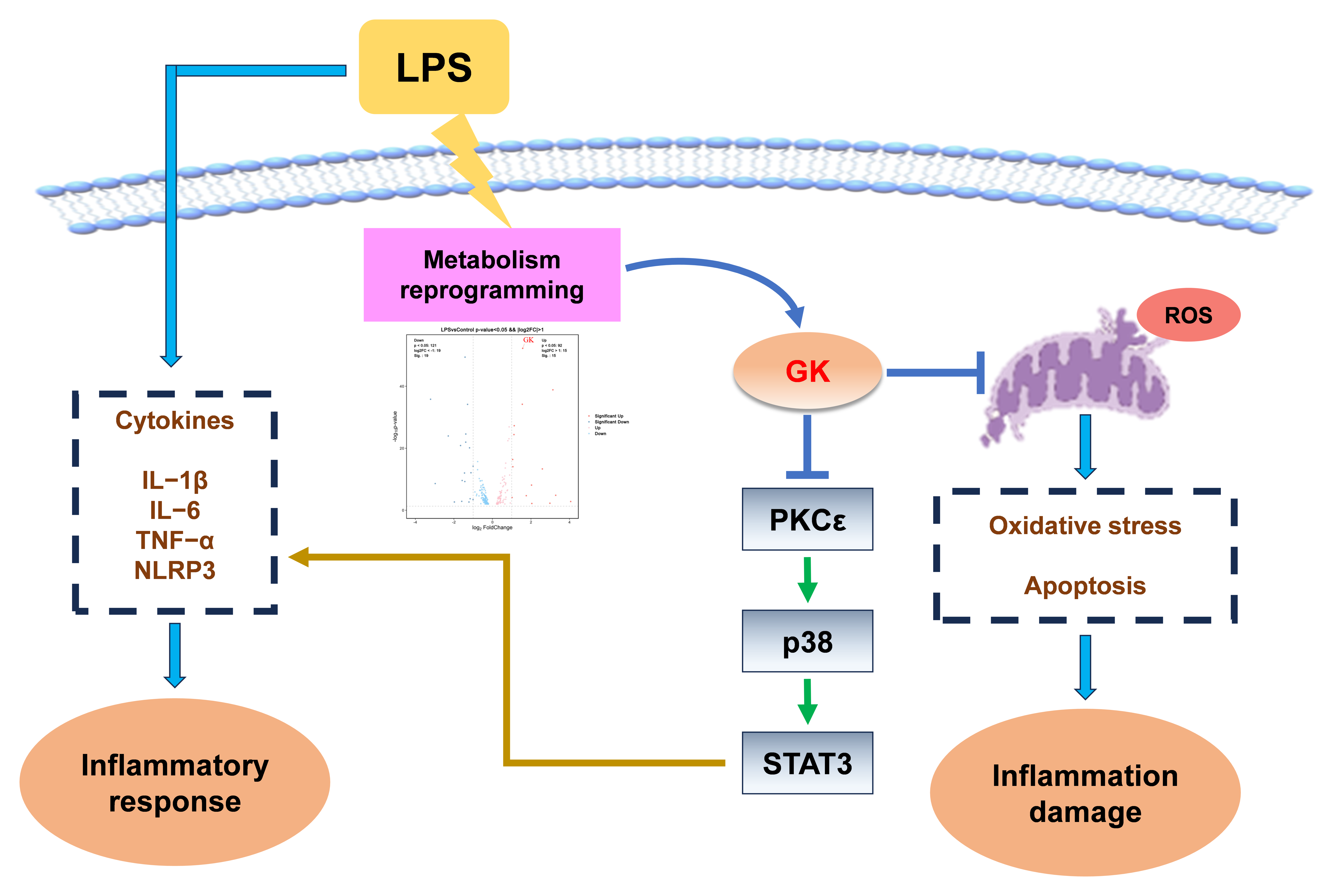
A Novel Function of Glycerol Kinase Alleviates LPS-Induced Inflammatory Responses by the p38/STAT3 Pathway and Mitigates ROS Generation in Kupffer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Transcriptomic Analysis
2.3. Metabolomic Analysis
2.4. Isolation of Primary KCs
2.5. Total Protein Extraction and Western Blot Analysis
2.6. Total RNA Isolation and Quantitative Polymerase Chain Reaction (PCR)
2.7. Fluorescence Microscopy
2.8. Flow Cytometry
2.9. Detection of MDA and GSH Contents
2.10. Statistical Analysis
3. Results
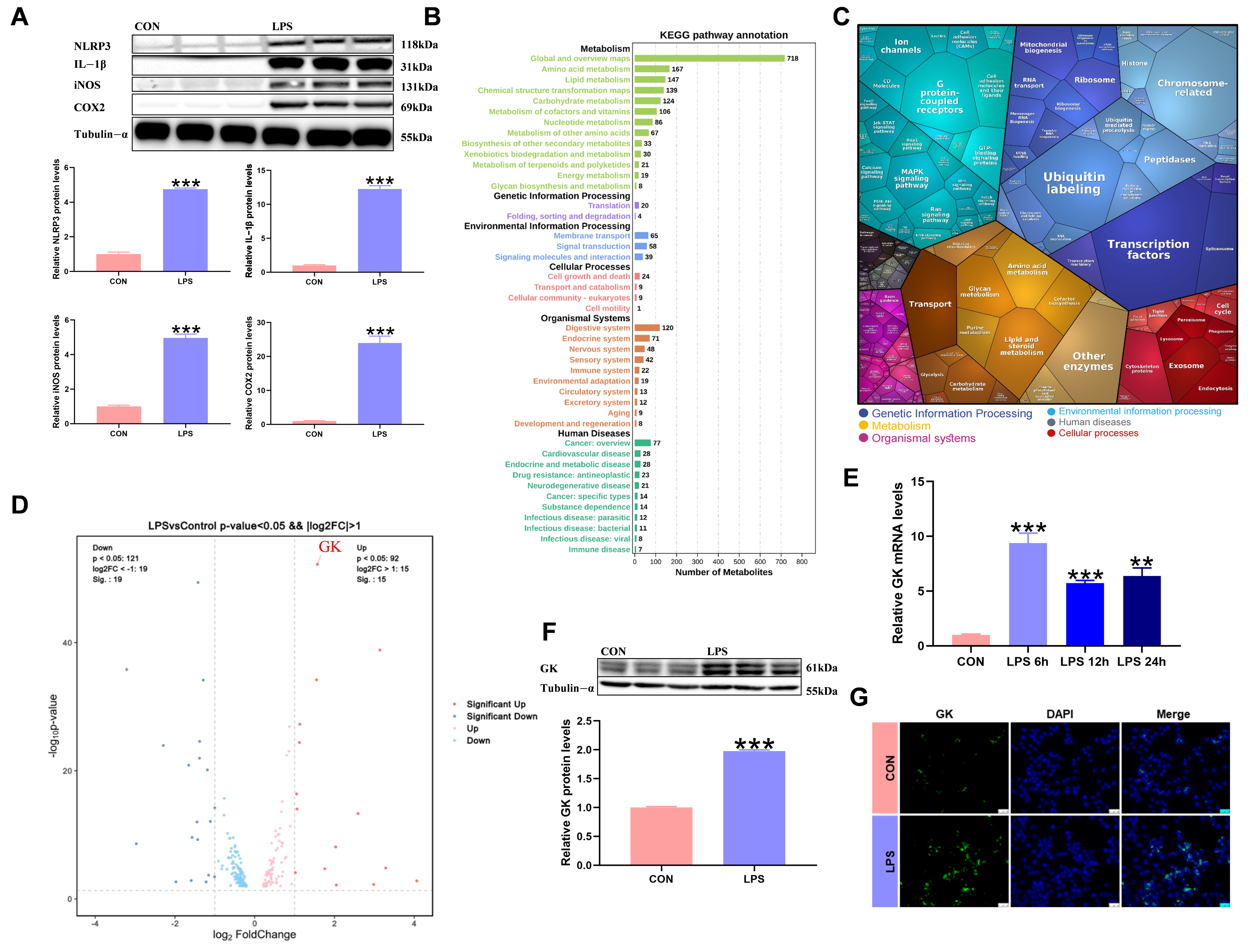
3.1. LPS Triggers Metabolic Reprogramming of KCs and Is Accompanied by Significant Upregulation of GK
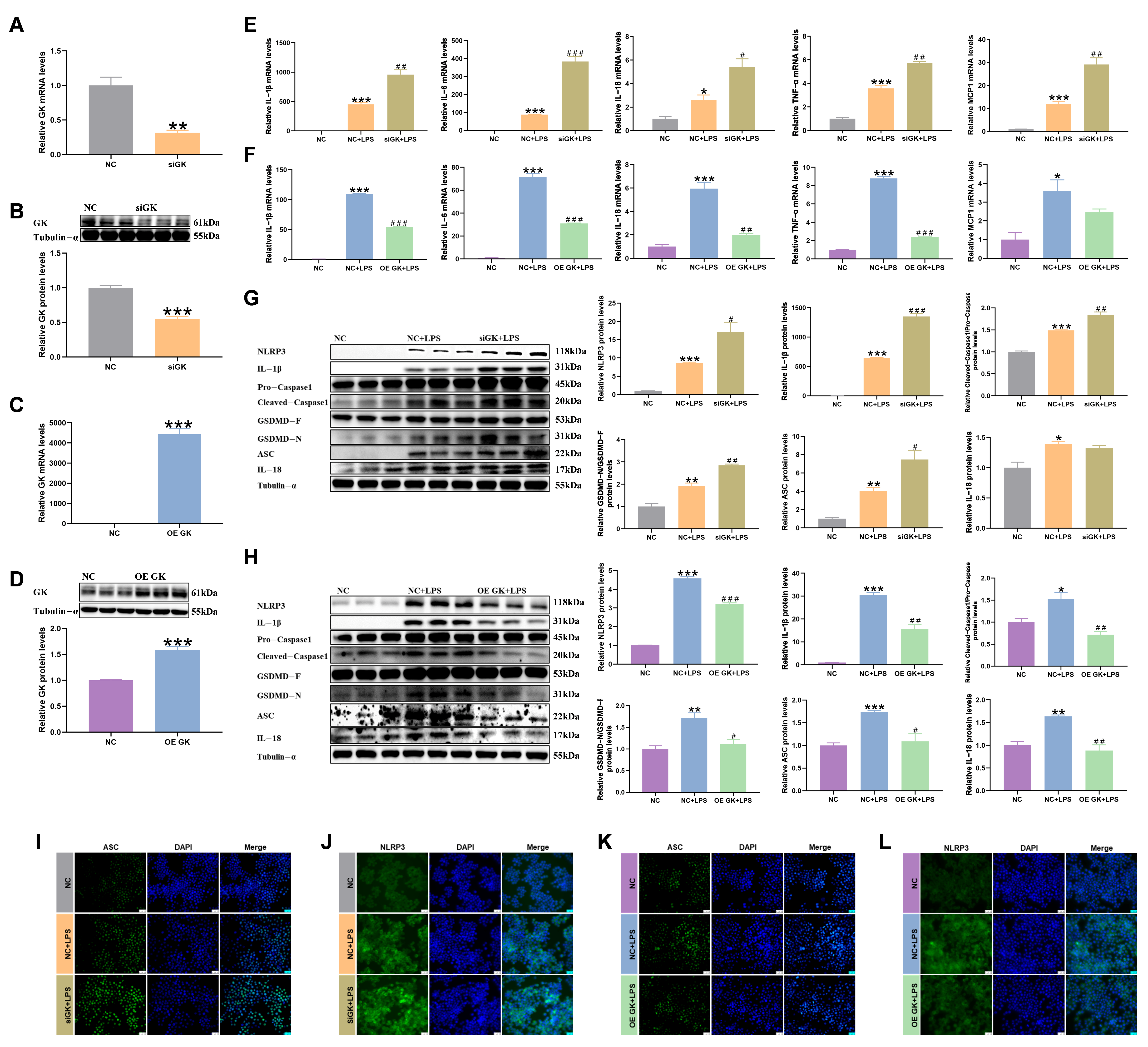
3.2. GK Improved the LPS-Induced Inflammatory Response of KCs
3.3. GK Inhibited iNOS and COX2 Expression in LPS-Stimulated KCs
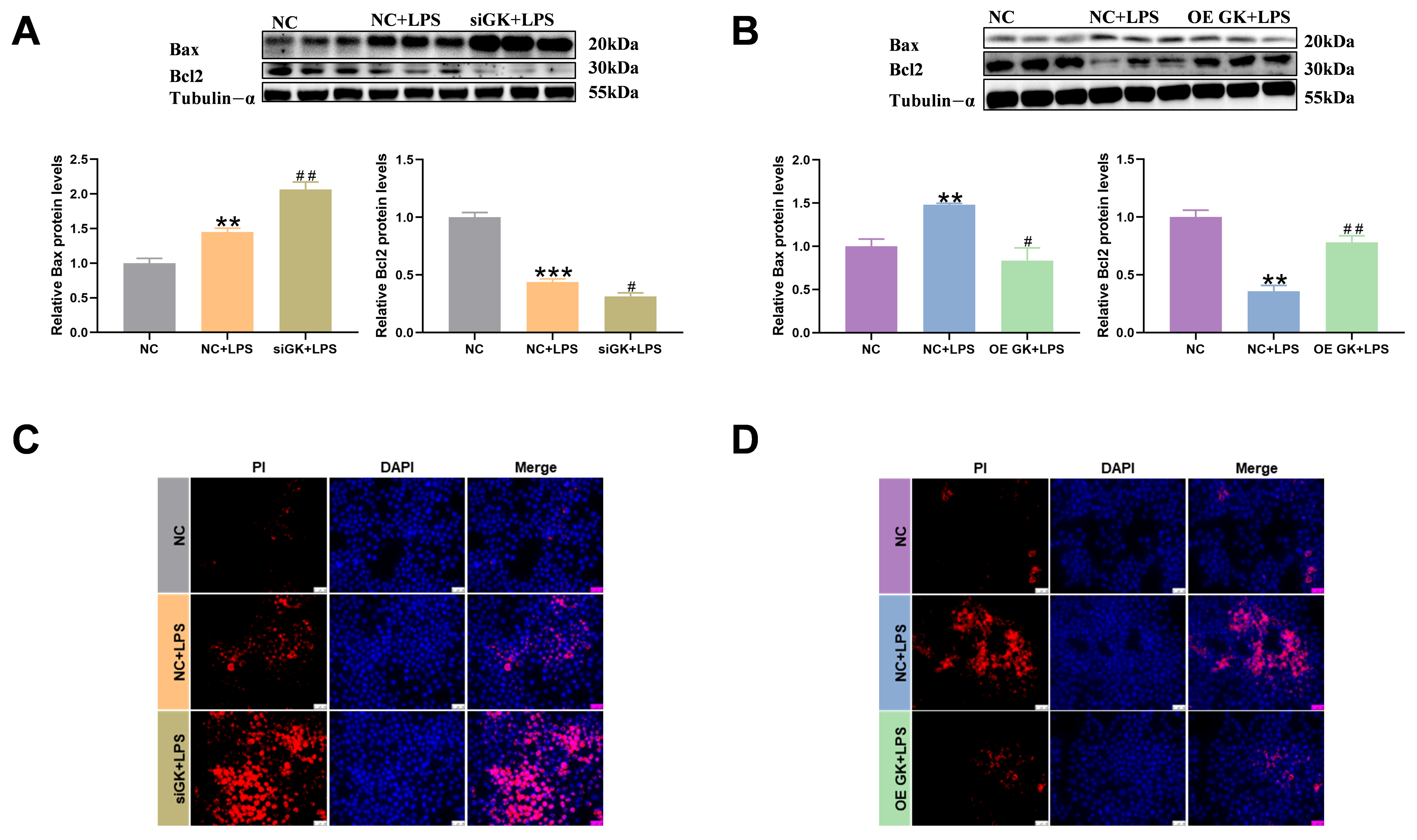
3.4. GK Significantly Decreased Apoptosis Level of KCs
3.5. GK Significantly Alleviates Oxidative Stress of KCs Induced by LPS
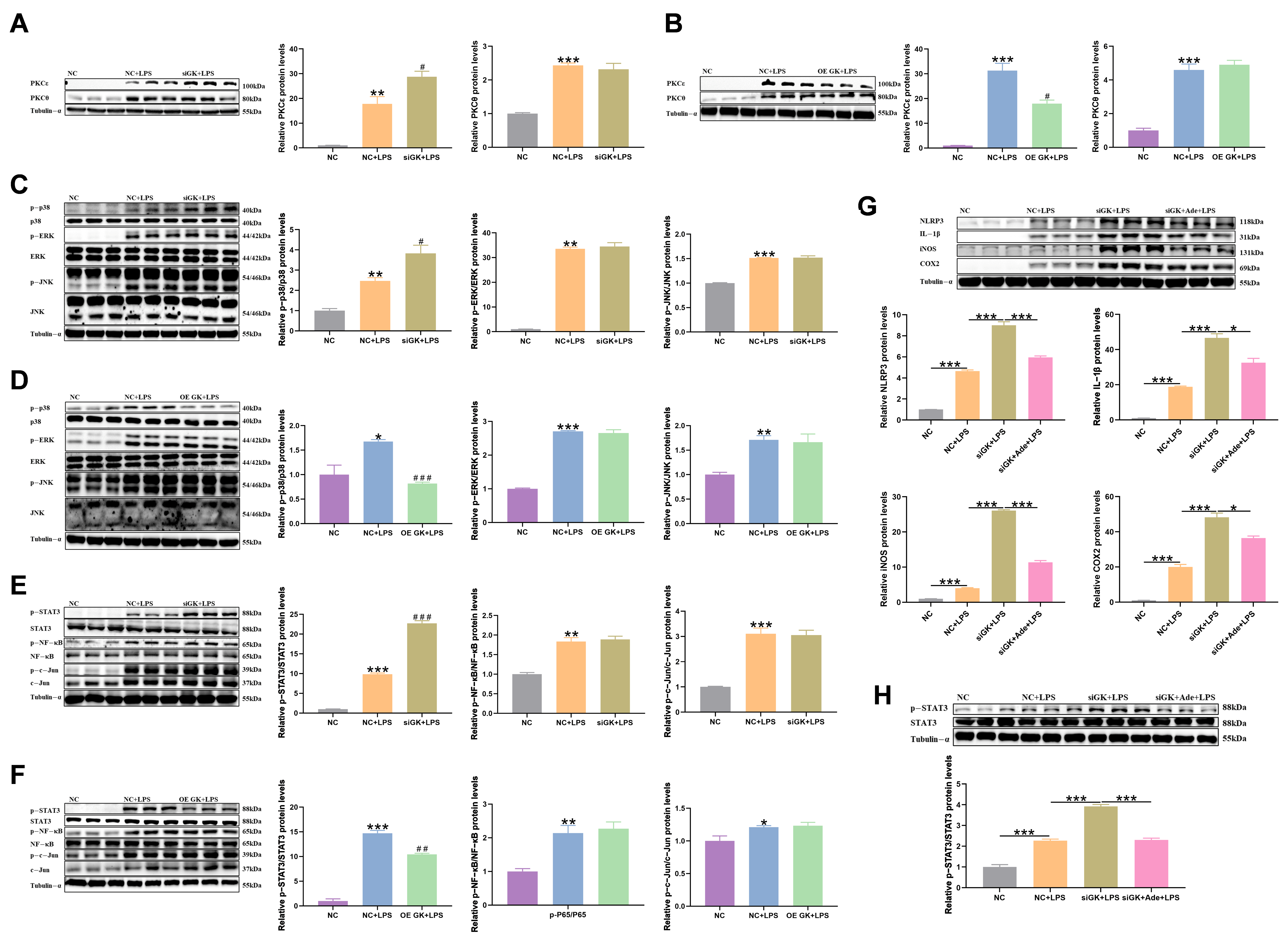
3.6. GK Influences Inflammation in KCs via Inhibiting PKCε/p38/STAT3 Pathway
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Bax | Bcl2-associated x protein |
| Bcl2 | Apoptosis regulator Bcl-2 |
| COX2 | Cyclooxygenase2 |
| c-Jun | Jun activation domain-binding protein |
| ERK | Extracellular-regulated protein kinase |
| GK | Glycerol kinase |
| HO-1 | Heme oxygenase 1 |
| IL | Interleukin |
| iNOS | Inducible nitric oxide synthase |
| JNK | Stress-activated protein kinase 1 |
| LPS | Lipopolysaccharide |
| NLRP3 | NOD-like receptor pyrin domain-containing protein 3 |
| NF-κB | Nuclear factor kappa-B |
| p38 | Cytokine-suppressive anti-inflammatory drug-binding protein |
| PI | Propidium iodide |
| SOD1 | Superoxide dismutase 1 |
| PKC | Protein kinase C |
| ROS | Reactive oxygen species |
| STAT3 | Signal transducer and activator of transcription 3 |
References
- Faas, M.; Ipseiz, N.; Ackermann, J.; Culemann, S.; Gruneboom, A.; Schroder, F.; Rothe, T.; Scholtysek, C.; Eberhardt, M.; Bottcher, M.; et al. IL-33-induced metabolic reprogramming controls the differentiation of alternatively activated macrophages and the resolution of inflammation. Immunity 2021, 54, 2531–2546. [Google Scholar] [CrossRef]
- Ma, J.; Wei, K.; Liu, J.; Tang, K.; Zhang, H.; Zhu, L.; Chen, J.; Li, F.; Xu, P.; Chen, J.; et al. Glycogen metabolism regulates macrophage-mediated acute inflammatory responses. Nat. Commun. 2020, 11, 1769. [Google Scholar] [CrossRef] [PubMed]
- Mocholi, E.; Corrigan, E.; Chalkiadakis, T.; Gulersonmez, C.; Stigter, E.; Vastert, B.; van Loosdregt, J.; Prekovic, S.; Coffer, P.J. Glycolytic reprogramming shapes the histone acetylation profile of activated CD4(+) T cells in juvenile idiopathic arthritis. Cell Rep. 2025, 44, 115287. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Li, Y.; Zong, Y.; Suo, X.; Jia, Y.; Gao, M.; Yang, X. GPAT3 regulates the synthesis of lipid intermediate LPA and exacerbates Kupffer cell inflammation mediated by the ERK signaling pathway. Cell Death Dis. 2023, 14, 208. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, C.; Zou, M.; Wang, D.; Sheng, R.; Zhan, M.; Chen, Q.; Yang, W.; Liu, X.; Xu, S. Inhibiting mitochondrial inflammation through Drp1/HK1/NLRP3 pathway: A mechanism of alpinetin attenuated aging-associated cognitive impairment. Phytother. Res. 2023, 37, 2454–2471. [Google Scholar] [CrossRef]
- Chen, J.; Pan, M.; Wang, J.; Zhang, M.; Feng, M.; Chai, X.; Zhang, Q.; Sun, Y. Hydroxysafflor yellow A protects against colitis in mice by suppressing pyroptosis via inhibiting HK1/NLRP3/GSDMD and modulating gut microbiota. Toxicol. Appl. Pharmacol. 2023, 467, 116494. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, C.; Lv, N.; Liang, Z.; Ma, T.; Cheng, H.; Xia, Y.; Shi, L. AdMSC-derived exosomes alleviate acute lung injury via transferring mitochondrial component to improve homeostasis of alveolar macrophages. Theranostics 2022, 12, 2928–2947. [Google Scholar] [CrossRef]
- Peng, J.; Tang, R.; He, J.; Yu, Q.; Wang, D.; Qi, D. S1PR3 inhibition protects against LPS-induced ARDS by inhibiting NF-kappaB and improving mitochondrial oxidative phosphorylation. J. Transl. Med. 2024, 22, 535. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomark Res. 2021, 9, 1. [Google Scholar] [CrossRef]
- Sun, X.R.; Yao, Z.M.; Chen, L.; Huang, J.; Dong, S.Y. Metabolic reprogramming regulates microglial polarization and its role in cerebral ischemia reperfusion. Fundam. Clin. Pharmacol. 2023, 37, 1065–1078. [Google Scholar] [CrossRef]
- Wu, H.; Zhao, Q.; Dong, L.; Wu, Y.; Zheng, C.; Wu, T.; Ma, D.; Xie, Y.; Wang, Y. Miconazole attenuates LPS-induced lung inflammation by modulating alveolar macrophage polarization via promoting lipid metabolic reprogramming. Inflamm. Res. 2025, 74, 113. [Google Scholar] [CrossRef]
- Wen, Y.; Wang, Y.; Zhuang, Y.; Xu, G.; Zhang, Y.; Zhang, S.; Xu, A.; Chen, X.; Li, K.; Cai, W.; et al. Wuwei Ganlu and Myricetin alleviate rheumatoid arthritis by inhibiting M1 macrophage polarization through modulation of SHBG/SREBP1-mediated lipid metabolism. Phytomedicine 2025, 146, 157167. [Google Scholar] [CrossRef]
- He, Q.; Yin, J.; Zou, B.; Guo, H. WIN55212-2 alleviates acute lung injury by inhibiting macrophage glycolysis through the miR-29b-3p/FOXO3/PFKFB3 axis. Mol. Immunol. 2022, 149, 119–128. [Google Scholar] [CrossRef]
- Rosas-Ballina, M.; Guan, X.L.; Schmidt, A.; Bumann, D. Classical Activation of Macrophages Leads to Lipid Droplet Formation Without de novo Fatty Acid Synthesis. Front. Immunol. 2020, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, Y.; Yang, L.; Chang, N.; Li, L. Monocyte-derived Kupffer cells dominate in the Kupffer cell pool during liver injury. Cell Rep. 2023, 42, 113164. [Google Scholar] [CrossRef] [PubMed]
- Golovko, M.Y.; Hovda, J.T.; Cai, Z.J.; Craigen, W.J.; Murphy, E.J. Tissue-dependent alterations in lipid mass in mice lacking glycerol kinase. Lipids 2005, 40, 287–293. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, N.K.; Rahib, L.; Shin, C.; Fang, Z.; Horvath, S.; Dean, J.; Liao, J.C.; McCabe, E.R.; Dipple, K.M. Targeted disruption of glycerol kinase gene in mice: Expression analysis in liver shows alterations in network partners related to glycerol kinase activity. Hum. Mol. Genet. 2006, 15, 405–415. [Google Scholar] [CrossRef]
- Huq, A.H.; Lovell, R.S.; Ou, C.N.; Beaudet, A.L.; Craigen, W.J. X-linked glycerol kinase deficiency in the mouse leads to growth retardation, altered fat metabolism, autonomous glucocorticoid secretion and neonatal death. Hum. Mol. Genet. 1997, 6, 1803–1809. [Google Scholar] [CrossRef]
- Rathnasiri, A.; Senarathne, U.; Arunath, V.; Hoole, T.; Kumarasiri, I.; Muthukumarana, O.; Jasinge, E.; Mettananda, S. A rare co-occurrence of duchenne muscular dystrophy, congenital adrenal hypoplasia and glycerol kinase deficiency due to Xp21 contiguous gene deletion syndrome: Case report. BMC Endocr. Disord. 2021, 21, 214. [Google Scholar] [CrossRef]
- Wikiera, B.; Jakubiak, A.; Zimowski, J.; Noczynska, A.; Smigiel, R. Complex glycerol kinase deficiency—X-linked contiguous gene syndrome involving congenital adrenal hypoplasia, glycerol kinase deficiency, muscular Duchenne dystrophy and intellectual disability (IL1RAPL gene deletion). Pediatr. Endocrinol. Diabetes Metab. 2012, 18, 153–157. [Google Scholar]
- Assis, A.P.; Silva, K.E.; Lautherbach, N.; Morgan, H.J.N.; Garofalo, M.A.R.; Zanon, N.M.; Navegantes, L.C.C.; Chaves, V.E.; Kettelhut, I.D.C. Glucocorticoids decrease thermogenic capacity and increase triacylglycerol synthesis by glycerokinase activation in the brown adipose tissue of rats. Lipids 2022, 57, 313–325. [Google Scholar] [CrossRef]
- Miao, L.; Yang, Y.; Liu, Y.; Lai, L.; Wang, L.; Zhan, Y.; Yin, R.; Yu, M.; Li, C.; Yang, X.; et al. Glycerol kinase interacts with nuclear receptor NR4A1 and regulates glucose metabolism in the liver. FASEB J. 2019, 33, 6736–6747. [Google Scholar] [CrossRef]
- Miao, L.; Su, F.; Yang, Y.; Liu, Y.; Wang, L.; Zhan, Y.; Yin, R.; Yu, M.; Li, C.; Yang, X.; et al. Glycerol kinase enhances hepatic lipid metabolism by repressing nuclear receptor subfamily 4 group A1 in the nucleus. Biochem. Cell Biol. 2020, 98, 370–377. [Google Scholar] [CrossRef]
- Iwase, M.; Tokiwa, S.; Seno, S.; Mukai, T.; Yeh, Y.S.; Takahashi, H.; Nomura, W.; Jheng, H.F.; Matsumura, S.; Kusudo, T.; et al. Glycerol kinase stimulates uncoupling protein 1 expression by regulating fatty acid metabolism in beige adipocytes. J. Biol. Chem. 2020, 295, 7033–7045. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, S.; Yang, M.; Zhao, Y.; Chen, X.; Zhang, F.; Li, N.; Yao, P.; Zhu, T.; Mei, H.; Wang, S.; et al. MicroRNA-451 Negatively Regulates Hepatic Glucose Production and Glucose Homeostasis by Targeting Glycerol Kinase-Mediated Gluconeogenesis. Diabetes 2016, 65, 3276–3288. [Google Scholar] [CrossRef]
- Zeng, N.; Jiang, H.; Fan, Q.; Wang, T.; Rong, W.; Li, G.; Li, R.; Xu, D.; Guo, T.; Wang, F.; et al. Aberrant expression of miR-451a contributes to 1,2-dichloroethane-induced hepatic glycerol gluconeogenesis disorder by inhibiting glycerol kinase expression in NIH Swiss mice. J. Appl. Toxicol. 2018, 38, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Ostlund, A.K.; Gohring, U.; Krause, J.; Brdiczka, D. The binding of glycerol kinase to the outer membrane of rat liver mitochondria: Its importance in metabolic regulation. Biochem. Med. 1983, 30, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Kurokawa, M.; Ishibashi, S. Binding and function of mitochondrial glycerol kinase in comparison with those of mitochondrial hexokinase. Arch. Biochem. Biophys. 1985, 237, 135–141. [Google Scholar] [CrossRef]
- Chen, J.; Deng, X.; Liu, Y.; Tan, Q.; Huang, G.; Che, Q.; Guo, J.; Su, Z. Kupffer Cells in Non-alcoholic Fatty Liver Disease: Friend or Foe? Int. J. Biol. Sci. 2020, 16, 2367–2378. [Google Scholar] [CrossRef]
- Wiszniewski, M.; Mori, D.; Sanchez Puch, S.I.; Martinez Calejman, C.; Cymeryng, C.B.; Repetto, E.M. Divergent Hepatic and Adipose Tissue Effects of Kupffer Cell Depletion in a Male Rat Model of Metabolic-Associated Steatohepatitis. Biology 2025, 14, 1058. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, R.; Xu, Z.; Ke, Y.; Sun, R.; Yang, H.; Zhang, X.; Zhen, X.; Zheng, L.T. Early glycolytic reprogramming controls microglial inflammatory activation. J. Neuroinflammation 2021, 18, 129. [Google Scholar] [CrossRef]
- Fan, G.; Li, Y.; Chen, J.; Zong, Y.; Yang, X. DHA/AA alleviates LPS-induced Kupffer cells pyroptosis via GPR120 interaction with NLRP3 to inhibit inflammasome complexes assembly. Cell Death Dis. 2021, 12, 73. [Google Scholar] [CrossRef]
- Wang, C.; Yang, T.; Xiao, J.; Xu, C.; Alippe, Y.; Sun, K.; Kanneganti, T.D.; Monahan, J.B.; Abu-Amer, Y.; Lieberman, J.; et al. NLRP3 inflammasome activation triggers gasdermin D-independent inflammation. Sci. Immunol. 2021, 6, eabj3859. [Google Scholar] [CrossRef]
- Shen, W.; Ma, X.; Shao, D.; Wu, X.; Wang, S.; Zheng, J.; Lv, Y.; Ding, X.; Ma, B.; Yan, Z. Neutrophil Extracellular Traps Mediate Bovine Endometrial Epithelial Cell Pyroptosis in Dairy Cows with Endometritis. Int. J. Mol. Sci. 2022, 23, 14013. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Ran, X.; Wang, D.; Zheng, X.; Zhang, M.; Yu, B.; Sun, Y.; Wu, J. Mettl14 mediates the inflammatory response of macrophages in atherosclerosis through the NF-kappaB/IL-6 signaling pathway. Cell Mol. Life Sci. 2022, 79, 311. [Google Scholar] [CrossRef]
- Fang, Y.; Yang, L.; He, J. Plantanone C attenuates LPS-stimulated inflammation by inhibiting NF-kappaB/iNOS/COX-2/MAPKs/Akt pathways in RAW 264.7 macrophages. Biomed. Pharmacother. 2021, 143, 112104. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xu, J.; Zhang, Z.; Shao, Z.; Meng, D. Barrigenol-like triterpenoid saponins from the husks of Xanthoceras sorbifolia bunge and their anti-inflammatory activity by inhibiting COX-2 and iNOS expression. Phytochemistry 2022, 204, 113430. [Google Scholar] [CrossRef]
- Su, H.H.; Yen, J.C.; Liao, J.M.; Wang, Y.H.; Liu, P.H.; MacDonald, I.J.; Tsai, C.F.; Chen, Y.H.; Huang, S.S. In situ slow-release recombinant growth differentiation factor 11 exhibits therapeutic efficacy in ischemic stroke. Biomed. Pharmacother. 2021, 144, 112290. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, D.; Ahmad, R.; Nayeem, S. Molecular interplay promotes amelioration by quercetin during experimental hepatic inflammation in rodents. Int. J. Biol. Macromol. 2022, 222, 2936–2947. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Li, Y.; Liu, Y.; Suo, X.; Jia, Y.; Yang, X. Gondoic acid alleviates LPS-induced Kupffer cells inflammation by inhibiting ROS production and PKCtheta/ERK/STAT3 signaling pathway. Int. Immunopharmacol. 2022, 111, 109171. [Google Scholar] [CrossRef]
- Wang, Y.; Xi, W.; Zhang, X.; Bi, X.; Liu, B.; Zheng, X.; Chi, X. CTSB promotes sepsis-induced acute kidney injury through activating mitochondrial apoptosis pathway. Front. Immunol. 2022, 13, 1053754. [Google Scholar] [CrossRef]
- Li, Y.; Sun, G.; Wang, L. MiR-21 participates in LPS-induced myocardial injury by targeting Bcl-2 and CDK6. Inflamm. Res. 2022, 71, 205–214. [Google Scholar] [CrossRef]
- Fu, Y.P.; Yuan, H.; Xu, Y.; Liu, R.M.; Luo, Y.; Xiao, J.H. Protective effects of Ligularia fischeri root extracts against ulcerative colitis in mice through activation of Bcl-2/Bax signalings. Phytomedicine 2022, 99, 154006. [Google Scholar] [CrossRef]
- Su, Y.; Yin, X.; Huang, X.; Guo, Q.; Ma, M.; Guo, L. Astragaloside IV ameliorates sepsis-induced myocardial dysfunction by regulating NOX4/JNK/BAX pathway. Life Sci. 2022, 310, 121123. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, N.K.; Dong, J.; Aten, J.E.; Horvath, S.; Rahib, L.; Ornelas, L.; Dipple, K.M.; McCabe, E.R. Weighted gene co-expression network analysis identifies biomarkers in glycerol kinase deficient mice. Mol. Genet. Metab. 2009, 98, 203–214. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.; Mu, W.; Barry, J.; Han, A.; Carpenter, R.L.; Jiang, B.H.; Peiper, S.C.; Mahoney, M.G.; Aplin, A.E.; et al. Reactive oxygen species reprogram macrophages to suppress antitumor immune response through the exosomal miR-155-5p/PD-L1 pathway. J. Exp. Clin. Cancer Res. 2022, 41, 41. [Google Scholar] [CrossRef]
- Jeon, H.; Huynh, D.T.N.; Baek, N.; Nguyen, T.L.L.; Heo, K.S. Ginsenoside-Rg2 affects cell growth via regulating ROS-mediated AMPK activation and cell cycle in MCF-7 cells. Phytomedicine 2021, 85, 153549. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, L.; Wang, J.; Chen, H.; Lin, Y.; Wang, F.; Wang, L.; Chen, J.; Liu, J.; Zhang, X. Luteolin, as a bidirectional ROS regulator, elevates mouse beige adipocyte browning. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2025, 1870, 159620. [Google Scholar] [CrossRef]
- Luo, X.; Bao, X.; Weng, X.; Bai, X.; Feng, Y.; Huang, J.; Liu, S.; Jia, H.; Yu, B. The protective effect of quercetin on macrophage pyroptosis via TLR2/Myd88/NF-kappaB and ROS/AMPK pathway. Life Sci. 2022, 291, 120064. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, Y.; Yi, J.; Zhao, Z.; Ye, R. Hyperglycemia modulates M1/M2 macrophage polarization via reactive oxygen species overproduction in ligature-induced periodontitis. J. Periodontal Res. 2021, 56, 991–1005. [Google Scholar] [CrossRef]
- Cheng, Y.; Gu, W.; Wu, X.; Tian, W.; Mu, Z.; Ye, Y.; Chao, H.; Bao, Z. Allicin alleviates traumatic brain injury-induced neuroinflammation by enhancing PKC-delta-mediated mitophagy. Phytomedicine 2025, 139, 156500. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, L.; Peng, J.; Zhang, J.H.; Zhang, H. LJ529 attenuates mast cell-related inflammation via A(3)R-PKCepsilon-ALDH2 pathway after subarachnoid hemorrhage in rats. Exp. Neurol. 2021, 340, 113686. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Li, X.; Xue, C.; Zhang, L.; Wang, C.; Xu, X.; Shan, A. Astragalus polysaccharides alleviates LPS-induced inflammation via the NF-kappaB/MAPK signaling pathway. J. Cell Physiol. 2020, 235, 5525–5540. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Zhou, Y.; Zhu, C.; Ren, T.; Zhang, Y.; Xiao, L.; Fang, B. Ginsenoside Rg1 Mitigates Porcine Intestinal Tight Junction Disruptions Induced by LPS through the p38 MAPK/NLRP3 Inflammasome Pathway. Toxics 2022, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Chen, W.; Huang, L.; Wang, H.; Su, Y.; Liang, J.; Lian, H.; Xu, J.; Zhao, J.; Liu, Q. Alpinetin ameliorates bone loss in LPS-induced inflammation osteolysis via ROS mediated P38/PI3K signaling pathway. Pharmacol. Res. 2022, 184, 106400. [Google Scholar] [CrossRef]
- Chen, L.; Wang, H.; Xing, J.; Shi, X.; Huang, H.; Huang, J.; Xu, C. Silencing P2X7R Alleviates Diabetic Neuropathic Pain Involving TRPV1 via PKCepsilon/P38MAPK/NF-kappaB Signaling Pathway in Rats. Int. J. Mol. Sci. 2022, 23, 14141. [Google Scholar] [CrossRef]
- Liao, J.; Zhao, W.; Zhang, Y.; Zou, Z.; Zhang, Q.; Chen, D.; Du, B.; Li, P. Dendrobium officinale Kimura et Migo polysaccharide ameliorated DNFB-induced atopic dermatitis in mice associated with suppressing MAPK/NF-kappaB/STAT3 signaling pathways. J. Ethnopharmacol. 2024, 335, 118677. [Google Scholar] [CrossRef]
- Zhong, T.; Feng, M.; Su, M.; Wang, D.; Li, Q.; Jia, S.; Luo, F.; Wang, H.; Hu, E.; Yang, X.; et al. Qihuzha granule attenuated LPS-induced acute spleen injury in mice via Src/MAPK/Stat3 signal pathway. J. Ethnopharmacol. 2021, 281, 114458. [Google Scholar] [CrossRef]
- Xue, C.; Liu, S.X.; Hu, J.; Huang, J.; Liu, H.M.; Qiu, Z.X.; Huang, F. Corydalis saxicola Bunting total alkaloids attenuate paclitaxel-induced peripheral neuropathy through PKCepsilon/p38 MAPK/TRPV1 signaling pathway. Chin. Med. 2021, 16, 58. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, K.H.; Lee, S.M.; Park, S.J.; Lee, S.; Cha, R.H.; Lee, J.W.; Kim, D.K.; Kim, Y.S.; Ye, S.K.; et al. STAT3 blockade ameliorates LPS-induced kidney injury through macrophage-driven inflammation. Cell Commun. Signal. 2024, 22, 476. [Google Scholar] [CrossRef]
- Hwang, S.; Park, J.; Koo, S.Y.; Lee, S.Y.; Jo, Y.; Ryu, D.; Go, H.; Lee, C.W. The ubiquitin ligase Pellino1 targets STAT3 to regulate macrophage-mediated inflammation and tumor development. Nat. Commun. 2025, 16, 1256. [Google Scholar] [CrossRef] [PubMed]


| Antibodies | Company | Item No. |
|---|---|---|
| Tubulin-α | Bioworld (Nanjing, China) | BS1699 |
| GK | Abcam (Cambridgeshire, UK) | ab228615 |
| IL-1β | Abcam (Cambridgeshire, UK) | ab254360 |
| NLRP3 | Abcam (Cambridgeshire, UK) | ab283819 |
| Caspase1 | AdipoGen Life Sciences (San Diego, USA) | AG-20B-0042 |
| GSDMD | Abcam (Cambridgeshire, UK) | ab219800 |
| ASC | Cell Signalling Technology (Boston, USA) | #67824 |
| PKCθ | Bioworld (Nanjing, China) | BS3248 |
| PKCε | Bioworld (Nanjing, China) | BS6704 |
| p-p38 | Proteintech (Wuhan; China) | 28796-1-AP |
| p38 | Bioworld (Nanjing, China) | MB66552 |
| p-ERK | Cell Signalling Technology (Boston, USA) | 4370 |
| ERK | Cell Signalling Technology (Boston, USA) | 4695 |
| p-JNK | Cell Signalling Technology (Boston, USA) | 4668 |
| JNK | Cell Signalling Technology (Boston, USA) | 9252 |
| p-NF-κB | Affinity (Ohio, USA) | AF2006 |
| NF-κB | Proteintech (Wuhan; China) | 10745-1-AP |
| p-c-Jun | Santa (Texas, USA) | sc-822 |
| c-Jun | Affinity (Ohio, USA) | AF6090 |
| p-STAT3 | Affinity (Ohio, USA) | AF3294 |
| STAT3 | Proteintech (Wuhan; China) | 10253-2-AP |
| Bax | Bioworld (Nanjing, China) | BS6420 |
| Bcl2 | Bioworld (Nanjing, China) | CAS7511 |
| iNOS | Affinity (Ohio, USA) | AF0199 |
| COX2 | Abcam (Cambridgeshire, UK) | ab15191 |
| HO-1 | Proteintech (Wuhan; China) | 10701-1-AP |
| SOD1 | Bioworld (Nanjing, China) | BS6057 |
| Gene | Gene ID | Forward (5′ to 3′) | Reverse (3′ to 5′) |
|---|---|---|---|
| IL-6 | 16193 | CCAAGAGGTGAGTGCTTCCC | CTGTTGTTCAGACTCTCTCCCT |
| IL-1β | 16176 | GCAACTGTTCCTGAACTCAACT | ATCTTTTGGGGTCCGTCAACT |
| IL-18 | 16173 | GACTCTTGCGTCAACTTCAAGG | CAGGCTGTCTTTTGTCAACGA |
| TNF-α | 21926 | GACGTGGAACTGGCAGAAGAG | TTGGTGGTTTGTGAGTGTGAG |
| iNOS | 18126 | GTTCTCAGCCCAACAATACAAGA | GTGGACGGGTCGATGTCAC |
| COX2 | 19225 | TTCAACACACTCTATCACTGGC | AGAAGCGTTTGCGGTACTCAT |
| GK | 14933 | TGAACCTGAGGATTTGTCAGC | CCATGTGGAGTAACGGATTTCG |
| PPIA | 268373 | GGGTTCCTCCTTTCACAGA | CCATCCAGCCATTCAGTC |
| NQO1 | 18104 | AGGATGGGAGGTACTCGAATC | AGGCGTCCTTCCTTATATGCTA |
| SOD1 | 20655 | AACCAGTTGTGTTGTCAGGAC | CCACCATGTTTCTTAGAGTGAGG |
| HO-1 | 15368 | AAGCCGAGAATGCTGAGTTCA | GCCGTGTAGATATGGTACAAGGA |
| GPX1 | 14775 | AGTCCACCGTGTATGCCTTCT | GAGACGCGACATTCTCAATGA |
| NRF2 | 18024 | TCTTGGAGTAAGTCGAGAAGTGT | GTTGAAACTGAGCGAAAAAGGC |
| GST1 | 56615 | ATGCCACCATACACCATTGTC | GGGAGCTGCCCATACAGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, X.; Wang, D.; Fan, G.; Yang, X. A Novel Function of Glycerol Kinase Alleviates LPS-Induced Inflammatory Responses by the p38/STAT3 Pathway and Mitigates ROS Generation in Kupffer Cells. Antioxidants 2025, 14, 1174. https://doi.org/10.3390/antiox14101174
Li Y, Zhang X, Wang D, Fan G, Yang X. A Novel Function of Glycerol Kinase Alleviates LPS-Induced Inflammatory Responses by the p38/STAT3 Pathway and Mitigates ROS Generation in Kupffer Cells. Antioxidants. 2025; 14(10):1174. https://doi.org/10.3390/antiox14101174
Chicago/Turabian StyleLi, Yanfei, Xu Zhang, Danping Wang, Guoqiang Fan, and Xiaojing Yang. 2025. "A Novel Function of Glycerol Kinase Alleviates LPS-Induced Inflammatory Responses by the p38/STAT3 Pathway and Mitigates ROS Generation in Kupffer Cells" Antioxidants 14, no. 10: 1174. https://doi.org/10.3390/antiox14101174
APA StyleLi, Y., Zhang, X., Wang, D., Fan, G., & Yang, X. (2025). A Novel Function of Glycerol Kinase Alleviates LPS-Induced Inflammatory Responses by the p38/STAT3 Pathway and Mitigates ROS Generation in Kupffer Cells. Antioxidants, 14(10), 1174. https://doi.org/10.3390/antiox14101174






