HDGF Protects Retinal Pigment Epithelium from Glyoxal-Induced Ferroptosis via SIRT1/PGC-1α/Nrf2 Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Treatment
2.2. Cell Viability Assay
2.3. Lactate Dehydrogenase (LDH) Assay
2.4. Mitochondrial Membrane Potential (MMP) Detection
2.5. Apoptosis Assay (Annexin V and PI)
2.6. Measurement of Intracellular ROS Level (DCF-DA) and Mitochondrial Superoxide (MitoSOX Red)
2.7. Intracellular Iron Measurement (FerroOrange)
2.8. Lipid Peroxidation Assay (BODIPY 581/591 C11)
2.9. RNA Extraction and Quantitative Polymerase Chain Reaction (qPCR)
2.10. Western Blot
2.11. Immunofluorescence Staining
2.12. Total and Reduced Glutathione Assay
2.13. Statistical Analysis
3. Results
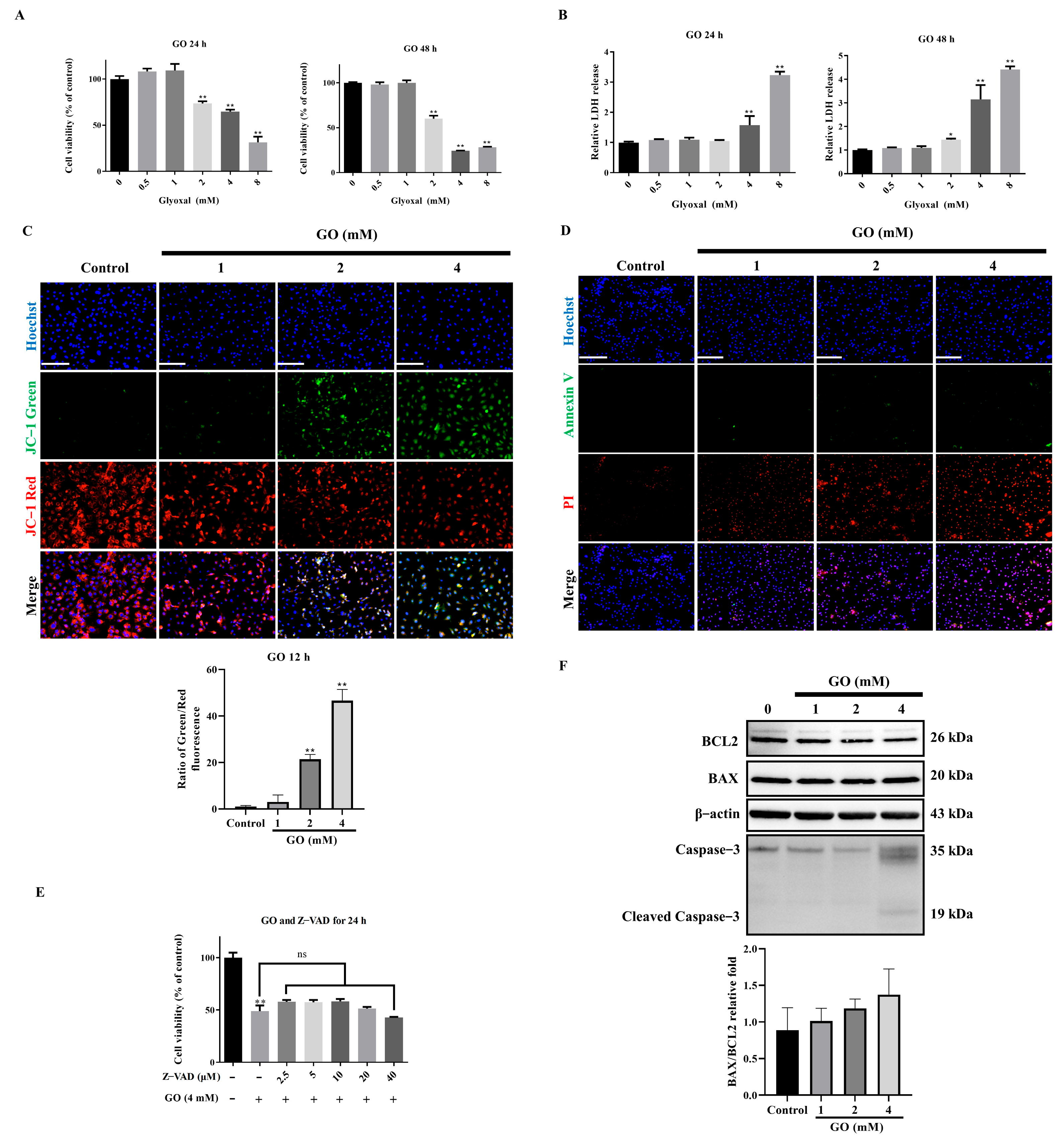
3.1. Non-Apoptotic Cell Death Induced by GO Exposure
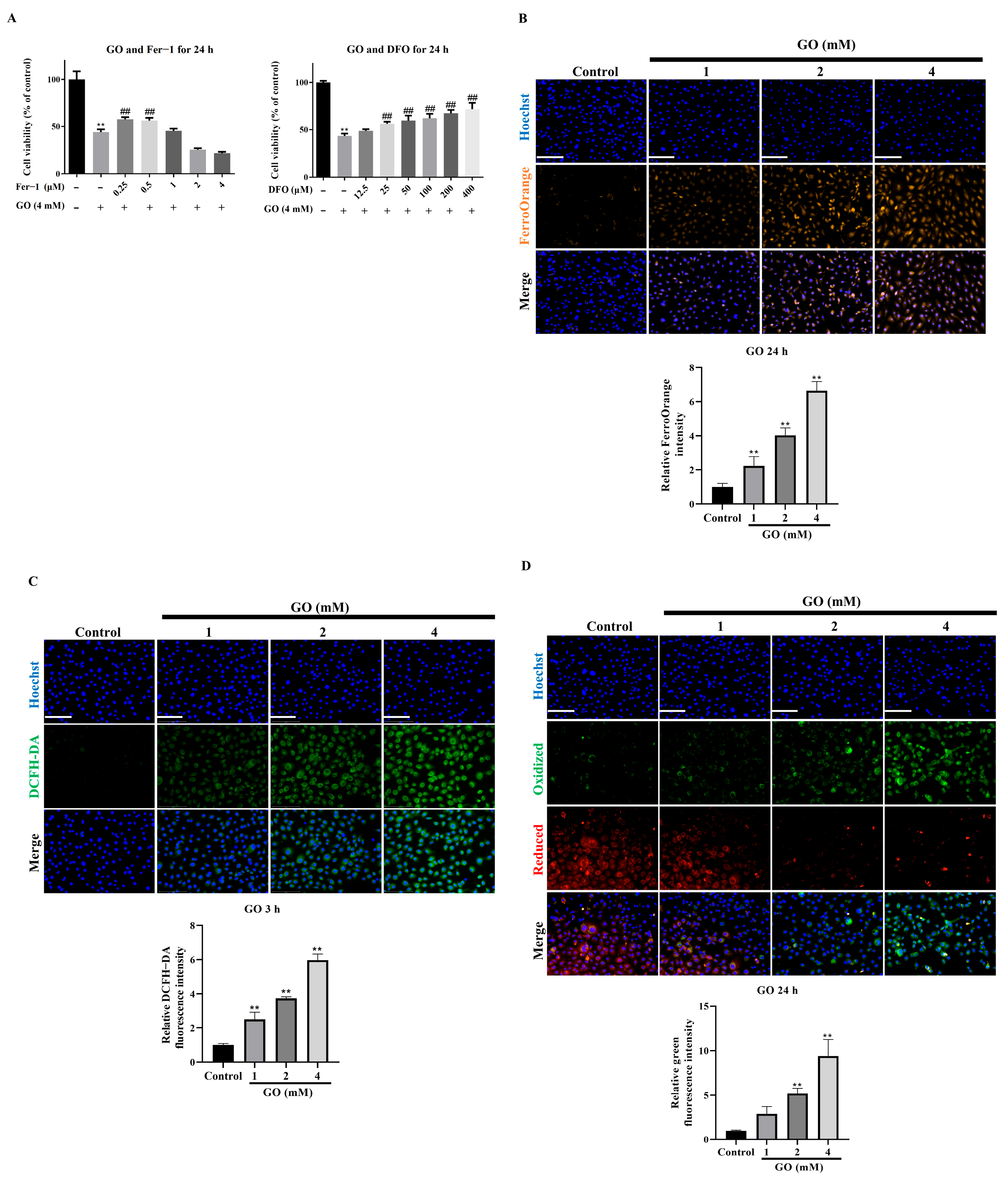
3.2. Ferroptosis Mediates GO-Induced RPE Cell Death
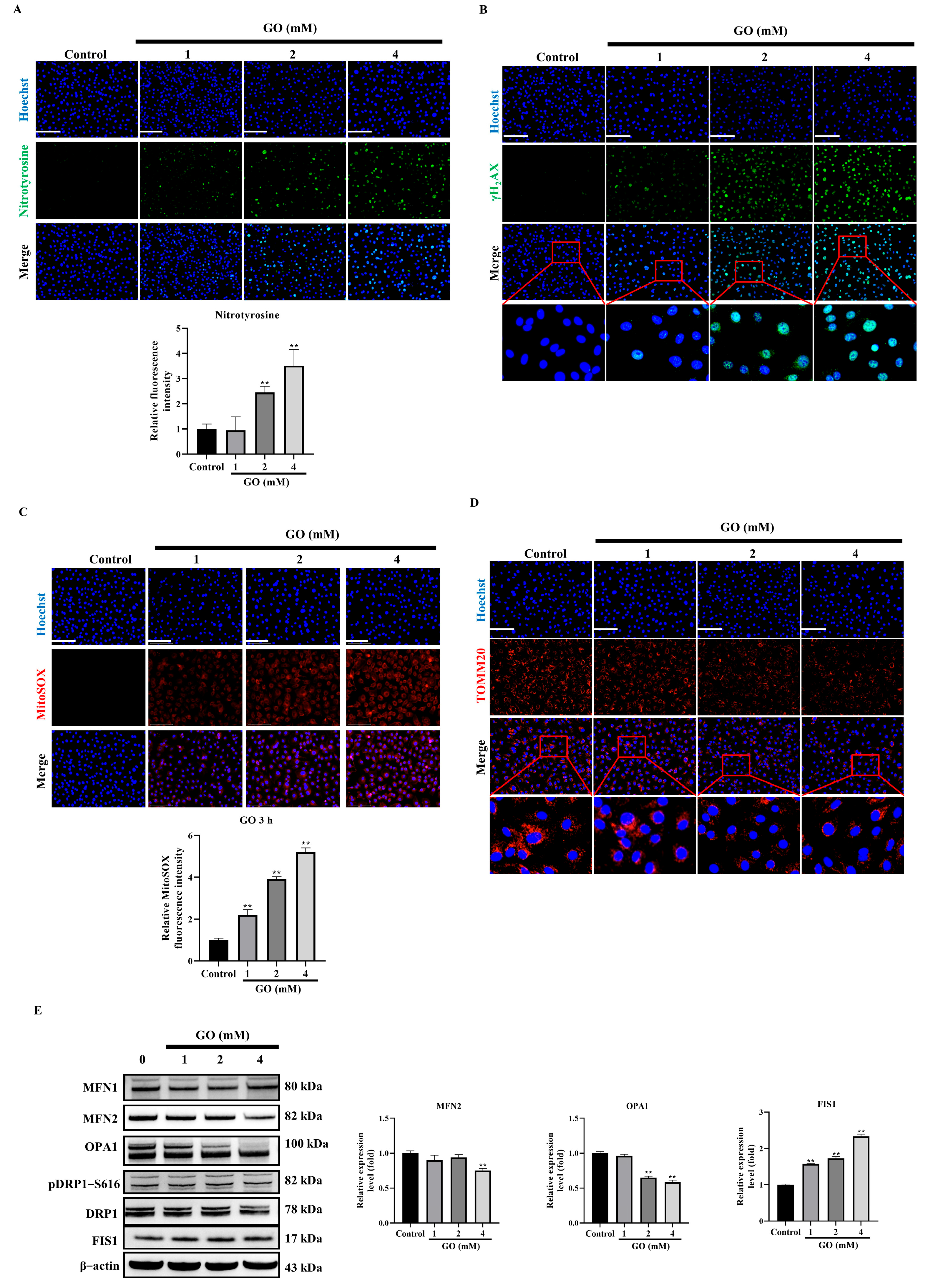
3.3. GO Induces Oxidative Stress and Mitochondrial Impairment in RPE Cells
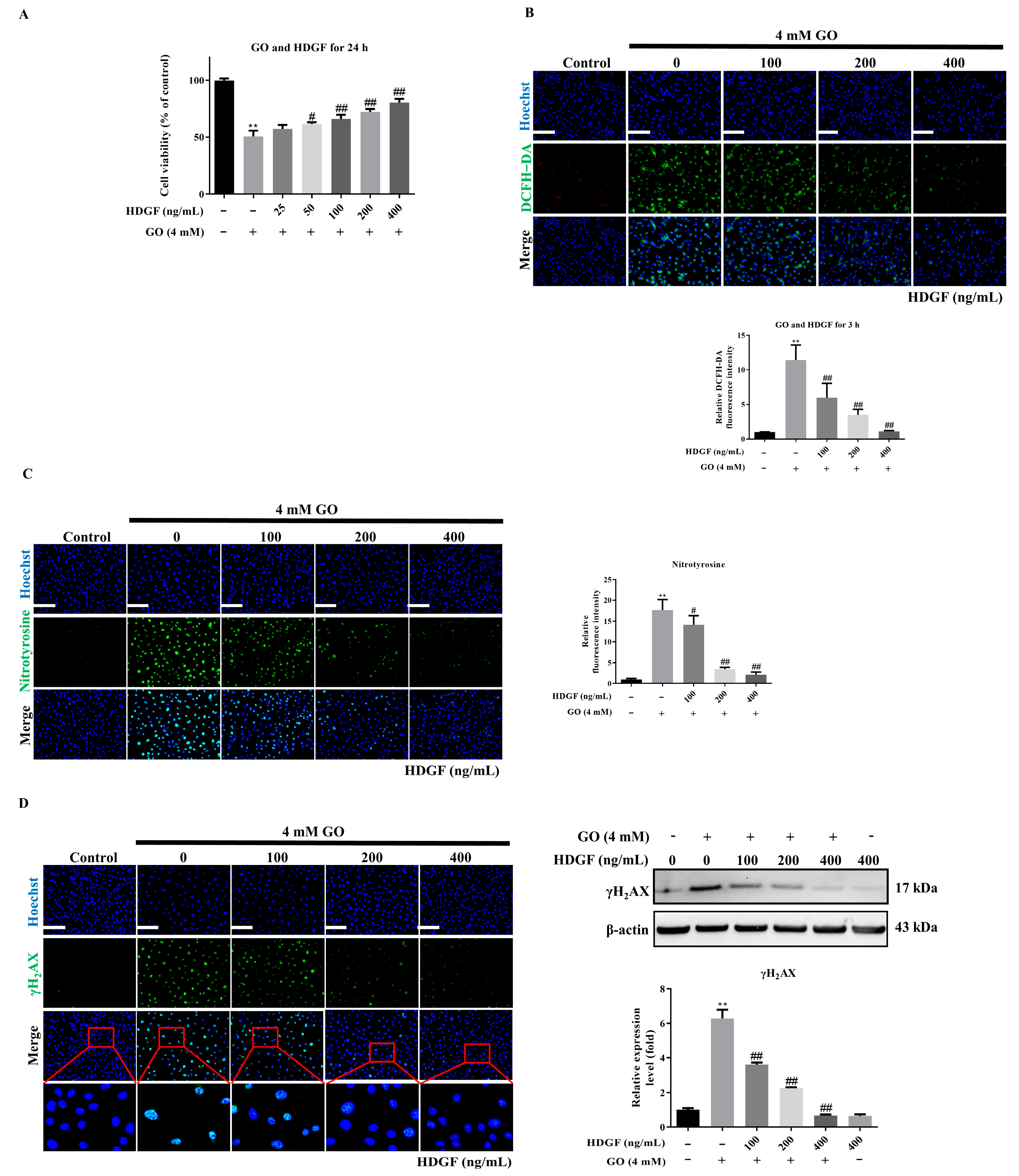
3.4. HDGF Protects RPE Cells Against GO-Induced Oxidative Damage
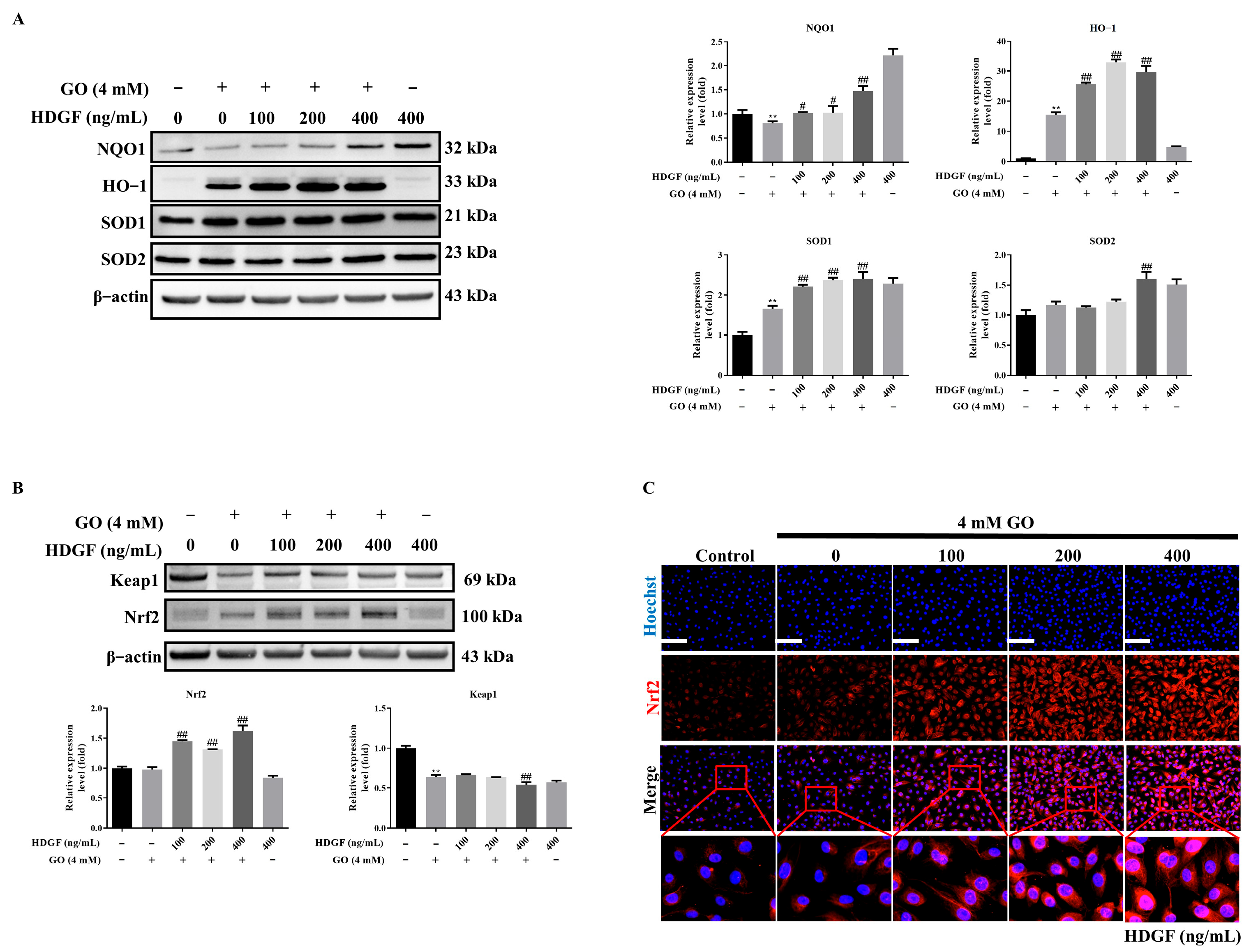
3.5. HDGF Regulates Nrf2 and Antioxidant Enzymes in GO-Treated RPE Cells
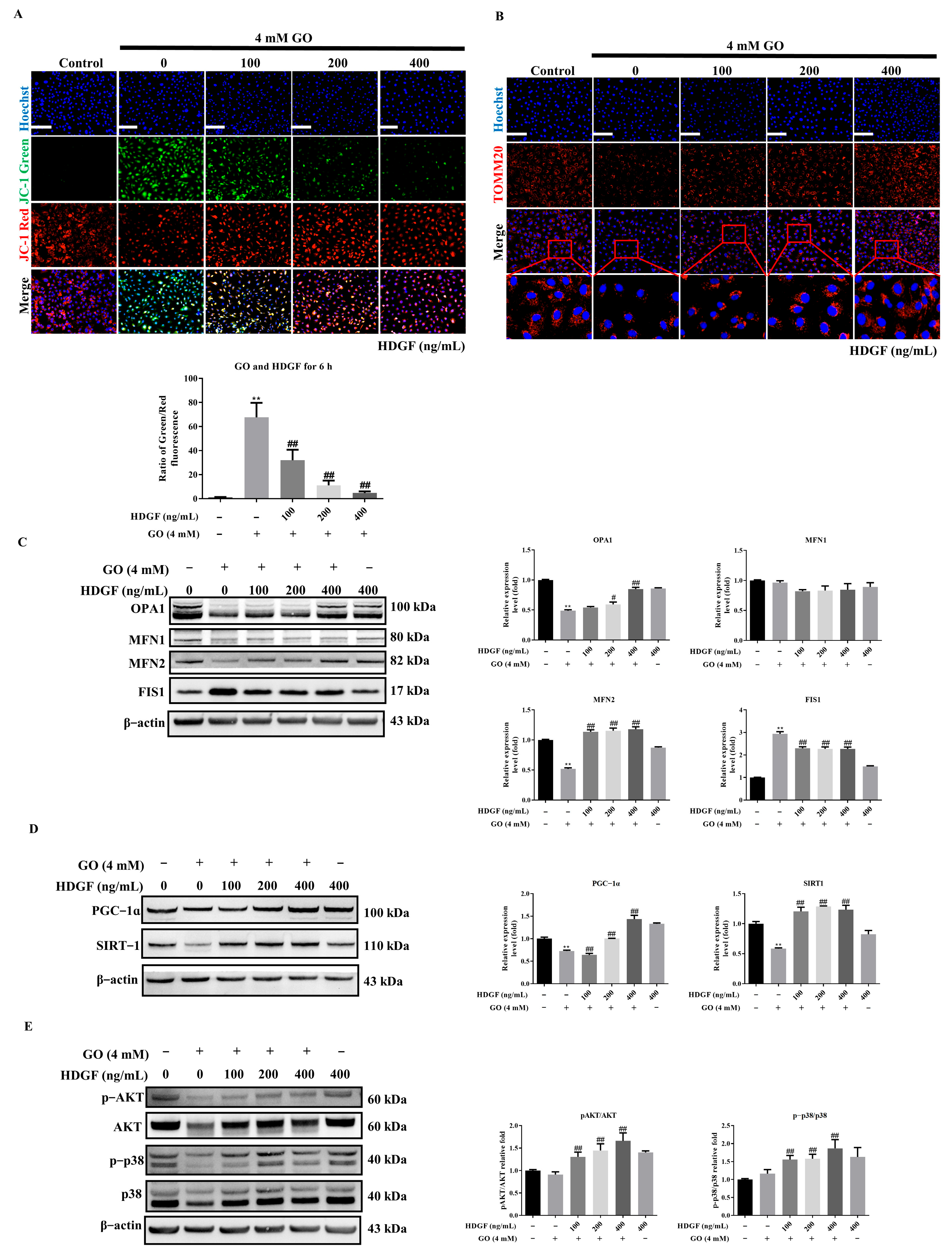
3.6. HDGF Preserves Mitochondrial Function Through SIRT1/PGC-1α Signaling in GO-Treated RPE Cells
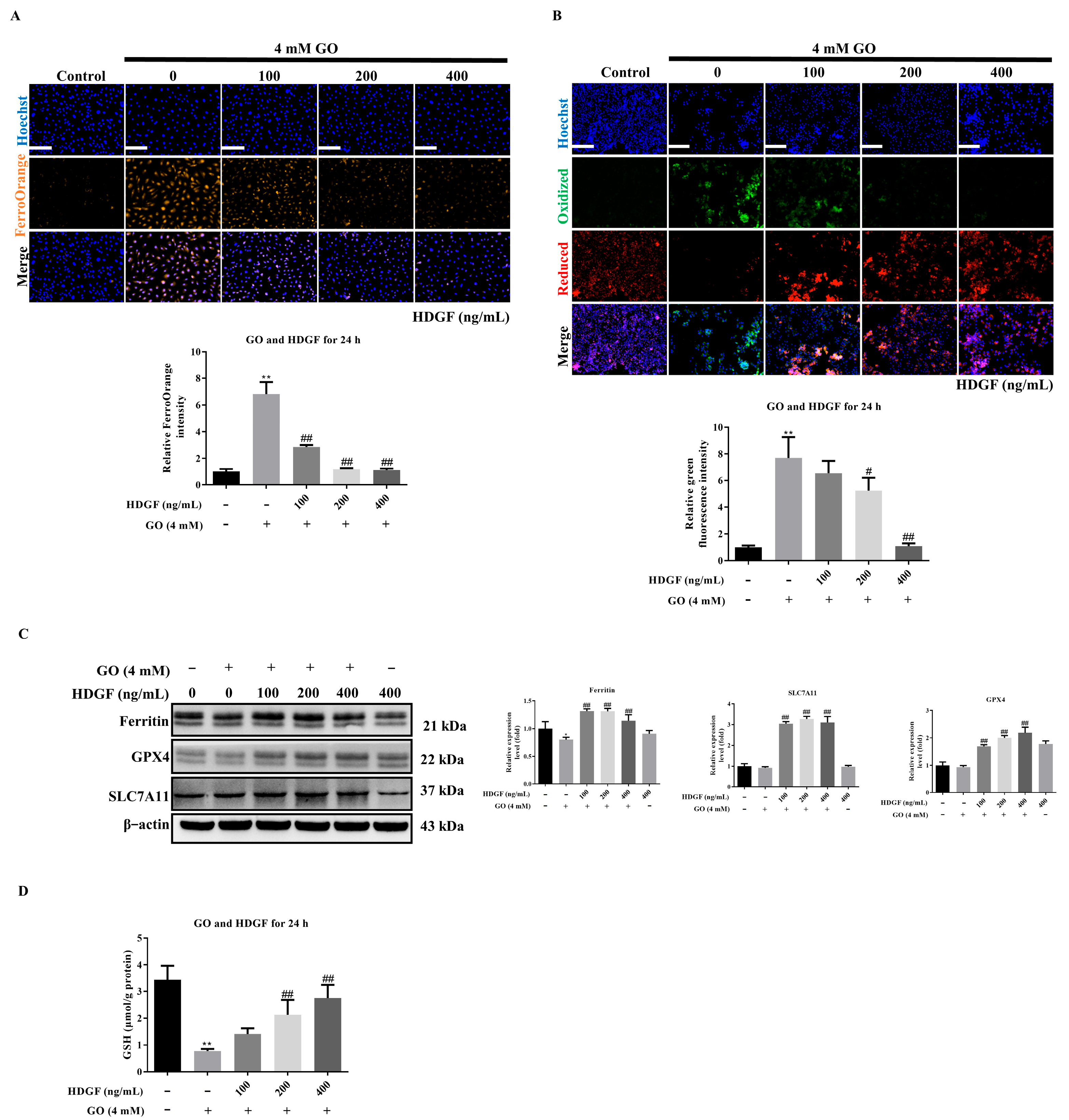
3.7. HDGF Against GO-Induced Ferroptosis in ARPE-19 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMD | Age-related macular degeneration |
| RPE | Retinal pigment epithelium |
| GO | Glyoxal |
| tBHP | Tert-butyl hydroperoxide |
| GSH | Glutathione |
| HDGF | Hepatoma-derived growth factor |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| PS | Penicillin/Streptomycin |
| FBS | Fetal bovine serum |
| LDH | Lactate dehydrogenase |
| MMP | Mitochondrial membrane potential |
| PBS | Phosphate-buffered saline |
| DMSO | Dimethyl sulfoxide |
| PI | Propidium iodide |
| qPCR | Quantitative polymerase chain reaction |
| TBS | Tris-buffered saline |
| HRP | Horseradish peroxidase |
| DFO | Deferoxamine mesylate |
| Fer-1 | Ferrostatin-1 |
| Fe2+ | Ferrous iron |
| MFN2 | Mitofusin 2 |
| H2O2 | Hydrogen peroxide |
| AGEs | Advanced glycation end-products |
| WB | Western Blot |
| IF | Immunofluorescence |
References
- Śpiewak, D.; Drzyzga, Ł.; Dorecka, M.; Wyględowska-Promieńska, D. Summary of the Therapeutic Options for Patients with Dry and Neovascular AMD. J. Clin. Med. 2024, 13, 4227. [Google Scholar] [CrossRef] [PubMed]
- Chiba, C. The Retinal Pigment Epithelium: An Important Player of Retinal Disorders and Regeneration. Exp. Eye Res. 2014, 123, 107–114. [Google Scholar] [CrossRef]
- Kandarakis, S.A.; Piperi, C.; Topouzis, F.; Papavassiliou, A.G. Emerging Role of Advanced Glycation-End Products (AGEs) in the Pathobiology of Eye Diseases. Prog. Retin. Eye Res. 2014, 42, 85–102. [Google Scholar] [CrossRef]
- Yoon, K.D.; Yamamoto, K.; Ueda, K.; Zhou, J.; Sparrow, J.R. A Novel Source of Methylglyoxal and Glyoxal in Retina: Implications for Age-Related Macular Degeneration. PLoS ONE 2012, 7, e41309. [Google Scholar] [CrossRef]
- Roehlecke, C.; Valtink, M.; Frenzel, A.; Goetze, D.; Knels, L.; Morawietz, H.; Funk, R.H.W. Stress Responses of Human Retinal Pigment Epithelial Cells to Glyoxal. Graefes Arch. Clin. Exp. Ophthalmol. 2016, 254, 2361–2372. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liu, Z.; Liao, H.; Chen, Z.-S.; Qin, B. Ferroptosis as a Potential Therapeutic Target for Age-Related Macular Degeneration. Drug Discov. Today 2024, 29, 103920. [Google Scholar] [CrossRef] [PubMed]
- Totsuka, K.; Ueta, T.; Uchida, T.; Roggia, M.F.; Nakagawa, S.; Vavvas, D.G.; Honjo, M.; Aihara, M. Oxidative Stress Induces Ferroptotic Cell Death in Retinal Pigment Epithelial Cells. Exp. Eye Res. 2019, 181, 316–324. [Google Scholar] [CrossRef]
- Liu, B.; Wang, W.; Shah, A.; Yu, M.; Liu, Y.; He, L.; Dang, J.; Yang, L.; Yan, M.; Ying, Y.; et al. Sodium Iodate Induces Ferroptosis in Human Retinal Pigment Epithelium ARPE-19 Cells. Cell Death Dis. 2021, 12, 230. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Pawlowska, E.; Szczepanska, J.; Jablkowska, A.; Blasiak, J. Role of Mitochondrial DNA Damage in ROS-Mediated Pathogenesis of Age-Related Macular Degeneration (AMD). Int. J. Mol. Sci. 2019, 20, 2374. [Google Scholar] [CrossRef]
- Mehrzadi, S.; Hemati, K.; Reiter, R.J.; Hosseinzadeh, A. Mitochondrial Dysfunction in Age-Related Macular Degeneration: Melatonin as a Potential Treatment. Expert Opin. Ther. Targets 2020, 24, 359–378. [Google Scholar] [CrossRef]
- Duarte, F.V.; Amorim, J.A.; Palmeira, C.M.; Rolo, A.P. Regulation of Mitochondrial Function and Its Impact in Metabolic Stress. Curr. Med. Chem. 2015, 22, 2468–2479. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.-J.; Moon, C.; Kim, S.Y.; Park, M.H.; Bae, Y.C.; Chun, M.-H.; Moon, J.-I. Glutathione depletion induces differential apoptosis in cells of mouse retina, in vivo. Neurosci. Lett. 2007, 417, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Kim, S.Y.; Moon, C.; Bae, Y.C.; Moon, J.-I.; Moon, C. Differential Cell Death and Bcl-2 Expression in the Mouse Retina after Glutathione Decrease by Systemic D,L-Buthionine Sulphoximine Administration. Mol. Cells 2013, 35, 235–242. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, Y.; Wang, C.; Liu, Y. Glutathione Depletion Induces Ferroptosis, Autophagy, and Premature Cell Senescence in Retinal Pigment Epithelial Cells. Cell Death Dis. 2018, 9, 753. [Google Scholar] [CrossRef]
- Zhou, Z.; Yamamoto, Y.; Sugai, F.; Yoshida, K.; Kishima, Y.; Sumi, H.; Nakamura, H.; Sakoda, S. Hepatoma-Derived Growth Factor Is a Neurotrophic Factor Harbored in the Nucleus*. J. Biol. Chem. 2004, 279, 27320–27326. [Google Scholar] [CrossRef]
- Marubuchi, S.; Okuda, T.; Tagawa, K.; Enokido, Y.; Horiuchi, D.; Shimokawa, R.; Tamura, T.; Qi, M.-L.; Eishi, Y.; Watabe, K.; et al. Hepatoma-Derived Growth Factor, a New Trophic Factor for Motor Neurons, Is up-Regulated in the Spinal Cord of PQBP-1 Transgenic Mice before Onset of Degeneration. J. Neurochem. 2006, 99, 70–83. [Google Scholar] [CrossRef]
- Hollander, A.; D’Onofrio, P.M.; Magharious, M.M.; Lysko, M.D.; Koeberle, P.D. Quantitative Retinal Protein Analysis after Optic Nerve Transection Reveals a Neuroprotective Role for Hepatoma-Derived Growth Factor on Injured Retinal Ganglion Cells. Investig. Ophthalmol. Vis. Sci. 2012, 53, 3973–3989. [Google Scholar] [CrossRef]
- Yang, Y.; Lin, Y.; Han, Z.; Wang, B.; Zheng, W.; Wei, L. Ferroptosis: A Novel Mechanism of Cell Death in Ophthalmic Conditions. Front. Immunol. 2024, 15, 1440309. [Google Scholar] [CrossRef]
- Su, L.-J.; Zhang, J.-H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.-Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxidative Med. Cell. Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef]
- Knutti, D.; Kressler, D.; Kralli, A. Regulation of the Transcriptional Coactivator PGC-1 via MAPK-Sensitive Interaction with a Repressor. Proc. Natl. Acad. Sci. USA 2001, 98, 9713–9718. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Monks, B.; Ge, Q.; Birnbaum, M.J. Akt/PKB Regulates Hepatic Metabolism by Directly Inhibiting PGC-1α Transcription Coactivator. Nature 2007, 447, 1012–1016. [Google Scholar] [CrossRef]
- Zhao, T.; Guo, X.; Sun, Y. Iron Accumulation and Lipid Peroxidation in the Aging Retina: Implication of Ferroptosis in Age-Related Macular Degeneration. Aging Dis. 2021, 12, 529. [Google Scholar] [CrossRef]
- Błasiak, J.; Skłodowska, A.; Ulińska, M.; Szaflik, J.P. Iron and Age-Related Macular Degeneration. Klin. Ocz. Acta Ophthalmol. Pol. 2009, 111, 174–177. [Google Scholar]
- He, X.; Hahn, P.; Iacovelli, J.; Wong, R.; King, C.; Bhisitkul, R.; Massaro-Giordano, M.; Dunaief, J.L. Iron Homeostasis and Toxicity in Retinal Degeneration. Prog. Retin. Eye Res. 2007, 26, 649–673. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Fisher, C.R.; Kowluru, R.A. Mitochondrial Defects Drive Degenerative Retinal Diseases. Trends Mol. Med. 2020, 26, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Bilbao-Malavé, V.; González-Zamora, J.; de la Puente, M.; Recalde, S.; Fernandez-Robredo, P.; Hernandez, M.; Layana, A.G.; Saenz de Viteri, M. Mitochondrial Dysfunction and Endoplasmic Reticulum Stress in Age Related Macular Degeneration, Role in Pathophysiology, and Possible New Therapeutic Strategies. Antioxidants 2021, 10, 1170. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Hsieh, M.-C.; Wu, H.-J.; Wu, W.-C.; Kao, Y.-H. Methylglyoxal, a Reactive Glucose Metabolite, Enhances Autophagy Flux and Suppresses Proliferation of Human Retinal Pigment Epithelial ARPE-19 Cells. Toxicol. Vitr. 2015, 29, 1358–1368. [Google Scholar] [CrossRef]
- Cantó, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK Regulates Energy Expenditure by Modulating NAD+ Metabolism and SIRT1 Activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef]
- Álvarez-Guardia, D.; Palomer, X.; Coll, T.; Davidson, M.M.; Chan, T.O.; Feldman, A.M.; Laguna, J.C.; Vázquez-Carrera, M. The P65 Subunit of NF-κB Binds to PGC-1α, Linking Inflammation and Metabolic Disturbances in Cardiac Cells. Cardiovasc. Res. 2010, 87, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.-W.; Zhao, G.-J.; Li, X.-L.; Hong, G.-L.; Li, M.-F.; Qiu, Q.-M.; Wu, B.; Lu, Z.-Q. SIRT1 Exerts Protective Effects against Paraquat-Induced Injury in Mouse Type II Alveolar Epithelial Cells by Deacetylating NRF2 in Vitro. Int. J. Mol. Med. 2016, 37, 1049–1058. [Google Scholar] [CrossRef]
- Velmurugan, S.; Pauline, R.; Chandrashekar, G.; Kulanthaivel, L.; Subbaraj, G.K. Understanding the Impact of the Sirtuin 1 (SIRT1) Gene on Age-Related Macular Degeneration: A Comprehensive Study. Niger. Postgrad. Med. J. 2024, 31, 93. [Google Scholar] [CrossRef] [PubMed]
- Di Gregorio, J.; Zerti, D.; Carozza, G.; Capozzo, A.; Flati, V.; Feligioni, M.; Maccarone, R. Hormetic Effects of Curcumin in RPE Cells: SIRT1 and Caspase-3 Inactivation with Implications for AMD. Int. J. Mol. Sci. 2025, 26, 8555. [Google Scholar] [CrossRef]
- Puigserver, P.; Rhee, J.; Lin, J.; Wu, Z.; Yoon, J.C.; Zhang, C.Y.; Krauss, S.; Mootha, V.K.; Lowell, B.B.; Spiegelman, B.M. Cytokine Stimulation of Energy Expenditure through P38 MAP Kinase Activation of PPARgamma Coactivator-1. Mol. Cell 2001, 8, 971–982. [Google Scholar] [CrossRef]
- Sano, M.; Tokudome, S.; Shimizu, N.; Yoshikawa, N.; Ogawa, C.; Shirakawa, K.; Endo, J.; Katayama, T.; Yuasa, S.; Ieda, M.; et al. Intramolecular Control of Protein Stability, Subnuclear Compartmentalization, and Coactivator Function of Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1alpha. J. Biol. Chem. 2007, 282, 25970–25980. [Google Scholar] [CrossRef]
- Fan, M.; Rhee, J.; St-Pierre, J.; Handschin, C.; Puigserver, P.; Lin, J.; Jäeger, S.; Erdjument-Bromage, H.; Tempst, P.; Spiegelman, B.M. Suppression of Mitochondrial Respiration through Recruitment of P160 Myb Binding Protein to PGC-1α: Modulation by P38 MAPK. Genes Dev. 2004, 18, 278–289. [Google Scholar] [CrossRef]
- Deng, X.; Li, Y.; Gu, S.; Chen, Y.; Yu, B.; Su, J.; Sun, L.; Liu, Y. P53 Affects PGC1α Stability Through AKT/GSK-3β to Enhance Cisplatin Sensitivity in Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 1252. [Google Scholar] [CrossRef]
- Larson-Casey, J.L.; Gu, L.; Davis, D.; Cai, G.-Q.; Ding, Q.; He, C.; Carter, A.B. Post-Translational Regulation of PGC-1α Modulates Fibrotic Repair. FASEB J. 2021, 35, e21675. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, M.; Jin, H. PI3K/AKT Phosphorylation Activates ERRα by Upregulating PGC-1α and PGC-1β in Gallbladder Cancer. Mol. Med. Rep. 2021, 24, 613. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowski, A.M.; Bell, B.A.; Song, Y.; Anderson, B.D.; Conomikes, A.; Petruconis, C.; Dunaief, J.L. Inducible RPE-Specific GPX4 Knockout Causes Oxidative Stress and Retinal Degeneration with Features of Age-Related Macular Degeneration. Exp. Eye Res. 2024, 247, 110028. [Google Scholar] [CrossRef]
- Ueta, T.; Azuma, K.; Shiraya, T.; Aihara, M. Impact of GPx4 Deficiency on RPE: Insights into Lipid Peroxidation-Driven Degeneration. Investig. Ophthalmol. Vis. Sci. 2024, 65, 1287. [Google Scholar]
- Poltorack, C.D.; Dixon, S.J. Understanding the Role of Cysteine in Ferroptosis: Progress & Paradoxes. FEBS J. 2022, 289, 374–385. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-D.; Tsai, R.-K.; Wen, Y.-T.; Liu, P.-K. HDGF Protects Retinal Pigment Epithelium from Glyoxal-Induced Ferroptosis via SIRT1/PGC-1α/Nrf2 Pathway. Antioxidants 2025, 14, 1434. https://doi.org/10.3390/antiox14121434
Lin H-D, Tsai R-K, Wen Y-T, Liu P-K. HDGF Protects Retinal Pigment Epithelium from Glyoxal-Induced Ferroptosis via SIRT1/PGC-1α/Nrf2 Pathway. Antioxidants. 2025; 14(12):1434. https://doi.org/10.3390/antiox14121434
Chicago/Turabian StyleLin, Heng-Dao, Rong-Kung Tsai, Yao-Tseng Wen, and Pei-Kang Liu. 2025. "HDGF Protects Retinal Pigment Epithelium from Glyoxal-Induced Ferroptosis via SIRT1/PGC-1α/Nrf2 Pathway" Antioxidants 14, no. 12: 1434. https://doi.org/10.3390/antiox14121434
APA StyleLin, H.-D., Tsai, R.-K., Wen, Y.-T., & Liu, P.-K. (2025). HDGF Protects Retinal Pigment Epithelium from Glyoxal-Induced Ferroptosis via SIRT1/PGC-1α/Nrf2 Pathway. Antioxidants, 14(12), 1434. https://doi.org/10.3390/antiox14121434







