Abstract
Liver segmentation is a crucial step in surgical planning from computed tomography scans. The possibility to obtain a precise delineation of the liver boundaries with the exploitation of automatic techniques can help the radiologists, reducing the annotation time and providing more objective and repeatable results. Subsequent phases typically involve liver vessels’ segmentation and liver segments’ classification. It is especially important to recognize different segments, since each has its own vascularization, and so, hepatic segmentectomies can be performed during surgery, avoiding the unnecessary removal of healthy liver parenchyma. In this work, we focused on the liver segments’ classification task. We exploited a 2.5D Convolutional Neural Network (CNN), namely V-Net, trained with the multi-class focal Dice loss. The idea of focal loss was originally thought as the cross-entropy loss function, aiming at focusing on “hard” samples, avoiding the gradient being overwhelmed by a large number of falsenegatives. In this paper, we introduce two novel focal Dice formulations, one based on the concept of individual voxel’s probability and another related to the Dice formulation for sets. By applying multi-class focal Dice loss to the aforementioned task, we were able to obtain respectable results, with an average Dice coefficient among classes of 82.91%. Moreover, the knowledge of anatomic segments’ configurations allowed the application of a set of rules during the post-processing phase, slightly improving the final segmentation results, obtaining an average Dice coefficient of 83.38%. The average accuracy was close to 99%. The best model turned out to be the one with the focal Dice formulation based on sets. We conducted the Wilcoxon signed-rank test to check if these results were statistically significant, confirming their relevance.
1. Introduction
Computed Tomography (CT) is one of the most adopted imaging methodologies to accomplish different medical tasks. If combined with intelligent systems based on artificial intelligence, its range of applicability includes, but is not limited to, Computer-Aided Diagnosis (CAD) systems and intelligent applications supporting surgical procedures [1,2].
Liver segmentation is a pivotal task in abdominal radiology, as it is essential for several clinical tasks, including surgery, radiotherapy, and objective quantification [3]. In addition, several pieces of clinical evidence over the last few decades have demonstrated the necessity to go beyond the conception of the liver as a single object (from a morphological perspective), but rather to consider the liver as formed by functional segments [4]. Couinaud’s classification [5] of liver segments is the most widely adopted system for this purpose, since it is especially useful in surgery setups. The separation of these anatomical areas can be exploited for tumor resections in Computer-Assisted Surgery (CAS) systems [6], allowing surgeons to focus on different segments. In this way, a surgeon can avoid removing healthy liver parenchyma, by performing a segmentectomy, also reducing the risk of complications for the patient [7].
However, different other tissues and organs, such as the kidney and spleen, are in the abdominal area. Since the tissues in such an area are characterized by similar intensity levels when acquired by CT or other imaging systems in general, their segmentation with automatic methods is not a simple task. In fact, the segmentation of organs in the abdominal area is mostly tackled by employing manual or semi-automatic approaches [8,9]. To do this, there is a multitude of traditional image processing algorithms for biomedical image segmentation, including thresholding algorithms, Region-Growing (RG) algorithms, level sets, and graph cuts.
Deep Learning (DL) is an emerging paradigm that is changing the world of medical imaging. DL consists of the application of hierarchical computational models that can directly learn from data, creating representations with multiple levels of abstraction [10]. Thanks to the natural proneness of DL architectures to process grid-like data, such as images or multi-dimensional signals, they are extensively investigated in image classification and segmentation tasks; in fact, several works have already demonstrated their capabilities to outperform the state-of-the-art in such tasks, making them valid systems to be applied also in the medical domain [1,11,12,13,14].
DL architectures may be employed for both classification and segmentation tasks, i.e., the delineation of areas or Regions of Interest (ROIs) within images or volumes. In this regard, it is necessary to distinguish between semantic segmentation [15,16] and object detection [17,18] approaches implemented using DL methodologies. The first aims at producing a dense, pixel- (or voxel-) level segmentation mask, whereas the latter aims at individuating the bounding boxes of the regions of interest [19,20]. In many contexts, precise segmentation masks are more useful than bounding boxes, especially in the medical domain, but they require more human effort for labeling the ground-truth dataset required to train and validate the models (since a domain expert must manually produce precise voxel-level masks to create the dataset themselves).
In this work, we performed the segmentation of the liver segments. This task may be performed directly or by performing a prior segmentation of the liver parenchyma. Although literature exists for both approaches, our experiments revealed that liver parenchyma segmentation is not needed to train a multi-class classification DL model on liver segments, since applying liver masks to the volumes reduces the context. Nonetheless, other authors found this step useful [21].
1.1. Deep Learning in Radiology
Radiology involves the exploitation of 3D images that result from non-invasive techniques such as Magnetic Resonance Imaging (MRI) or CT, allowing studying the anatomical structures inside the body. When there is the need to delineate the boundary of organs or lesions, e.g., in CAS or radiomics [22] workflows, the manual delineation of boundaries performed by trained radiologists is considered the gold standard. Unfortunately, this operation is tedious and can lead to errors, since it involves the annotation of a volume composed of many slices. Lastly, inter- and intra-observer variability are well-known problems in the medical imaging landscape, further showing the limitations of the classical workflow [23].
In this context, the design and implementation of automatic systems, often based on DL architectures, capable of performing organ and lesion segmentation are very appreciated, since they can reduce the workload of radiologists and also increase the robustness of the findings [24]. The application of artificial intelligence methodologies in the radiological workflow is leading to a huge impact in the field, with a special regard for oncological applications [25].
Important radiological tasks that can be efficiently met by DL methodologies include classification, the detection of diseases or lesions, the quantification of radiographic characteristics, and image segmentation [11,14,26,27].
In this work, the possibility to automatically delineate the boundaries of liver segments can be effectively exploited by radiologists and surgeons to improve the surgery workflow with CASs for liver tumor resections.
The interested reader is referred to the works of Liu et al. [28] and Litjens et al. [29] for a wider perspective on the architectures that can be exploited to solve clinical problems in radiology with deep learning.
1.2. Liver Parenchyma Segmentation
Prencipe et al. developed an RG algorithm for liver and spleen segmentation [30]. The crucial point of segmentation algorithms based on RG is the individuation of a suitable criterion for growing the segmentation mask including only those pixels belonging to the area of interest. In particular, the authors devised an algorithm that, starting from an initial seed point, created a tridimensional segmentation mask adopting two utility data structures, namely the Moving Average Seed Heat map (MASH) and the Area Union Map (AUM), with the positive effect of avoiding the choice of the subsequent seeds from unsuitable locations while propagating the mask.
There is a realm of other techniques proposed for liver segmentation. Contrast enhancement and cropping were used for pre-processing by Bevilacqua et al. [11]. Then, local thresholding, the extraction of the largest connected component, and the adoption of operators from a mathematical morphology were applied to obtain the 2D segmentation. Finally, the obtained mask was broadcast upward and downward to process the entire volume.
In [31], an automatic 3D segmentation approach was presented, where the seed was chosen via the minimization of an objective function and the homogeneity criteria based on the Euclidean distance of the texture features related to the voxels. The homogeneity criteria can also depend on the difference between the currently segmented area and the pixel intensity [32,33,34] or pixel gradient [35,36]. There exist also other approaches based on the adoption of a suitable homogeneity criteria [37,38].
As useful pre-processing techniques, it is worth noting the adoption of the Non-Sub-sampled Contourlet Transform (NSCT) to enhance the liver’s edges [39] and the implementation of a contrast stretch algorithm and an atlas intensity distribution to create voxel probability maps [40]. For the post-processing step, we note the adoption of an entropy filter for finding the best structural element [41] or the adoption of the GrowCut algorithm [42,43]. Other approaches that involve image processing techniques both for pre- and post-processing can be found in [12,44].
1.3. Liver Segments’ Classification
There are some approaches proposed in the literature for liver segments’ classification. Most of the published research tries to obtain the segments from an accurate modeling of the liver vessels.
Oliveira et al. employed a geometric fit algorithm, based on the least-squares method, to generate planes for separating the liver into its segments [45].
Yang et al. exploited a semi-automatic approach for calculating the segments [6]. The user has to input the root points for the branches of the portal vein and the hepatic veins, and then, a Nearest Neighbor Approximation (NNA) algorithm is implemented to assign each liver voxel to a specific segment. The classification was very straightforward: the voxel belonged to the segment whose branch was nearest to that voxel.
Among the approaches that aim to directly classify liver segments, it is worth noting the work of Tian et al. [21], who offered the first publicly available dataset for liver segments’ classification. They realized an architecture based on U-Net [46], but with considerations for the Global and Local Context (GLC), resulting in a model that they referred to as GLC U-Net.
Yan et al. collected a huge CT dataset, composed of 500 cases, to realize CAD systems targeted at the liver region [47]. The ComLiver datasets include labels for liver parenchyma, vessels, and Couinaud segments. For the task of performing Couinaud segmentation, the authors compared only 3D CNN models, namely Fully Convolutional Networks (FCNs) [48], U-Net [46,49], U-Net++ [50], nnU-Net [51], Attention U-Net (AU-Net) [52], and Parallel Reverse Attention Network (PraNet) [53].
In this work, we introduce two possible definitions of the Focal Dice Loss (), and we compare them with a definition already provided by Wang et al. [54]. One of the proposed definitions is similar to that of Wang et al., whereas the other one is more adherent to the original formulation provided by Lin et al. [55], considering the modulating factor on the single voxel and not on the whole Dice loss. In particular, we trained a 2.5D CNN model, the V-Net architecture, originally proposed for 3D segmentation by Milletari et al. [56], but in a multi-class classification fashion. We also implemented and tested a rule-based post-processing to correct the segmentation errors.
2. Materials and Methods
2.1. Dataset
To train and validate our models, we used the Medical Segmentation Decathlon Task 08 (MSD 08 dataset [57]). This dataset is composed of 443 portal venous phase CT scans. The pixel spacing in the direction is in the range , whereas the thickness lies between . We considered liver segments’ annotations (n = 193) realized in a studio by Lenovo [21]. We exploited 168 CT scans as the training set and 25 CT scans as the validation set. For the selected training set, the frequencies of class values for each segment are represented in Table 1. Some examples from the MSD 08 dataset with the annotations of the Lenovo team are depicted in Figure 1. As pre-processing, images were clipped to the Hounsfield Unit (HU) range and then rescaled in the range , as already performed in [14].

Table 1.
Class distribution for the selected training set (n = 168) from MSD 08 dataset.
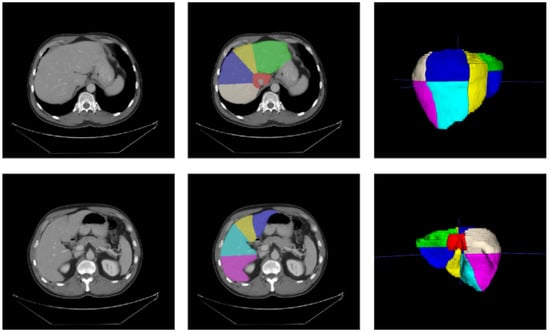
Figure 1.
Medical Segmentation Decathlon Task 08 [57], with annotations from Tian et al. [21]. Left: original slices; middle: slices with segments’ annotations; right: meshes’ reconstructions of annotations.
2.2. Convolutional Neural Network
Encoder–decoder architectures (especially those that are U-shaped) have drawn much attention in the biomedical semantic segmentation domain [28]. Important examples are the U-Net model for 2D image segmentation [46], its 3D counterpart [49], and the V-Net model [56]. Compared to U-Net, V-Net employs down-convolutions instead of max pooling and residual connections inside the encoding or decoding path. Applications of 3D V-Net include prostate segmentation [56] and vertebrae segmentation [58]. A 2.5D variant of V-Net has been successfully applied for liver and vessels’ segmentation [14,59] and lung parenchyma, together with COVID-19 lesions’ segmentation [22].
In this work, we implemented the 2.5D architecture, but in a multi-class classification fashion, with a different loss function (focal Dice loss). The scheme of the employed architecture is depicted in Figure 2.
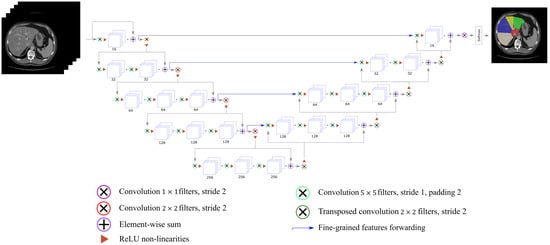
Figure 2.
V-Net for multi-class segments’ classification.
For the liver segments’ classification task, we randomly sampled 2.5D, i.e., 2D multi-channel, patches composed of 5 slices from volumes of the training set, as performed in Altini et al. [14]. We trained the V-Net model for 1000 epochs, for each of the proposed loss function configurations reported in Section 3. The learning rate started from 0.01 and was reduced by a factor of 10 every 166 epochs. The Adam optimizer [60], with a batch size of 4, was used to carry out the training process.
2.2.1. Focal Loss
In order to ease the convergence process and to perform the desired optimization of the task, the loss function has to be selected accurately. Lin et al. introduced the concept of focal loss [55]. The main idea is that, for detectors (but we can extend this consideration to semantic segmentation classifiers), a mechanism is needed to focus on hard samples (i.e., samples difficult to classify), avoiding that the gradient is overwhelmed by the larger number of falsenegatives. According to the convention introduced in [55], we can define the Cross-Entropy (CE) loss as:
where is the binary ground-truth label and is the probability estimated from the model when . Lin et al. introduced for notational convenience:
so that we can rewrite . A weighted version of the cross-entropy can be introduced. For instance, let be a weighting factor in the range , so that is adopted for class and is exploited for class . Typically, may be chosen by the inverse class frequency [55]. Introducing , analogously to , it is possible to define the -balanced CE loss as:
In order to redefine the cross-entropy loss, Lin et al. added a modulating factor to the CE loss, where can be found with cross-validation, and it is called the focusing parameter.
Then, the CE Focal Loss (FL) is:
The authors noted that this formulation has two main properties:
- The loss is unaffected when there is a misclassification with a small ; meanwhile, if is large (near 1), the loss for well-classified examples is reduced. This means that easy examples are not penalized, if compared to “hard” examples;
- The rate with which easy examples are down-weighted is governed by the focusing parameter . For , the FL corresponds to the standard CE, whereas for larger values of , there is an increase in the modulating effect. Lin et al. found as the best value in their experiments [55].
Furthermore, can be weighted, so that the -balanced FL can be defined as:
According to [55], this yields a slight improvement in accuracy, over the naive non--balanced form.
2.2.2. Focal Dice Loss
After the introduction of the FL, Wang et al. proposed the Focal Dice Loss () and effectively employed it for brain tumor segmentation [54].
The Dice coefficient () is usually defined for sets (let B be the binarized prediction and G the ground-truth volume) as:
We define this as the “hard” Dice coefficient , in the sense that it is defined for binary data only. Let us consider the estimated probability predicted volume P, composed of N voxels. Each element ranges in , whereas each belongs to . Then, the “soft” Dice coefficient can be defined as:
where is a term for numerical stability, avoiding divisions by zero or by very small values. In our experiments, we set . The “hard” Dice coefficient is useful for assessing the final results, whereas the “soft” formulation is needed during the optimization process, where a differentiable function is compulsory.
Then, we can introduce a “soft” Dice coefficient loss simply as:
These definitions hold for the binary classification case, but they can be easily generalized for classes. For each value of t, we can compute the binary , so that the weighted multi-Dice loss () can be written as:
where is the weighting parameter for the class t and is the binary soft Dice loss for class t. A common way to tune the parameters is to exploit the logarithm of the inverse class frequency, as reported in Table 1. Nonetheless, in our experiments, this did not result in improvements, so we chose the version with all , with . Starting from this definition, Wang et al. defined their FDL as:
where is the “soft” Dice coefficient for class t, and a factor , with , is applied as the exponent of for each class. According to Wang et al., their formulation has three properties:
- is basically unaffected by pixels, which are misclassified to class t with a large . Conversely, is significantly decreased when is small;
- The rate with which better-segmented classed are weighted lower is governed by the parameter . For , we have that corresponds to . Increasing , the network is forced to focus on classes with bad segmentation;
- Since is based on the definition of the Dice coefficient, it focuses on the relevant metric for assessing brain tumor segmentation (the case studied in [54]).
In this paper, we defined two versions of the FDL. The first one is similar to the one proposed by Wang et al.:
with .
The second one is based on the concept of the focal loss of Lin et al. [55]. First, we define the “soft” Focal Dice Coefficient (FDC), which can be formulated as:
Then, we calculated the FDL as:
We excluded class t associated with the background, and we divided the sum in Equations (11) and (13) by . In our case, we note that the numerator sums only the terms that have a . This means that we can have two possible categories of values for :
- A high value of , associated with true positives;
- A low value of , associate to false negatives.
For misclassifications, our goal was to reduce the total value of , penalizing the values. Since values lie in the range , we added a modulating factor . When , we can see that the reduction of is greater than when . This means that a voxel with a low value provides a minor contribution to the after the modulation. We can see how the value affects the values in Figure 3.
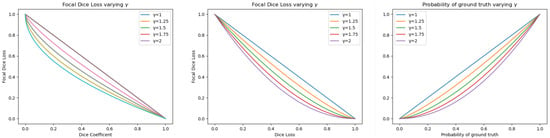
Figure 3.
Left: values of as a function of the Dice coefficient varying . Center: values of as a function of the Dice loss. Right: values of varying in the formulation.
2.3. Post-Processing
The post-processing consisted of two phases: in the first, we retained only the largest connected component for each segment; in the second, we eliminated all segments’ inconsistencies, i.e., we avoided the presence, on the same axial slice, of inconsistent segments (from an anatomical perspective). To introduce this concept, we realized the inconsistency matrix reported in Table 2.

Table 2.
Segments’ inconsistency table. If the presence of the pair of segments in the same axial slice is possible, we inserted a “1” into the matrix, otherwise a “0”.
This table was inspired by the liver anatomy. Considering only slices on the axial plane, we can divide the liver into three regions: (1) above the left branch of the portal vein, (2) below the right branch of the portal vein, and (3) between the two branches. For each region, as it is possible to see from Figure 4, the allowed segments are:
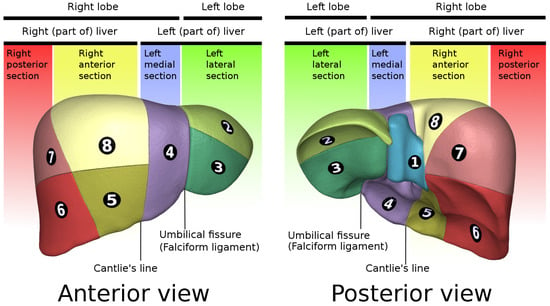
Figure 4.
Liver segments’ anatomy. Data generated by the Database Center for Life Science (DBCLS) https://dbcls.rois.ac.jp/index-en.html, last accessed: 17 January 2022; data provided by BodyParts3D https://lifesciencedb.jp/bp3d/?lng=en, last accessed: 17 January 2022 [61].
- Segments II, IV, VII, and VIII;
- Segments III, IV, V, and VI;
- Segments III, IV, VII, and VIII.
From these observations, we derived these substitution rules:
- Left Branch (LB): above the LB, all the voxels classified as Segment III are relabeled as Segment II; below the LB, all the voxels classified as Segment II are relabeled as Segment III;
- Right Branch (RB):
- ̵
- Above the RB, all the voxels classified as Segment V are relabeled as Segment VIII, and below the RB, Segment VIII labels are reassigned to Segment V;
- ̵
- Above the RB, all the voxels classified as Segment VI are relabeled as Segment VII; below the RB, all the voxels classified as Segment VII are relabeled as Segment VI.
In order to define and localize in the CT volume the two aforementioned branches, i.e., the LB and RB, of the portal vein, we implemented a majority criterion based on the classification of the voxels. The first slice where the sum of the counts of the voxels belonging to Segments III, IV, V, and VI was less than the sum of the counts of the remaining voxels in the first slice above the RB.
Similarly, the first slice where the sum of the counts of the voxels from Segments II, IV, VII, and VIII was higher than the sum of the counts of the remaining voxels in the first slice above the LB.
3. Experimental Results
In order to assess the performances of the different classifiers obtained by varying the formulation and the associated modulation factor , we assessed the “hard” Dice coefficient for each class t, the average Dice coefficient across all classes, the mean accuracy , and the confusion matrix, in order to see the relationships of the errors from different segments. We compared the presented by Wang et al. with the ones presented in this paper. We considered five values for the modulation parameter: . The case is equivalent to using the Dice coefficient as defined in Equation (7). For each experiment, we checked if the application of the post-processing was beneficial or not.
Experimental results are reported in Table 3, Table 4, Table 5 and Table 6, for models trained with , , , and , respectively. Figure 5 shows the segmentation obtained for a single CT scan, adopting with .

Table 3.
Results for the models trained with the formulation, as defined in Equation (11). Comparisons for the different liver segments. PP stands for Post-Processing. is the “hard” Dice coefficient for class t. It is reported as a percentage. is the arithmetic mean of . is the standard deviation of . The average accuracy is denoted as .

Table 4.
Results for the models trained with the formulation, as defined in Equation (13). Comparisons for the different liver segments. PP stands for Post-Processing. is the “hard” Dice coefficient for class t. It is reported as a percentage. is the arithmetic mean of . is the standard deviation of . The average accuracy is denoted as .

Table 5.
Results for the models trained with the formulation, as defined in Equation (10). Comparisons for the different liver segments. PP stands for Post-Processing. is the “hard” Dice coefficient for class t. It is reported as a percentage. is the arithmetic mean of . is the standard deviation of . The average accuracy is denoted as .

Table 6.
Results for the models trained with the Dice loss, which corresponds to the case of for all the previously defined focal Dice losses. Comparisons for the different liver segments. PP stands for Post-Processing. is the “hard” Dice coefficient for class t. It is reported as a percentage. is the arithmetic mean of . is the standard deviation of . The average accuracy is denoted as .
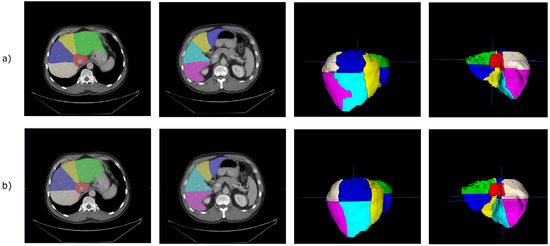
Figure 5.
(a) Prediction obtained from the 2.5D V-Net trained with our formulation of the focal Dice loss. For these images, the model was trained with with . (b) Corresponding ground-truth.
In the case of , the best model was the one with after post-processing. A possible justification is that larger values of result in flattening too much the loss of the volumetric predictions, so that those having a good Dice coefficient are too penalized.
For the FDLs defined in this paper, the model trained achieved the best performance also in the case , after post-processing, while the one trained with obtained the best performance in the configuration .
Concerning , in our experiments, we found that the model trained with could not generalize, and the for at least a class remained one (worst case) during the training. A possible explanation could be similar to the one provided for the from the paper of Wang et al. [54]. When you increase the value of , even the high values of are penalized, as we can see from Figure 3.
We tested if was better than the other formulations. To do this, we performed the Wilcoxon signed-rank test, obtaining that showed superior performances to with , to with , and to with . Considering, as usual, the significance threshold , the obtained results were statistically significant.
We also report the normalized confusion matrices in Figure 6. Elements in the main diagonal of the row-normalized and column-normalized confusion matrices are the precision(s) and recall(s) of the corresponding classes, respectively. From the confusion matrices, we can see that the most frequent errors arose between (in all the list here, the notation is predicted segment and ground-truth segment): Segment I and the background (this could be due to the fact that the CNN classifies the vena cava as Segment I, instead of background), Segment II and Segment III (this depends on the fact that the LB is predicted in a lower position than the real one), Segment IV and Segment II (they are adjacent on the axial plane), Segment VIII and Segment IV (they are adjacent on the axial plane), Segment V and Segment VI (they are adjacent on the axial plane), and Segment VIII and Segment VII (they are adjacent on the axial plane).
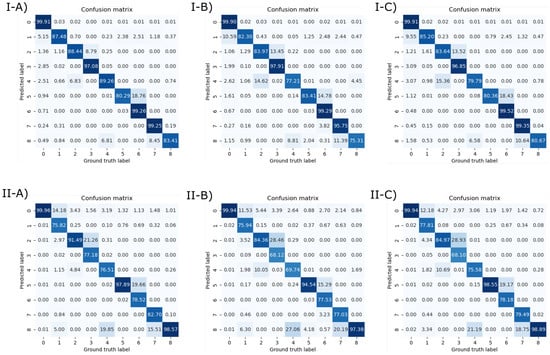
Figure 6.
(Top) Row-normalized confusion matrices. (Bottom) Column-normalized confusion matrices. Left: Confusion matrix for the model trained with . Center: Confusion matrix for the model trained with . Right: Confusion matrix for the model trained with .
Existing approaches are compared in Table 7. We note that existing works did not provide results (as ) for each segment. Different papers, such as [6,45], only proposed a qualitative assessment of the obtained results via expert radiologists, whereas Tian et al. [21] and Yan et al. [47] only reported the aggregated Dice coefficientper case across all classes.

Table 7.
Literature overview for liver segments’ classification. Quantitative results are not available for [6,45], since they only provided visual assessment of the segments’ classification.
4. Conclusions and Future Works
In this paper, we proposed two novel focal Dice loss formulations for liver segments’ classification and compared them with the previous one presented in the literature. The problem of correctly identifying liver segments is pivotal in surgical planning, since it can allow more precise surgery, as performing segmentectomies. In fact, according to the Couinaud model [5], hepatic vessels are the anatomic boundaries of the liver segments. Therefore, in current hepatic surgery, the identification of the liver vessel tree can avoid the unnecessary removal of healthy liver parenchyma, thus lowering the risk of complications that can arise from larger resections [7]. Despite this, in the literature, many authors that focused on liver segments’ classification methods usually only provided visual assessment (as [6,45]), making it difficult to benchmark different algorithms for this task. Thanks to the work of Tian et al. [21], a huge dataset composed of CT scans with liver segments’ annotation is now publicly available, easing the development and evaluation of algorithms suited for the task. Our CNN-based approach allowed us to obtain reliable results, with the best formulation, with , obtaining an average Dice coefficient higher than 83% and an average accuracy higher than 98%. The adoption of a rule-based post-processing, which implements a mechanism for detecting and resolving inconsistencies across axial slice images, allowed slightly improving the obtained results for different models. Given that the difference between the models was exiguous, it may be necessary to exploit a different model to assess a clearer difference between these loss functions.
A limitation of this work is the absence of an external validation dataset, which is due to the facts that there is a lack of annotated datasets with liver segments and it is very expensive to create a local one labeled by radiologists. Furthermore, with more GPU memory available, more complex 3D architectures capable of handling a larger context can be exploited, whereas in this work, we limited this to five slices at once.
As future work, we note that another possible way to approach this task consists of determining regression planes using a 3D CNN. Even if there are classical approaches (as Oliveira et al. [45]) devoted to the estimation of these cutting planes, there are actually no DL-based approaches in this direction (Tian et al. [21] and Yan et al. [47], as us, developed a semantic segmentation model), which can be more resilient to overfitting and to the prediction of inconsistent segments’ classification.
Author Contributions
Conceptualization: B.P., N.A., A.B. and V.B.; methodology: B.P., N.A., A.B. and V.B.; software: B.P. and N.A.; visualization: B.P. and N.A.; writing–original draft: B.P., N.A. and G.D.C.; writing–revisions: all authors; supervision: A.B., A.G. and V.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset for liver segments is publicly accessible. CT Images are from Medical Segmentation Decathlon Task08 [57]; annotated ground-truths for liver segments are from a Lenovo research work [21].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brunetti, A.; Carnimeo, L.; Trotta, G.F.; Bevilacqua, V. Computer-assisted frameworks for classification of liver, breast and blood neoplasias via neural networks: A survey based on medical images. Neurocomputing 2019, 335, 274–298. [Google Scholar] [CrossRef]
- Pepe, A.; Trotta, G.F.; Mohr-Ziak, P.; Gsaxner, C.; Wallner, J.; Bevilacqua, V.; Egger, J. A marker-less registration approach for mixed reality–aided maxillofacial surgery: A pilot evaluation. J. Digit. Imaging 2019, 32, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xie, Q.; Zha, Y.; Wang, D. Fully automatic liver segmentation combining multi-dimensional graph cut with shape information in 3D CT images. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germain, T.; Favelier, S.; Cercueil, J.P.; Denys, A.; Krausé, D.; Guiu, B. Liver segmentation: Practical tips. Diagn. Interv. Imaging 2014, 95, 1003–1016. [Google Scholar] [CrossRef] [Green Version]
- Couinaud, C. Liver lobes and segments: Notes on the anatomical architecture and surgery of the liver. Presse Med. 1954, 62, 709–712. [Google Scholar]
- Yang, X.; Yang, D.J.; Hwang, H.P.; Yu, H.C.; Ahn, S.; Kim, B.W.W.; You, H. Segmentation of liver and vessels from CT images and classification of liver segments for preoperative liver surgical planning in living donor liver transplantation. Comput. Methods Programs Biomed. 2018, 158, 41–52. [Google Scholar] [CrossRef]
- Helling, T.S.; Blondeau, B. Anatomic segmental resection compared to major hepatectomy in the treatment of liver neoplasms. HPB 2005, 7, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.T.; Lei, C.C.; Hung, S.W. Computer-aided kidney segmentation on abdominal CT images. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 59–65. [Google Scholar] [CrossRef]
- Magistroni, R.; Corsi, C.; Marti, T.; Torra, R. A review of the imaging techniques for measuring kidney and cyst volume in establishing autosomal dominant polycystic kidney disease progression. Am. J. Nephrol. 2018, 48, 67–78. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Brunetti, A.; Trotta, G.G.F.; Dimauro, G.; Elez, K.; Alberotanza, V.; Scardapane, A. A novel approach for Hepatocellular Carcinoma detection and classification based on triphasic CT Protocol. In Proceedings of the 2017 IEEE Congress on Evolutionary Computation (CEC), Donostia, Spain, 5–8 June 2017; pp. 1856–1863. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Carnimeo, L.; Brunetti, A.; De Pace, A.; Galeandro, P.; Trotta, G.G.F.; Caporusso, N.; Marino, F.; Alberotanza, V.; Scardapane, A. Synthesis of a Neural Network Classifier for Hepatocellular Carcinoma Grading Based on Triphasic CT Images. In Communications in Computer and Information Science; Springer: Cham, Switzerland, 2017; Volume 709, pp. 356–368. [Google Scholar] [CrossRef]
- Altini, N.; Marvulli, T.M.; Caputo, M.; Mattioli, E.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Tommasi, S.; Bevilacqua, V.; De Summa, S.; et al. Multi-class Tissue Classification in Colorectal Cancer with Handcrafted and Deep Features. In Intelligent Computing Theories and Application; Huang, D.S., Jo, K.H., Li, J., Gribova, V., Bevilacqua, V., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 512–525. [Google Scholar]
- Altini, N.; Prencipe, B.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; Scardapane, A.; et al. A Tversky Loss-Based Convolutional Neural Network for Liver Vessels Segmentation; Springer: Cham, Switzerland, 2020; pp. 342–354. [Google Scholar] [CrossRef]
- Lateef, F.; Ruichek, Y. Survey on semantic segmentation using deep learning techniques. Neurocomputing 2019, 338, 321–348. [Google Scholar] [CrossRef]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Martinez-Gonzalez, P.; Garcia-Rodriguez, J. A survey on deep learning techniques for image and video semantic segmentation. Appl. Soft Comput. J. 2018, 70, 41–65. [Google Scholar] [CrossRef]
- Zhao, B.; Feng, J.; Wu, X.; Yan, S. A survey on deep learning-based fine-grained object classification and semantic segmentation. Int. J. Autom. Comput. 2017, 14, 119–135. [Google Scholar] [CrossRef]
- Zhao, Z.Q.Q.; Zheng, P.; Xu, S.t.T.; Wu, X. Object Detection With Deep Learning: A Review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altini, N.; Cascarano, G.D.; Brunetti, A.; De Feudis, D.I.; Buongiorno, D.; Rossini, M.; Pesce, F.; Gesualdo, L.; Bevilacqua, V. A Deep Learning Instance Segmentation Approach for Global Glomerulosclerosis Assessment in Donor Kidney Biopsies. Electronics 2020, 9, 1768. [Google Scholar] [CrossRef]
- Altini, N.; Cascarano, G.D.; Brunetti, A.; Marino, F.; Rocchetti, M.T.; Matino, S.; Venere, U.; Rossini, M.; Pesce, F.; Gesualdo, L.; et al. Semantic Segmentation Framework for Glomeruli Detection and Classification in Kidney Histological Sections. Electronics 2020, 9, 503. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Liu, L.; Shi, Z.; Xu, F. Automatic Couinaud Segmentation from CT Volumes on Liver Using GLC-UNet; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11861, pp. 274–282. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Altini, N.; Prencipe, B.; Brunetti, A.; Villani, L.; Sacco, A.; Morelli, C.; Ciaccia, M.; Scardapane, A. Lung Segmentation and Characterization in COVID-19 Patients for Assessing Pulmonary Thromboembolism: An Approach Based on Deep Learning and Radiomics. Electronics 2021, 10, 2475. [Google Scholar] [CrossRef]
- Hoyte, L.; Ye, W.; Brubaker, L.; Fielding, J.R.; Lockhart, M.E.; Heilbrun, M.E.; Brown, M.B.; Warfield, S.K. Segmentations of MRI images of the female pelvic floor: A study of inter- and intra-reader reliability. J. Magn. Reson. Imaging 2011, 33, 684–691. [Google Scholar] [CrossRef] [Green Version]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Tridandapani, S.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L.; Edu, H.H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep learning: A primer for radiologists. Radiographics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cheng, J.; Quan, Q.; Wu, F.X.; Wang, Y.P.; Wang, J. A survey on U-shaped networks in medical image segmentations. Neurocomputing 2020, 409, 244–258. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prencipe, B.; Altini, N.; Cascarano, G.D.; Guerriero, A.; Brunetti, A. A Novel Approach Based on Region Growing Algorithm for Liver and Spleen Segmentation from CT Scansitle; Springer International Publishing: Cham, Switzerland, 2020; Volume 2. [Google Scholar] [CrossRef]
- Gambino, O.; Vitabile, S.; Lo Re, G.; La Tona, G.; Librizzi, S.; Pirrone, R.; Ardizzone, E.; Midiri, M.; Re, L.G.; La Tona, G.; et al. Automatic volumetric liver segmentation using texture based region growing. In Proceedings of the 2010 International Conference on Complex, Intelligent and Software Intensive Systems, Krakow, Poland, 15–18 February 2010; pp. 146–152. [Google Scholar] [CrossRef]
- Mostafa, A.; Abd Elfattah, M.; Fouad, A.; Hassanien, A.E.; Hefny, H.; Kim, T.H.H.; Elfattah, M.A.; Fouad, A.; Hassanien, A.E.; Hefny, H.; et al. Region growing segmentation with iterative K-means for CT liver images. In Proceedings of the 2015 4th International Conference on Advanced Information Technology and Sensor Application (AITS), Harbin, China, 21–23 August 2015; Number V; pp. 88–91. [Google Scholar] [CrossRef]
- Arica, S.; Av\csar, T.S.; Erbay, G.; Avşar, T.S.; Erbay, G. A Plain Segmentation Algorithm Utilizing Region Growing Technique for Automatic Partitioning of computed Tomography Liver Images. In Proceedings of the 2018 Medical Technologies National Congress (TIPTEKNO), Magusa, Cyprus, 8–10 November 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Kumar, S.S.; Moni, R.S.; Rajeesh, J. Automatic segmentation of liver and tumor for CAD of liver. J. Adv. Inf. Technol. 2011, 2, 63–70. [Google Scholar] [CrossRef]
- Arjun, P.; Monisha, M.K.; Mullaiyarasi, A.; Kavitha, G. Analysis of the liver in CT images using an improved region growing technique. In Proceedings of the 2015 International Conference on Industrial Instrumentation and Control (ICIC), Pune, India, 28–30 May 2015; pp. 1561–1566. [Google Scholar]
- Lu, X.; Wu, J.; Ren, X.; Zhang, B.; Li, Y. The study and application of the improved region growing algorithm for liver segmentation. Optik 2014, 125, 2142–2147. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, W.; Yu, H.; Huang, J. Based on pre-treatment and region growing segmentation method of liver. In Proceedings of the 2010 3rd International Congress on Image and Signal Processing, Yantai, China, 16–18 October 2010; Volume 3, pp. 1338–1341. [Google Scholar]
- Huang, J.; Qu, W.; Meng, L.; Wang, C. Based on statistical analysis and 3D region growing segmentation method of liver. In Proceedings of the 2011 3rd International Conference on Advanced Computer Control, Harbin, China, 18–20 January 2011; pp. 478–482. [Google Scholar]
- Lakshmipriya, B.; Jayanthi, K.; Pottakkat, B.; Ramkumar, G. Liver Segmentation using Bidirectional Region Growing with Edge Enhancement in NSCT Domain. In Proceedings of the 2018 IEEE International Conference on System, Computation, Automation and Networking (ICSCA), Pondicherry, India, 6–7 July 2018; pp. 1–5. [Google Scholar]
- Rafiei, S.; Karimi, N.; Mirmahboub, B.; Najarian, K.; Felfeliyan, B.; Samavi, S.; Soroushmehr, S.M.R. Liver Segmentation in Abdominal CT Images Using Probabilistic Atlas and Adaptive 3D Region Growing. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 6310–6313. [Google Scholar]
- Elmorsy, S.A.; Abdou, M.A.; Hassan, Y.F.; Elsayed, A. K3. A region growing liver segmentation method with advanced morphological enhancement. In Proceedings of the 2015 32nd National Radio Science Conference (NRSC), Giza, Egypt, 24–26 March 2015; pp. 418–425. [Google Scholar]
- Vezhnevets, V.; Konouchine, V. GrowCut- Interactive multi-label N-D image segmentation by cellular automata. In Proceedings of the GraphiCon 2005 - International Conference on Computer Graphics and Vision, Novosibirsk Akademgorodok, Russia, 20–24 June 2005; pp. 1–7. [Google Scholar]
- Czipczer, V.; Manno-Kovacs, A. Automatic liver segmentation on CT images combining region-based techniques and convolutional features. In Proceedings of the 2019 International Conference on Content-Based Multimedia Indexing (CBMI), Dublin, Ireland, 4–6 September 2019; pp. 1–6. [Google Scholar]
- Xu, L.; Zhu, Y.; Zhang, Y.; Yang, H. Liver segmentation based on region growing and level set active contour model with new signed pressure force function. Optik 2020, 202, 163705. [Google Scholar] [CrossRef]
- Oliveira, D.A.A.; Feitosa, R.Q.; Correia, M.M. Segmentation of liver, its vessels and lesions from CT images for surgical planning. Biomed. Eng. Online 2011, 10, 30. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Qingsen, Y.; Bo, W.; Dong, G.; Dingwen, Z.; Yang, Y.; Zheng, Y.; Yanning, Z.; Javen, Q.S. A Comprehensive CT Dataset for Liver Computer Assisted Diagnosis. In Proceedings of the BMVC 2021—The 32nd British Machine Vision Conference, Online, 22–25 November 2021; pp. 1–13. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2015, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Çiçek, O.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9901 LNCS, pp. 424–432. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2020; pp. 263–273. [Google Scholar]
- Wang, P.; Chung, A.C. Focal Dice loss and image dilation for brain tumor segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11045 LNCS, pp. 119–127. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2017; Volume 2017, pp. 2999–3007. [Google Scholar] [CrossRef] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.A.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef] [Green Version]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Altini, N.; De Giosa, G.; Fragasso, N.; Coscia, C.; Sibilano, E.; Prencipe, B.; Hussain, S.M.; Brunetti, A.; Buongiorno, D.; Guerriero, A.; et al. Segmentation and Identification of Vertebrae in CT Scans Using CNN, k-Means Clustering and k-NN. Informatics 2021, 8, 40. [Google Scholar] [CrossRef]
- Altini, N.; Prencipe, B.; Cascarano, G.D.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; et al. Liver, Kidney and Spleen Segmentation from CT scans and MRI with Deep Learning: A Survey. Neurocomputing 2022. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- BodyParts3D, © The Database Center for Life Science licensed under CC Attribution-Share Alike 2.1 Japan. Available online: https://lifesciencedb.jp/bp3d/?lng=en (accessed on 17 January 2022).
- Heimann, T.; van Ginneken, B.; Styner, M.M.A.; Arzhaeva, Y.; Aurich, V.; Bauer, C.; Beck, A.; Becker, C.; Beichel, R.; Bekes, G.; et al. Comparison and Evaluation of Methods for Liver Segmentation From CT Datasets. IEEE Trans. Med. Imaging 2009, 28, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).