, we performed an untargeted metabolomics study comprising 268 CRC patients and 353 controls using independent discovery and replication sets from two European cohorts, the ColoCare Study from Germany and CORSA. The aim of this study was to identify circulating plasma metabolites associated with CRC and to improve knowledge regarding CRC etiology. Multiple logistic regression models were used to test the association between disease state and metabolic features. We identified 691 statistically significant features in the discovery cohort. Testing the second cohort narrowed it to 97. These corresponded to 28 metabolites, of which 15 could be identified [11].
Furthermore, we performed a targeted metabolomics profiling within the MetaboCCC consortium aiming to identify biomarkers related to CRC progression. We investigated plasma concentrations of 130 metabolites from 744 CRC patients (stages I-IV) from two Dutch cohorts (COLON, EnCoRe), the ColoCare study from Germany, and CORSA. Our results suggest that metabolic pathway involving citrulline, histidine, and other molecules that have been previously implicated in CRC development may also be linked to progression [12].
4. Discussion
CORSA has multiple strengths bringing together a multidisciplinary team of molecular biologists, clinicians, laboratory scientists, and statisticians to foster research on CRC. The close collaboration with clinicians and clinical centers allows a regular and comprehensive clinical follow-up as well as the automated abstraction of data such as survival information. The multicenter cohort standardized the recruitment of patients, with uniform protocols for the collection, processing, and management of biospecimens. Participants were mainly Caucasian and geographically and ethnically uniform. The comprehensive biological samples and harmonized data provides the basis for national and international cooperation projects on CRC research. Therefore, CORSA represents an active and leading player in cooperation, support actions, and consortia. So far, CORSA contributed to multiple international consortia such as Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) [23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49], COlorectal cancer GENeTics (COGENT) and COnsortium of METabolomics Studies (COMETS) [13]. Furthermore, CORSA was a partner in two ERANET-TRANSCAN-funded projects: MetaboCCC [11,12,44,45,46,50,51], and “Biomarkers related to folate-dependent metabolism in colorectal cancer recurrence and survival” (FOCUS) [35,36]. These interdisciplinary projects bring together multiple European CRC cohorts, including CORSA, the ColoCare study of the German Cancer Research Center in Heidelberg [14], the EnCoRe study of Maastricht University, the Netherlands [15], as well as the COLON study of Wageningen University, the Netherlands [16]. In the course of this collaboration study, data have been pooled and harmonized to increase the statistical power for joint analysis.
A major strength of the biobank is the colonoscopy-negative control group, known to be free of polyps and CRC because all participants underwent a complete colonoscopy within the screening project B-PREDICT.
Nevertheless, some limitations have to be mentioned. CORSA high-risk and low-risk adenomas as well as controls are mainly recruited within the screening program B-PREDICT from centers in Burgenland rather than from centers in Vienna. The reason is that in Vienna no organized CRC screening program has been performed so far; therefore, no colonoscopy-negative controls can be recruited. In Austria, the organization of CRC screening programs is not regulated nationwide; it is a matter of the federal states. Within CORSA, no regularly invited, targeted recruitment at defined follow-up time points is performed.
A new research focus of CORSA besides genomics and metabolomics is the gut microbiome. Recently, the project “Gut MICRObiome-based approach for incorporating new biomarkers into COLOrectal cancer screening” (MICROCOLO) was funded by the Austrian Research Promotion Agency (FFG). There is evidence that changes in the gut microbiome occur during different stages of colorectal neoplasia, supporting an etiologic and diagnostic role for the microbiome. Harnessing knowledge of the microbiota may lead to new preventive strategies and diagnostics. myBioma GmbH, our company partner within this FFG-funded project, will use their already established data processing and statistical analysis workflow to process the resulting microbiome data. Predictive microbiome-based signatures specific for CRC and high-risk adenomas will be defined by multivariate statistical modelling. The combination of conventional screening methods such as FIT with microbiome-based methods is a promising tool for early detection of CRC and could improve diagnostic accuracy.
5. Conclusions
The CORSA biobank, comprising genomic DNA, plasma, fecal samples, and a comprehensive CORSA database, represents a valuable resource for ongoing and future OMICS-based CRC projects. CORSA is open for cooperative research projects; expressions of interest are more than welcome.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology10080722/s1, Table S1: Complete list of CORSA publications (as of June 2021).
Author Contributions
A.G. is responsible for the conception and design of CORSA, obtaining of funding and drafting the manuscript. A.B. was involved in statistical analysis and data interpretation. S.B. was mainly involved in the analysis of metabolomics and genomics datasets and in the management of the CORSA database, including the recruitment of patients, and collection of clinical data. All authors have read and agreed to the published version of the manuscript.
Funding
Analyses or measurements conducted within the CORSA study have been supported by funding from FFG (Bridge, Project number: 829675), FWF (I1542-B-13), ERA-NET on Translational Cancer Research (TRANSCAN) (I1578-B19), TRANSCAN (I2104-B26), and the Medical University of Vienna. CORSA did not receive any funding awarding only the study itself.
Institutional Review Board Statement
Using samples from the CORSA biobank is subject to strict ethical and legal regulations. Ethics approval, data transfer agreement (DTA) and/or material transfer agreement (MTA) is necessary for each study. All CORSA subjects have provided written informed consent and the study was approved by the Ethics Committee of the Medical University of Vienna (EK 1160/2016) and the Ethics Committee of the Federal State Burgenland (EK 33/2010).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Paper concept proposals of interest for cooperation studies are welcomed. Please send a short proposal to the corresponding author, Andrea Gsur (andrea.gsur@meduniwien.ac.at). The proposal must include a working title, author information, scientific rationale and objectives, study design (study data, primary analysis plan), and material (genomic DNA or plasma) or data requested.
Acknowledgments
We acknowledge the contribution to CORSA of all participants who have provided samples and questionnaires. We thank the supporting recruitment teams of all study sites. The authors would also like to thank all physicians (Michael Micksche, Karl Mach, Gernot Leeb, Azita Deutinger-Permoon, Wilfried Horvath, Paul A. Gabriel, Gerhard Böhm, Anton Stift, Thomas Bachleitner-Hofmann, Judith Karner-Hanusch, Johannes Längle, Michael Bergmann, Josef Karner, Stefan Stättner, Marie C. Singer, Katharina Kosma, Gerhard Halter, Thomas Grünberger, Wolfgang Hulla, Harald Kirschner, Werner Weiss, Thomas Pulgram, Christian Madl and Friedrich Längle) and all technicians that have been involved in the CORSA recruitment. Furthermore, we want to acknowledge the MetaboCCC consortium and the COST Actions (BMI1206 and CA17118).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fearon, E.R. Molecular Genetics of Colorectal Cancer. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 479–507. [Google Scholar] [CrossRef]
- O’Connell, J.B.; Maggard, M.A.; Ko, C.Y. Colon Cancer Survival Rates With the New American Joint Committee on Cancer Sixth Edition Staging. J. Natl. Cancer Inst. 2004, 96, 1420–1425. [Google Scholar] [CrossRef]
- Armaghany, T.; Wilson, J.D.; Chu, Q.; Mills, G. Genetic Alterations in Colorectal Cancer. Gastrointest. Cancer Res. 2012, 5, 19–27. [Google Scholar]
- Roselló, S.; Simón, S.; Cervantes, A. Programmed colorectal cancer screening decreases incidence and mortality. Transl. Gastroenterol. Hepatol. 2019, 4, 84. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, B.; Malaspina, M.; Fraser, C.G.; Tintori, B.; Carlani, A.; D’Angelo, V.; Galeazzi, P.; Di Dato, E.; Mariotti, L.; Bulletti, S.; et al. A comparative effectiveness trial of two faecal immunochemical tests for haemoglobin (FIT). Assessment of test performance and adherence in a single round of a population-based screening programme for colorectal cancer. Gut 2018, 67, 485–496. [Google Scholar] [CrossRef]
- Feik, E.; Baierl, A.; Hieger, B.; Fuhrlinger, G.; Pentz, A.; Stattner, S.; Weiss, W.; Pulgram, T.; Leeb, G.; Mach, K.; et al. Association of IGF1 and IGFBP3 polymorphisms with colo-rectal polyps and colorectal cancer risk. Cancer Causes Control 2010, 21, 91–97. [Google Scholar] [CrossRef] [PubMed]
- White, E.; Patterson, R.E.; Kristal, A.; Thornquist, M.; King, I.; Shattuck, A.L.; Evans, I.; Satia-Abouta, J.; Littman, A.J.; Potter, J. VITamins And Lifestyle Cohort Study: Study Design and Characteristics of Supplement Users. Am. J. Epidemiol. 2004, 159, 83–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nothlings, U.; Hoffmann, K.; Bergmann, M.M.; Boeing, H. Fitting portion sizes in a self-administered food frequency question-naire. J. Nutr. 2007, 137, 2781–2786. [Google Scholar] [CrossRef] [PubMed]
- Hofer, P.; Hagmann, M.; Brezina, S.; Dolejsi, E.; Mach, K.; Leeb, G.; Baierl, A.; Buch, S.; Sutterlüty-Fall, H.; Karner-Hanusch, J.; et al. Bayesian and frequentist analysis of an Austrian genome-wide association study of colorectal cancer and advanced adenomas. Oncotarget 2017, 8, 98623–98634. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, A.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Bachleitner-Hofmann, T.; Bergmann, M.M.; Boehm, J.; Brenner, H.; Chang-Claude, J.; van Duijnhoven, F.J.B.; et al. Plasma metabolites associated with colorectal cancer: A discovery-replication strategy. Int. J. Cancer 2019, 145, 1221–1231. [Google Scholar] [CrossRef] [Green Version]
- Geijsen, A.J.; Van Roekel, E.H.; Van Duijnhoven, F.J.; Achaintre, D.; Bachleitner-Hofmann, T.; Baierl, A.; Bergmann, M.M.; Boehm, J.; Bours, M.J.; Brenner, H.; et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int. J. Cancer 2020, 146, 3256–3266. [Google Scholar] [CrossRef] [Green Version]
- Playdon, M.C.; Joshi, A.D.; Tabung, F.K.; Cheng, S.; Henglin, M.; Kim, A.; Lin, T.; Van Roekel, E.H.; Huang, J.; Krumsiek, J.; et al. Metabolomics Analytics Workflow for Epidemiological Research: Perspectives from the Consortium of Metabolomics Studies (COMETS). Metabolites 2019, 9, 145. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, C.M.; Gigic, B.; Böhm, J.; Ose, J.; Viskochil, R.; Schneider, M.; Colditz, G.A.; Figueiredo, J.C.; Grady, W.M.; Li, C.I.; et al. The ColoCare Study: A Paradigm of Transdisciplinary Science in Colorectal Cancer Outcomes. Cancer Epidemiol. Biomark. Prev. 2019, 28, 591–601. [Google Scholar] [CrossRef] [Green Version]
- van Roekel, E.H.; Bours, M.J.; de Brouwer, C.P.; Ten Napel, H.; Sanduleanu, S.; Beets, G.L.; Kant, J.I.; Weijenberg, M.P. The applicability of the international classification of functioning, disability, and health to study lifestyle and quality of life of colorectal cancer survivors. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1394–1405. [Google Scholar] [CrossRef] [Green Version]
- Winkels, R.M.; Heine-Bröring, R.C.; van Zutphen, M.; van Harten-Gerritsen, S.; Kok, D.E.; van Duijnhoven, F.J.; Kampman, E. The COLON study: Colorectal cancer: Longitudinal, Observational study on Nutritional and lifestyle factors that may influence colorectal tumour recurrence, survival and quality of life. BMC Cancer 2014, 14, 374. [Google Scholar] [CrossRef] [Green Version]
- Gsur, A.; Bernhart, K.; Baierl, A.; Feik, E.; Führlinger, G.; Höfer, P.; Leeb, G.; Mach, K.; Micksche, M. No association of XRCC1 polymorphisms Arg194Trp and Arg399Gln with colorectal cancer risk. Cancer Epidemiol. 2011, 35, e38–e41. [Google Scholar] [CrossRef]
- Hofer, P.; Baierl, A.; Feik, E.; Führlinger, G.; Leeb, G.; Mach, K.; Holzmann, K.; Micksche, M.; Gsur, A. MNS16A tandem repeats minisatellite of human telomerase gene: A risk factor for colorectal cancer. Carcinogenesis 2011, 32, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Hofer, P.; Baierl, A.; Bernhart, K.; Leeb, G.; Mach, K.; Micksche, M.; Gsur, A. Association of genetic variants of human telomerase with colorectal polyps and colorectal cancer risk. Mol. Carcinog. 2012, 51, E176–E182. [Google Scholar] [CrossRef]
- Heinzle, C.; Gsur, A.; Hunjadi, M.; Erdem, Z.; Gauglhofer, C.; Stättner, S.; Karner, J.; Klimpfinger, M.; Wrba, F.; Reti, A.; et al. Differential Effects of Polymorphic Alleles of FGF Receptor 4 on Colon Cancer Growth and Metastasis. Cancer Res. 2012, 72, 5767–5777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zöchmeister, C.; Brezina, S.; Höfer, P.; Baierl, A.; Bergmann, M.M.; Bachleitner-Hofmann, T.; Karner-Hanusch, J.; Stift, A.; Gerger, A.; Leeb, G.; et al. Leukocyte telomere length throughout the continuum of colorectal carcinogenesis. Oncotarget 2018, 9, 13582–13592. [Google Scholar] [CrossRef] [Green Version]
- Jiraskova, K.; Hughes, D.J.; Brezina, S.; Gumpenberger, T.; Veskrnova, V.; Buchler, T.; Schneiderova, M.; Levy, M.; Liska, V.; Vodenkova, S.; et al. Functional Polymorphisms in DNA Re-pair Genes Are Associated with Sporadic Colorectal Cancer Susceptibility and Clinical Outcome. Int. J. Mol. Sci. 2018, 20, 97. [Google Scholar] [CrossRef] [Green Version]
- Luna Coronell, J.A.; Sergelen, K.; Hofer, P.; Gyurjan, I.; Brezina, S.; Hettegger, P.; Leeb, G.; Mach, K.; Gsur, A.; Weinhäusel, A. The Immunome of Colon Cancer: Functional In Silico Analysis of Antigenic Proteins Deduced from IgG Microarray Profiling. Genom. Proteom. Bioinform. 2018, 16, 73–84. [Google Scholar] [CrossRef]
- Huyghe, J.R.; Bien, S.A.; Harrison, T.A.; Kang, H.M.; Chen, S.; Schmit, S.L.; Conti, D.V.; Qu, C.; Jeon, J.; Edlund, C.K.; et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 2019, 51, 76–87. [Google Scholar] [CrossRef]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; Orlando, G.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafmayer, C.; Harrison, J.W.; Buch, S.; Lange, C.; Reichert, M.C.; Hofer, P.; Cossais, F.; Kupcinskas, J.; von Schönfels, W.; Schniewind, B.; et al. Genome-wide association analysis of diverticular disease points towards neuromuscular, connective tissue and epithelial pathomechanisms. Gut 2019, 68, 854–865. [Google Scholar] [CrossRef] [Green Version]
- Schmit, S.L.; Edlund, C.K.; Schumacher, F.R.; Gong, J.; Harrison, T.A.; Huyghe, J.R.; Qu, C.; Melas, M.; Van Den Berg, D.J.; Wang, H.; et al. Novel Common Genetic Susceptibility Loci for Colorectal Cancer. J. Natl. Cancer Inst. 2019, 111, 146–157. [Google Scholar] [CrossRef]
- Cornish, A.J.; Law, P.J.; Timofeeva, M.; Palin, K.; Farrington, S.M.; Palles, C.; Jenkins, M.A.; Casey, G.; Brenner, H.; Chang-Claude, J.; et al. Modifiable pathways for colorectal cancer: A mendelian randomisation analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Holowatyj, A.N.; Gigic, B.; Herpel, E.; Scalbert, A.; Schneider, M.; Ulrich, C.M.; MetaboCCC Consortium; ColoCare Study. Distinct Molecular Phenotype of Sporadic Colo-rectal Cancers Among Young Patients Based on Multiomics Analysis. Gastroenterology 2020, 158, 1155–1158.e2. [Google Scholar] [CrossRef]
- Archambault, A.N.; Su, Y.-R.; Jeon, J.; Thomas, M.; Lin, Y.; Conti, D.V.; Win, A.K.; Sakoda, L.C.; Lansdorp-Vogelaar, I.; Peterse, E.F.; et al. Cumulative Burden of Colorectal Cancer–Associated Genetic Variants Is More Strongly Associated With Early-Onset vs. Late-Onset Cancer. Gastroenterology 2020, 158, 1274–1286.e12. [Google Scholar] [CrossRef]
- Lu, Y.; Kweon, S.-S.; Cai, Q.; Tanikawa, C.; Shu, X.-O.; Jia, W.-H.; Xiang, Y.-B.; Huyghe, J.R.; Harrison, T.A.; Kim, J.; et al. Identification of Novel Loci and New Risk Variant in Known Loci for Colorectal Cancer Risk in East Asians. Cancer Epidemiol. Biomark. Prev. 2020, 29, 477–486. [Google Scholar] [CrossRef]
- Murphy, N.; Carreras-Torres, R.; Song, M.; Chan, A.T.; Martin, R.M.; Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Bradbury, K.E.; et al. Circulating Levels of Insulin-like Growth Factor 1 and Insulin-like Growth Factor Binding Protein 3 Associate With Risk of Colorectal Cancer Based on Sero-logic and Mendelian Randomization Analyses. Gastroenterology 2020, 158, 1300–1312.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Martin, R.M.; Lewis, S.J.; Kazmi, N.; Robinson, T.M.; Albanes, D.; Aleksandrova, K.; et al. Physical activity and risks of breast and colorectal cancer: A Mendelian randomisation analysis. Nat. Commun. 2020, 11, 597. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, S.H.; Harrison, T.A.; Phipps, A.I.; Steinfelder, R.; Trinh, Q.M.; Qu, C.; Banbury, B.L.; Georgeson, P.; Grasso, C.S.; Giannakis, M.; et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat. Commun. 2020, 11, 3644. [Google Scholar] [CrossRef]
- Thomas, M.; Sakoda, L.C.; Hoffmeister, M.; Rosenthal, E.A.; Lee, J.K.; van Duijnhoven, F.J.; Platz, E.A.; Wu, A.H.; Dampier, C.H.; de la Chapelle, A.; et al. Genome-wide Modeling of Polygenic Risk Score in Colorectal Cancer Risk. Am. J. Hum. Genet. 2020, 107, 432–444. [Google Scholar] [CrossRef]
- Koole, J.L.; Bours, M.J.L.; Geijsen, A.J.M.R.; Gigic, B.; Ulvik, A.; Kok, D.E.; Brezina, S.; Ose, J.; Baierl, A.; Böhm, J.; et al. Circulating B-vitamin biomarkers and B-vitamin supplement use in relation to quality of life in patients with colorectal cancer: Results from the FOCUS consortium. Am. J. Clin. Nutr. 2021, 113, 1468–1481. [Google Scholar] [CrossRef]
- Geijsen, A.J.M.R.; Ulvik, A.; Gigic, B.; Kok, D.E.; van Duijnhoven, F.J.B.; Holowatyj, A.N.; Brezina, S.; van Roekel, E.H.; Baierl, A.; Bergmann, M.M.; et al. Circulating Folate and Folic Acid Concentrations: Associations With Colorectal Cancer Recurrence and Survival. JNCI Cancer Spectr. 2020, 4, pkaa051. [Google Scholar] [CrossRef] [PubMed]
- Jarvik, G.P.; Wang, X.; Fontanillas, P.; Kim, E.; Chanprasert, S.; Gordon, A.S.; Bastarache, L.; Kowdley, K.V.; Harrison, T.; Rosenthal, E.A.; et al. Hemochromatosis risk genotype is not associated with colorectal cancer or age at its diagnosis. Hum. Genet. Genom. Adv. 2020, 1, 100010. [Google Scholar] [CrossRef]
- Guo, X.; Lin, W.; Wen, W.; Huyghe, J.; Bien, S.; Cai, Q.; Harrison, T.; Chen, Z.; Qu, C.; Bao, J.; et al. Identifying Novel Susceptibility Genes for Colorectal Cancer Risk From a Transcriptome-Wide Association Study of 125,478 Subjects. Gastroenterology 2021, 160, 1164–1178.e6. [Google Scholar] [CrossRef] [PubMed]
- Nounu, A.; Greenhough, A.; Heesom, K.J.; Richmond, R.C.; Zheng, J.; Weinstein, S.J.; Albanes, D.; Baron, J.A.; Hopper, J.L.; Figueiredo, J.C.; et al. A Combined Proteomics and Mendelian Randomization Approach to Investigate the Effects of Aspirin-Targeted Proteins on Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2021, 30, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Holowatyj, A.N.; Haffa, M.; Lin, T.; Scherer, D.; Gigic, B.; Ose, J.; Warby, C.A.; Himbert, C.; Abbenhardt-Martin, C.; Achaintre, D.; et al. Multi-omics analysis reveals adipose-tumor crosstalk in colorectal cancer patients. Cancer Prev. Res. 2020, 13, 817–828. [Google Scholar] [CrossRef]
- Bull, C.J.; Bell, J.A.; Murphy, N.; Sanderson, E.; Smith, G.D.; Timpson, N.J.; Banbury, B.L.; Albanes, D.; Berndt, S.I.; Bézieau, S.; et al. Adiposity, metabolites, and colorectal cancer risk: Mendelian randomization study. BMC Med. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tsilidis, K.K.; Papadimitriou, N.; Dimou, N.; Gill, D.; Lewis, S.J.; Martin, R.M.; Murphy, N.; Markozannes, G.; Zuber, V.; Cross, A.J.; et al. Genetically predicted circulating concentrations of micronutrients and risk of colorectal cancer among individuals of European descent: A Mendelian randomization study. Am. J. Clin. Nutr. 2021, 113, 1490–1502. [Google Scholar] [CrossRef]
- Huyghe, R.J.; Harrison, A.T.; Bien, A.S.; Hampel, H.; Figueiredo, C.J.; Schmit, L.S.; Conti, D.V.; Chen, S.; Qi, C.; Lin, Y.; et al. Genetic architectures of proximal and distal colorectal cancer are partly distinct. Gut 2021. [Google Scholar] [CrossRef] [PubMed]
- Geijsen, A.J.M.R.; Kok, D.E.; van Zutphen, M.; Keski-Rahkonen, P.; Achaintre, D.; Gicquiau, A.; Gsur, A.; Kruyt, F.M.; Ulrich, C.M.; Weijenberg, M.P.; et al. Diet quality indices and dietary patterns are associated with plasma metabolites in colorectal cancer patients. Eur. J. Nutr. 2021, 1–14. [Google Scholar] [CrossRef]
- Gumpenberger, T.; Brezina, S.; Keski-Rahkonen, P.; Baierl, A.; Robinot, N.; Leeb, G.; Habermann, N.; Kok, D.; Scalbert, A.; Ueland, P.-M.; et al. Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome between Colorectal Cancer and Colorectal Adenomas. Metabolites 2021, 11, 119. [Google Scholar] [CrossRef]
- Ose, J.; Gigic, B.; Brezina, S.; Lin, T.; Baierl, A.; Geijsen, A.; van Roekel, E.; Robinot, N.; Gicquiau, A.; Achaintre, D.; et al. Targeted plasma metabolic profiles and risk of recurrence in stage II and III colorectal cancer patients: Results from an international cohort consortium Polymorphisms within autophagy-related genes influence the risk of developing colorectal cancer: A meta-analysis of four large cohorts. Metabolites 2021, 11, 129. [Google Scholar] [CrossRef]
- Sainz, J.; García-Verdejo, F.; Martínez-Bueno, M.; Kumar, A.; Sánchez-Maldonado, J.; Díez-Villanueva, A.; Vodičková, L.; Vymetálková, V.; Sánchez, V.M.; Filho, M.D.S.; et al. Polymorphisms within Autophagy-Related Genes Influence the Risk of Developing Colorectal Cancer: A Meta-Analysis of Four Large Cohorts. Cancers 2021, 13, 1258. [Google Scholar] [CrossRef]
- Zheng, T.; Ellinghaus, D.; Juzenas, S.; Cossais, F.; Burmeister, G.; Mayr, G.; Jørgensen, I.F.; Teder-Laving, M.; Skogholt, A.H.; Chen, S.; et al. Genome-wide analysis of 944 133 individuals provides insights into the etiology of haemorrhoidal disease. Gut 2021. [Google Scholar] [CrossRef]
- Culliford, R.; Cornish, A.J.; Law, P.J.; Farrington, S.M.; Palin, K.; Jenkins, M.A.; Casey, G.; Hoffmeister, M.; Brenner, H.; Chang-Claude, J.; et al. Lack of an association between gallstone disease and bilirubin levels with risk of colorectal cancer: A Mendelian randomisation analysis. Br. J. Cancer 2021, 124, 1169–1174. [Google Scholar] [CrossRef]
- Papadimitriou, N.; Gunter, M.J.; Murphy, N.; Gicquiau, A.; Achaintre, D.; Brezina, S.; Gumpenberger, T.; Baierl, A.; Ose, J.; Geijsen, A.J.; et al. Circulating tryptophan metabolites and risk of colon cancer: Results from case-control and prospective cohort studies. Int. J. Cancer 2021. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).