Potential Rapid Quantification of Antioxidant Capacity of Olea europaea L. Leaves by Near-Infrared Spectroscopy Using Different Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Plant Material
2.3. Preparation of Extracts of Olea europaea L. Leaves
2.4. Antioxidant Activity
2.4.1. DPPH
2.4.2. ABTS
2.4.3. ORAC
2.5. Near-Infrared Spectroscopy Measurements
2.6. Datasets Distribution
2.7. Spectral Preprocessing
2.8. Model Evaluation
2.9. Reversed-Phase High-Performance Liquid Chromatography with Diode Array Detection (RP-HPLC-DAD) Conditions
3. Results
3.1. Antioxidant Potential of Olea europaea L. Leaves Across Sample Collection
3.2. Spectra Inspection
3.3. NIR Calibration and Validation of Antioxidant Capacity
3.4. NIR Prediction Models for Antioxidant Capacity
3.5. Correlation Between Phenolic Compounds and Antioxidant Capacity in NIR Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kostelenos, G.; Kiritsakis, A. Olive Tree History and Evolution. In Olives and Olive Oil as Functional Foods: Bioactivity, Chemistry and Processing; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 1–12. [Google Scholar] [CrossRef]
- Espeso, J.; Isaza, A.; Lee, J.Y.; Sörensen, P.M.; Jurado, P.; Avena-Bustillos, R.d.J.; Olaizola, M.; Arboleya, J.C. Olive Leaf Waste Management. Front. Sustain. Food Syst. 2021, 5, 660582. [Google Scholar] [CrossRef]
- Jabalbarezi Hukerdi, Y.; Fathi, M.H.; Rashidi, L.; Ganjkhanlou, M. The Study of Physicochemical Properties and Nutrient Composition of Mari Olive Leaf Cultivated in Iran. Nutr. Food Sci. Res. 2018, 5, 39–46. [Google Scholar] [CrossRef]
- Martínez-Navarro, E.M.; Cebrián-Tarancón, C.; Moratalla-López, N.; Lorenzo, C.; Alonso, G.L.; Salinas, R.M. Development and Validation of an HPLC-DAD Method for Determination of Oleuropein and Other Bioactive Compounds in Olive Leaf by-Products. J. Sci. Food Agric. 2021, 101, 1447–1453. [Google Scholar] [CrossRef]
- Pessoa, H.R.; Zago, L.; Difonzo, G.; Pasqualone, A.; Caponio, F.; Ferraz da Costa, D.C. Olive Leaves as a Source of Anticancer Compounds: In Vitro Evidence and Mechanisms. Molecules 2024, 29, 4249. [Google Scholar] [CrossRef] [PubMed]
- Bulotta, S.; Celano, M.; Lepore, S.M.; Montalcini, T.; Pujia, A.; Russo, D. Beneficial Effects of the Olive Oil Phenolic Components Oleuropein and Hydroxytyrosol: Focus on Protection against Cardiovascular and Metabolic Diseases. J. Transl. Med. 2014, 12, 219. [Google Scholar] [CrossRef]
- Cerri, L.; Parri, S.; Dias, M.C.; Fabiano, A.; Romi, M.; Cai, G.; Cantini, C.; Zambito, Y. Olive Leaf Extracts from Three Italian Olive Cultivars Exposed to Drought Stress Differentially Protect Cells against Oxidative Stress. Antioxidants 2024, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Apak, R.; Özyürek, M.; Güçlü, K.; Çapanoğlu, E. Antioxidant Activity/Capacity Measurement. 1. Classification, Physicochemical Principles, Mechanisms, and Electron Transfer (ET)-Based Assays. J. Agric. Food Chem. 2016, 64, 997–1027. [Google Scholar] [CrossRef]
- Can, A.; Ayvaz, H.; Pala, Ç.U.; Condelli, N.; Galgano, F.; Tolve, R. The Potential of near and Mid-Infrared Spectroscopy for Rapid Quantification of Oleuropein, Total Phenolics, Total Flavonoids and Antioxidant Activity in Olive Tree (Olea Europaea) Leaves. J. Food Meas. Charact. 2018, 12, 2747–2757. [Google Scholar] [CrossRef]
- Andersen, C.M.; Bro, R. Variable Selection in Regression—A Tutorial. J. Chemom. 2010, 24, 728–737. [Google Scholar] [CrossRef]
- Gerlach, R.W.; Kowalski, B.R.; Wold, H.O.A. Partial Least-Squares Path Modelling with Latent Variables. Anal. Chim. Acta 1979, 112, 417–421. [Google Scholar] [CrossRef]
- Mehany, T.; González-Sáiz, J.M.; Pizarro, C. The Quality Prediction of Olive and Sunflower Oils Using NIR Spectroscopy and Chemometrics: A Sustainable Approach. Foods 2025, 14, 2152. [Google Scholar] [CrossRef]
- Grassi, S.; Jolayemi, O.S.; Giovenzana, V.; Tugnolo, A.; Squeo, G.; Conte, P.; De Bruno, A.; Flamminii, F.; Casiraghi, E.; Alamprese, C. Near Infrared Spectroscopy as a Green Technology for the Quality Prediction of Intact Olives. Foods 2021, 10, 1042. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT-Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ou, B.; Hampsch-Woodill, M.; Flanagan, J.A.; Prior, R.L. High-Throughput Assay of Oxygen Radical Absorbance Capacity (ORAC) Using a Multichannel Liquid Handling System Coupled with a Microplate Fluorescence Reader in 96-Well Format. J. Agric. Food Chem. 2002, 50, 4437–4444. [Google Scholar] [CrossRef]
- Brescia, P.J. Determination of Antioxidant Potential Using an Oxygen Radical Absorbance Capacity (ORAC) Assay with SynergyTM H4; BioTek Instruments, Inc.: Winooski, VT, USA, 2012. [Google Scholar]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Wu, D.; Chen, J.; Lu, B.; Xiong, L.; He, Y.; Zhang, Y. Application of near Infrared Spectroscopy for the Rapid Determination of Antioxidant Activity of Bamboo Leaf Extract. Food Chem. 2012, 135, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zareef, M.; He, P.; Sun, H.; Chen, Q.; Li, H.; Ouyang, Q.; Guo, Z.; Zhang, Z.; Xu, D. Evaluation of Matcha Tea Quality Index Using Portable NIR Spectroscopy Coupled with Chemometric Algorithms. J. Sci. Food Agric. 2019, 99, 5019–5027. [Google Scholar] [CrossRef]
- Martínez-Navarro, M.E.; Kaparakou, E.H.; Kanakis, C.D.; Cebrián-Tarancón, C.; Alonso, G.L.; Salinas, M.R.; Tarantilis, P.A. Quantitative Determination of the Main Phenolic Compounds, Antioxidant Activity, and Toxicity of Aqueous Extracts of Olive Leaves of Greek and Spanish Genotypes. Horticulturae 2023, 9, 55. [Google Scholar] [CrossRef]
- Martínez-Navarro, E.M. Study for the Use of Olive Leaf for Its Oleuropein Content and Other Phenolic Compounds; UCLM: Albacete, Spain, 2022. [Google Scholar]
- Orak, H.H.; Karamać, M.; Amarowicz, R.; Orak, A.; Penkacik, K. Genotype-Related Differences in the Phenolic Compound Profile and Antioxidant Activity of Extracts from Olive (Olea europaea L.) Leaves. Molecules 2019, 24, 1130. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Xin, X.; Zhu, S.; Niu, E.; Wu, Q.; Li, T.; Liu, D. Changes in Phytochemical Profiles and Biological Activity of Olive Leaves Treated by Two Drying Methods. Front. Nutr. 2022, 9, 854680. [Google Scholar] [CrossRef]
- Frumuzachi, O.; Gavrilaș, L.I.; Vodnar, D.C.; Rohn, S.; Mocan, A. Systemic Health Effects of Oleuropein and Hydroxytyrosol Supplementation: A Systematic Review of Randomized Controlled Trials. Antioxidants 2024, 13, 1040. [Google Scholar] [CrossRef]
- Magyari-Pavel, I.Z.; Moacă, E.A.; Avram, Ș.; Diaconeasa, Z.; Haidu, D.; Ștefănuț, M.N.; Rostas, A.M.; Muntean, D.; Bora, L.; Badescu, B.; et al. Antioxidant Extracts from Greek and Spanish Olive Leaves: Antimicrobial, Anticancer and Antiangiogenic Effects. Antioxidants 2024, 13, 774. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Sánchez-Villasclaras, S.; Pan, L.; Lan, W.; García-Martín, J.F. Advances in Vibrational Spectroscopic Techniques for the Detection of Bio-Active Compounds in Virgin Olive Oils: A Comprehensive Review. Foods 2024, 13, 3894. [Google Scholar] [CrossRef]
- Bellincontro, A.; Taticchi, A.; Servili, M.; Esposto, S.; Farinelli, D.; Mencarelli, F. Feasible Application of a Portable NIR-AOTF Tool for on-Field Prediction of Phenolic Compounds during the Ripening of Olives for Oil Production. J. Agric. Food Chem. 2012, 60, 2665–2673. [Google Scholar] [CrossRef]
- Bensehaila, S.; Ilias, F.; Saadi, F.; Zaouadi, N. Phenolic Compounds and Antimicrobial Activity of Olive (Olea europaea L.) Leaves. Asian J. Dairy Food Res. 2022, 41, 237–241. [Google Scholar] [CrossRef]
- Williams, P. The RPD Statistic: A Tutorial Note. NIR News 2010, 25, 22–26. [Google Scholar] [CrossRef]
- Moncada, G.W.; González Martín, M.I.; Escuredo, O.; Fischer, S.; Míguez, M. Multivariate Calibration by near Infrared Spectroscopy for the Determination of the Vitamin E and the Antioxidant Properties of Quinoa. Talanta 2013, 116, 65–70. [Google Scholar] [CrossRef]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the Most Common Pre-Processing Techniques for near-Infrared Spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Manley, M. Near-Infrared Spectroscopy and Hyperspectral Imaging: Non-Destructive Analysis of Biological Materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef]
- Prior, R.L.; Wu, X.; Schaich, K. Standardized Methods for the Determination of Antioxidant Capacity and Phenolics in Foods and Dietary Supplements. J. Agric. Food Chem. 2005, 53, 4290–4302. [Google Scholar] [CrossRef]
- Munteanu, I.G.; Apetrei, C. Analytical Methods Used in Determining Antioxidant Activity: A Review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef]
- Ou, B.; Hampsch-Woodill, M.; Prior, R.L. Development and Validation of an Improved Oxygen Radical Absorbance Capacity Assay Using Fluorescein as the Fluorescent Probe. J. Agric. Food Chem. 2001, 49, 4619–4626. [Google Scholar] [CrossRef]
- Ninfali, P.; Bacchiocca, M. Polyphenols and Antioxidant Capacity of Vegetables under Fresh and Frozen Conditions. J. Agric. Food Chem. 2003, 51, 2222–2226. [Google Scholar] [CrossRef]
- Talhaoui, N.; Taamalli, A.; Gómez-Caravaca, A.M.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Phenolic Compounds in Olive Leaves: Analytical Determination, Biotic and Abiotic Influence, and Health Benefits. Food Res. Int. 2015, 77, 92–108. [Google Scholar] [CrossRef]
- Valinger, D.; Kušen, M.; Jurinjak Tušek, A.; Panić, M.; Jurina, T.; Benković, M.; Radojčić Redovniković, I.; Gajdoš Kljusurić, J. Development of near Infrared Spectroscopy Models for Quantitative Prediction of the Content of Bioactive Compounds in Olive Leaves. Chem. Biochem. Eng. Q. 2018, 32, 535–543. [Google Scholar] [CrossRef]
- Pompeu, D.R.; Pissard, A.; Rogez, H.; Dupont, P.; Lateur, M.; Baeten, V. Estimation of Phenolic Compounds and Antioxidant Capacity in Leaves of Fruit Species Using Near-Infrared Spectroscopy and a Chemometric Approach [Estimation Des Composés Phénoliques et de La Capacité Antioxydante Des Feuilles d’espèces Fruitières Par Spectroscopie Proche Infrarouge et Approche Chimiométrique]. Biotechnol. Agron. Soc. Environ. 2021, 25, 109–119. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Li, F.; Zhang, Z.; Li, R.; Su, J.; Zhang, J.; Yang, B. Application of NIR and MIR Spectroscopy for Rapid Determination of Antioxidant Activity of Radix Scutellariae from Different Geographical Regions. Phytochem. Anal. 2016, 27, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Talavera, J.F.; Martínez-Navarro, M.E.; Alonso, G.L.; Sánchez-Gómez, R. Determination of the Main Phenolic Compounds of Olive (Olea europaea L.) Leaves by near Infrared Spectroscopy (NIR). Sci. Rep. 2025, 15, 29791. [Google Scholar] [CrossRef] [PubMed]
- Platzer, M.; Kiese, S.; Herfellner, T.; Schweiggert-Weisz, U.; Miesbauer, O.; Eisner, P. Common Trends and Differences in Antioxidant Activity Analysis of Phenolic Substances Using Single Electron Transfer Based Assays. Molecules 2021, 26, 1244. [Google Scholar] [CrossRef]
- Danet, A.F.; Danet, A.F. Recent Advances in Antioxidant Capacity Assays. In Antioxidants—Benefits, Sources, Mechanisms of Action; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Martínez-Zamora, L.; Peñalver, R.; Ros, G.; Nieto, G. Olive Tree Derivatives and Hydroxytyrosol: Their Potential Effects on Human Health and Its Use as Functional Ingredient in Meat. Foods 2021, 10, 2611. [Google Scholar] [CrossRef] [PubMed]
- Goulas, V.; Exarchou, V.; Troganis, A.N.; Psomiadou, E.; Fotsis, T.; Briasoulis, E.; Gerothanassis, I.P. Phytochemicals in Olive-Leaf Extracts and Their Antiproliferative Activity against Cancer and Endothelial Cells. Mol. Nutr. Food Res. 2009, 53, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Hostetler, G.L.; Ralston, R.A.; Schwartz, S.J. Flavones: Food Sources, Bioavailability, Metabolism, and Bioactivity. Adv. Nutr. 2017, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Romero-Márquez, J.M.; Navarro-Hortal, M.D.; Forbes-Hernández, T.Y.; Varela-López, A.; Puentes, J.G.; Sánchez-González, C.; Sumalla-Cano, S.; Battino, M.; García-Ruiz, R.; Sánchez, S.; et al. Effect of Olive Leaf Phytochemicals on the Anti-Acetylcholinesterase, Anti-Cyclooxygenase-2 and Ferric Reducing Antioxidant Capacity. Food Chem. 2024, 444, 138516. [Google Scholar] [CrossRef]
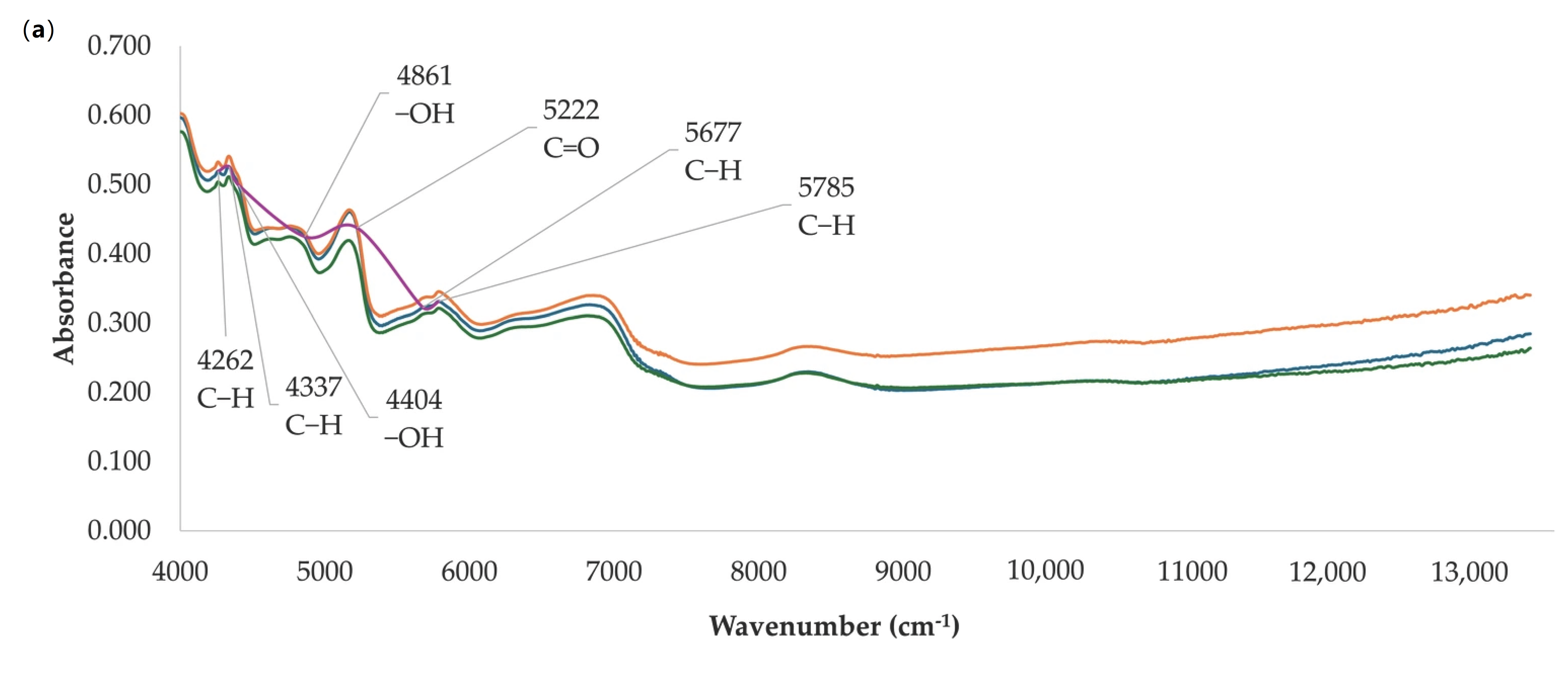
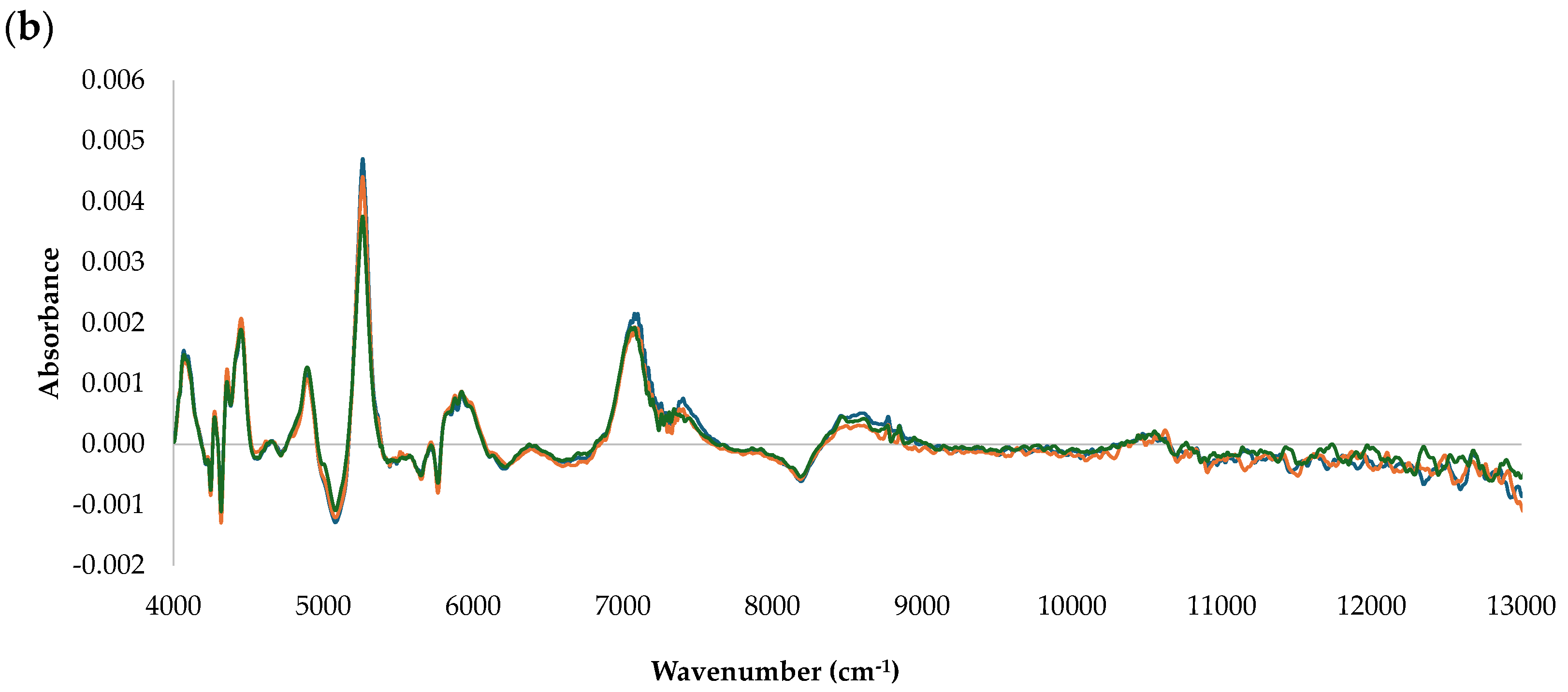
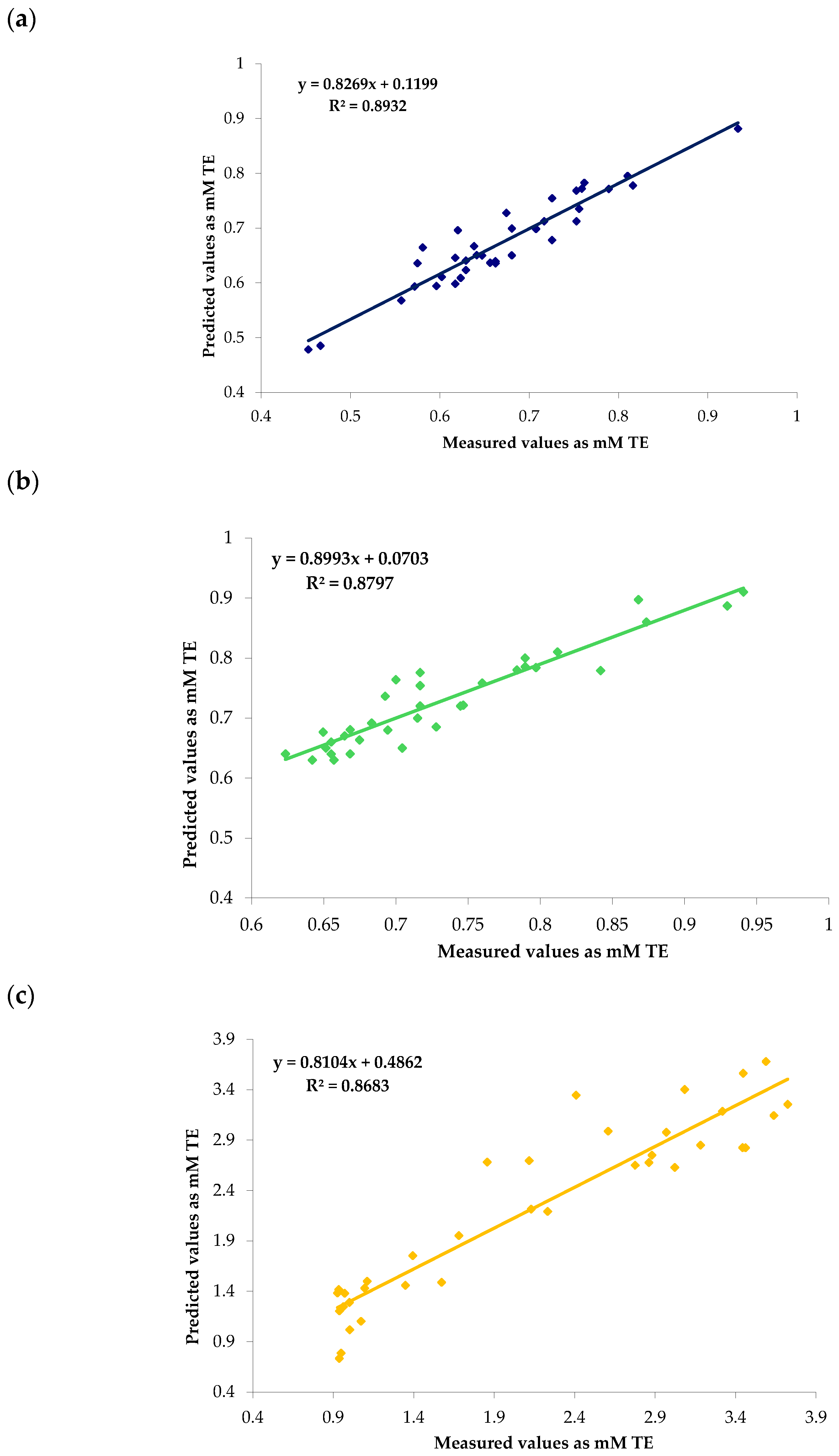
| Antioxidant Method | Data Sets | N | Range | Mean ± SD | CV (%) |
|---|---|---|---|---|---|
| DPPH | Set 1+2 | 120 | 0.42–0.96 | 0.69 ± 0.23 | 33.32 |
| Set 1 | 84 | 0.42–0.96 | 0.69 ± 0.22 | 32.74 | |
| Set 2 | 36 | 0.45–0.94 | 0.70 ± 0.20 | 31.33 | |
| ABTS | Set 1+2 | 120 | 0.60–0.99 | 0.74 ± 0.26 | 30.67 |
| Set 1 | 84 | 0.60–0.99 | 0.74 ± 0.25 | 30.43 | |
| Set 2 | 36 | 0.62–0.96 | 0.76 ± 0.23 | 29.14 | |
| ORAC | Set 1+2 | 120 | 0.94–4.10 | 2.72 ± 0.55 | 12.86 |
| Set 1 | 84 | 0.94–4.10 | 2.77 ± 0.56 | 12.12 | |
| Set 2 | 36 | 0.94–3.70 | 2.68 ± 0.52 | 12.56 |
| Antioxidant Method | Spectral Range (cm−1) | Pre-Process | Data Sets | R2 | RMSEC | RMSECV | RMSEP | RPD |
|---|---|---|---|---|---|---|---|---|
| DPPH | 13,333–4000 | MSC- 1.19.5 | Set 1+2 | 1.00 | 0.000 | |||
| Set 1 | 0.80 | 0.090 | 1.89 | |||||
| Set 2 | 0.82 | 0.087 | 2.02 | |||||
| SNV- 2.13.9 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.85 | 0.081 | 2.01 | |||||
| Set 2 | 0.86 | 0.078 | 2.30 | |||||
| MSC- 2.19.5 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.92 | 0.067 | 4.10 | |||||
| Set 2 | 0.90 | 0.073 | 3.01 | |||||
| SNV- 2.19.5 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.92 | 0.067 | 4.10 | |||||
| Set 2 | 0.90 | 0.073 | 3.01 | |||||
| ABTS | 13,333–4000 | MSC- 1.19.5 | Set 1+2 | 1.00 | 0.000 | |||
| Set 1 | 0.79 | 0.094 | 1.79 | |||||
| Set 2 | 0.81 | 0.091 | 1.92 | |||||
| SNV- 2.13.9 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.81 | 0.090 | 1.85 | |||||
| Set 2 | 0.82 | 0.087 | 2.13 | |||||
| SNV- 2.5.19 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.85 | 0.073 | 3.14 | |||||
| Set 2 | 0.88 | 0.064 | 2.90 | |||||
| MSC- 2.5.19 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.85 | 0.073 | 3.14 | |||||
| Set 2 | 0.88 | 0.064 | 2.90 | |||||
| ORAC | 13,333–4000 | MSC- 1.19.5 | Set 1+2 | 1.00 | 0.000 | |||
| Set 1 | 0.50 | 0.98 | 0.82 | |||||
| Set 2 | 0.52 | 0.99 | 0.90 | |||||
| SNV- 2.13.9 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.70 | 0.62 | 1.42 | |||||
| Set 2 | 0.73 | 0.55 | 1.63 | |||||
| SNV- 2.19.5 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.88 | 0.36 | 3.50 | |||||
| Set 2 | 0.87 | 0.47 | 2.69 | |||||
| MSC- 2.19.5 | Set 1+2 | 1.00 | 0.000 | |||||
| Set 1 | 0.88 | 0.36 | 3.50 | |||||
| Set 2 | 0.87 | 0.47 | 2.69 |
| DPPH | ABTS | ORAC | |
|---|---|---|---|
| Oleuropein | 0.383 ** | 0.011 | 0.524 ** |
| Hydroxytyrosol | 0.365 ** | −0.050 | 0.411 ** |
| Hydroxytyrosol hexoside | 0.125 | 0.207 * | −0.052 |
| Verbascoside | 0.007 | 0.244 ** | −0.165 |
| Apigenin-7-glucoside | 0.042 | −0.184 * | −0.095 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piqueras-García, M.; Escobar-Talavera, J.F.; Martínez-Navarro, M.E.; Alonso, G.L.; Sánchez-Gómez, R. Potential Rapid Quantification of Antioxidant Capacity of Olea europaea L. Leaves by Near-Infrared Spectroscopy Using Different Assays. Antioxidants 2025, 14, 1246. https://doi.org/10.3390/antiox14101246
Piqueras-García M, Escobar-Talavera JF, Martínez-Navarro ME, Alonso GL, Sánchez-Gómez R. Potential Rapid Quantification of Antioxidant Capacity of Olea europaea L. Leaves by Near-Infrared Spectroscopy Using Different Assays. Antioxidants. 2025; 14(10):1246. https://doi.org/10.3390/antiox14101246
Chicago/Turabian StylePiqueras-García, Manuel, Jorge F. Escobar-Talavera, María Esther Martínez-Navarro, Gonzalo L. Alonso, and Rosario Sánchez-Gómez. 2025. "Potential Rapid Quantification of Antioxidant Capacity of Olea europaea L. Leaves by Near-Infrared Spectroscopy Using Different Assays" Antioxidants 14, no. 10: 1246. https://doi.org/10.3390/antiox14101246
APA StylePiqueras-García, M., Escobar-Talavera, J. F., Martínez-Navarro, M. E., Alonso, G. L., & Sánchez-Gómez, R. (2025). Potential Rapid Quantification of Antioxidant Capacity of Olea europaea L. Leaves by Near-Infrared Spectroscopy Using Different Assays. Antioxidants, 14(10), 1246. https://doi.org/10.3390/antiox14101246








