Abstract
Mushroom ligninolytic enzymes are attractive biocatalysts that can degrade lignin through oxido-reduction. Laccase, lignin peroxidase, manganese peroxidase, and versatile peroxidase are the main enzymes that depolymerize highly complex lignin structures containing aromatic or aliphatic moieties and oxidize the subunits of monolignol associated with oxidizing agents. Among these enzymes, mushroom laccases are secreted glycoproteins, belonging to a polyphenol oxidase family, which have a powerful oxidizing capability that catalyzes the modification of lignin using synthetic or natural mediators by radical mechanisms via lignin bond cleavage. The high redox potential laccase within mediators can catalyze the oxidation of a wide range of substrates and the polymerization of lignin derivatives for value-added chemicals and materials. The chemoenzymatic process using mushroom laccases has been applied effectively for lignin utilization and the degradation of recalcitrant chemicals as an eco-friendly technology. Laccase-mediated grafting has also been employed to modify lignin and other polymers to obtain novel functional groups able to conjugate small and macro-biomolecules. In this review, the biochemical features of mushroom ligninolytic enzymes and their potential applications in catalytic reactions involving lignin and its derivatives to obtain value-added chemicals and novel materials in lignin valorization are discussed.
1. Introduction
Lignocellulose is a renewable bioresource used to produce bio-based fuel, chemicals, and materials. Lignocellulosic biomass is an abundant and renewable resource from plants such as soft grasses, hard woods, agricultural crops, and their waste byproducts, mainly composed of polysaccharides. These plant cell walls consist of a complex polyphenolic lignin combined with rigid cellulose amorphous hemicellulose structures [1,2]. Since plant cell walls are arranged with multi-layers consisting of lignin, cellulose, and hemicellulose, which have strong interactions with each other, the disturbance and breakdown of polymeric structures are the initial processes required for the utilization of subunit compounds such as sugar monomers, oligomers, and other aromatic derivatives [1,2]. Lignin binds to cellulose fibers to harden and strengthen plant cell walls. Cellulose is the main structural polysaccharide of the primary plant cell wall, and it accounts for 30–50% of the dry weight of lignocellulose. The second polysaccharide component of lignocellulose is hemicellulose, which accounts for 15–34% of the plant cell wall. The third main component is lignin. Conceptually, the microbial bioconversion of renewable chemicals from the biomass involves four main steps: physical and chemical pretreatment of biomass, depolymerization of the pretreated biomass, fermentation of the resulting sugars, and purification or distillation of the products [3,4]. Other processes directly depolymerize biomass under harsh physical and chemical reactions, and the lysed monomers or oligomers can then be converted to renewable products through chemical or biological catalysts [1,2,3,4]. Theoretically, the main three fractions (% dry wt), cellulose (30–50%), hemicellulose (15–34%), and lignin (9–32%), from renewable sources could be converted completely to fuels and chemicals within the concept of net-zero emissions technology [5]. Nevertheless, different physical or chemical pretreatment procedures are employed for various types of lignocellulosic biomass, which may influence the downstream process for the conversion efficiency and productivity of final products [5,6].
Over recent decades, the process of the extraction of cellulose and hemicellulose linked to lignin from different types of biomass has been well-established to obtain fermentable sugars for the production of cellulosic biofuels or bio-building-block substances for chemical polymers [1,2,4]. Since cellulose and hemicellulose consist of simple or uniform monosaccharide subunits, these fractions can be extracted easily as monomeric or oligomeric sugars through pretreatment processes rather than lignin-containing complex heteropolymers within aromatic structures [6,7,8]. Most of the conventional pretreatments comprised chemical methods combined with physical processes to enhance the cellulose content per gram of biomass or to extract hemicellulose fractions [6,7,8,9,10,11]. However, lignin fractions are released as byproducts of these pretreatment processes [7,10,12]. The high content lignin substances are then found in the wastewater generated by the pretreatment processes. Treatment of this wastewater accounted for approximately 20% of overall production costs [13]. The extracted lignin could be potentially utilizable for renewable chemicals to reduce non-utilized materials from biomass and overall production costs down for a sustainable green technology [14].
Lignin consists of aromatic heteropolymers with complex chemical linkages. The heteropolymeric structures contain hydroxyl, methoxyl, carbonyl, and carboxyl groups linked to aromatic or aliphatic moieties, with different molecular weights [15]. Lignin valorization is challenging due to the complexity of its structures compared with other cellulose and hemicellulose polymers [15,16]. The chemical bonds from the heterogeneous structures in lignin should be cleaved off by sophisticated catalysts with chemo-, regio-, and stereo-selectivity. Therefore, ligninolytic enzymes could be potential biocatalysts to convert lignin into valuable aromatic compounds [17,18,19]. Although plant lignin is resistant to biodegradation, many microorganisms, including fungi, secrete lignocellulotic enzymes to decay biomass for mineralization of the recalcitrant compounds [19]. These microorganisms, along with other degraders in nature, play a carbon-recycling role in ecosystems. These bacterial degradation ecosystems could potentially mimic lignin conversion into high-value aromatic compounds [16,17,19].
Ligninolytic enzymes that could be used for lignin valorization (i.e., that can selectively carry out regio- and stereo-specific bond formation under mild reaction conditions) have been highlighted as a promising method to overcome the limitations associated with classical chemical catalysts [17,18]. In this review, I describe the biochemical properties of ligninolytic enzymes reported from mushroom species and their applications for chemoenzymatic approaches to produce renewable aromatic compound derivatives.
2. Structure of Lignin for Valuable Aromatic Compounds
Lignin is one of the main components consisting of a plant biomass. Lignin has a unique structure of closely associated cellulose and hemicellulose, which resists the bio-degradation of plant cell walls to protect plants against environmental stress [19,20,21,22]. Lignin is a heterogeneous polymer containing various aromatic structures consisting of phenylpropane subunits, linked with carbon–carbon (C–C) or ether (C–O–C) into a complex three-dimensional structure [20,21,22]. The complex structure of lignin is initially biosynthesized from an aromatic amino acid phenylalanine or tyrosine. Then, the aromatic amino acid is converted into p-coumaryl alcohol via the phenylpropanoid pathway following deamination, ring hydroxylation, methylation, and carboxyl reduction. Following that, p-coumaryl alcohol (H-unit) is elaborated to coniferyl and sinapyl alcohols (G- and S-units). These phytochemical substances of the three alcohol types are called monolignols or lignols, which are the main building blocks of lignin structures [23,24,25] (Figure 1).
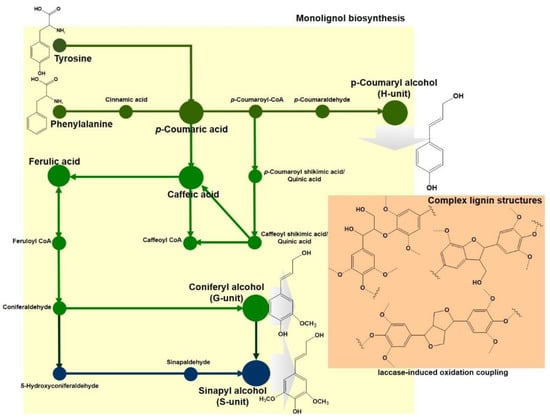
Figure 1.
Schematic representation of biosynthesis for three main monolignol subunit structures for lignin using two aromatic amino acid precursors. Moss green, green, and Prussian blue indicate the biosynthetic pathways for the H-, G-, and S-unit, respectively.
Interestingly, the biochemical synthesis of lignin is a unique polymerization through chemoenzymatic reactions, in which oxidases and peroxidases generate reactive quinone methide intermediates as lignin precursors from hydroxylated monolignols, p-hydroxyphenyl, guaiacyl, and a syringyl unit, and then these compounds are spontaneously polymerized [23,24,25]. Lignin polymers have been assumed as non-optically active chemicals since their monomers do not contain chiral structures. Nevertheless, the subunit structures of phenylpropanoid have two potential asymmetric carbons at the α- and β-positions of the side-chain moiety [23,26]. These two chiral centers generate structural variety of lignin in the monomer condensation process. In the polymerization reaction, benzylic hydroxyl groups in the β-aryl ether structural units in monolignols are lost, forming benzylic cation [26]. One of them is formed at the β-position by a β-O-4, β–β, β-5, or β-1 coupling reaction between new Cα–Cβ cross-linkage bonds from monolignols or the phenolic end of the growing polymer [23,26,27]. This polymerization of the precursors randomly occurs to form lignin complex structures. This reduces the enzyme accessibility and solubility for biodegradation, compared with the uniform structures of other cellulose and hemicellulose polymers in plant cell walls.
Although lignin, cellulose, and hemicellulose contents vary across plant species, plant cell walls usually contain around 20–35% lignin by weight [22,25]. Comparing lignin subunits in soft and hard woods, soft wood plants contain higher guaiacyl content and lower p-hydroxyphenyl unit content in their lignin structures [28]. Hardwood lignin primarily consists of guaiacyl and syringyl units in various proportions, together with minor levels of p-hydroxyphenyl units [28]. These different subunits of lignin contribute to differences in their physical properties.
3. Challenge of Lignin Fractionation and Depolymerization
To achieve lignin valorization, fractionation and depolymerization are still challenging research issues due to the complex mixture of lignin’s subunits and the oligomers extracted from its biomass. Some procedures, preparing klason, alkali, sulfide lignin, etc., are known to fractionate lignin from biomass [29,30]. However, separating certain types of β-aryl ether structural units or the monomerized monolignols from lignin remains a challenging task for lignin valorization to obtain products, such as hydrolyzed sugars or oligosaccharides from cellulose or hemicellulose. Nevertheless, two main resources are considered to obtain potentially available lignin as byproducts in the utilization of lignocellulose. One major source of lignin is the byproducts of the pulp and paper industry [21,30]. The kraft and sulfite processes generate kraft lignin and water-soluble lignosulfonates as a byproduct, respectively. These two methods can extract lignin by chemical treatments with sodium hydroxide and sodium sulfide for kraft lignin or using aqueous sulfur dioxide for lignosulfonate. The other main source is byproducts of lignocellulosic bio-ethanol production processes [14]. There are various chemical pretreatment processes used to extract cellulose and hemicellulose for fermentable sugars of ethanol production, and lignin is usually solubilized into wastewater or remains in the treated biomass [7,12,13]. The residual lignin in the biomass remains as a solid waste after fermentation and the product recovery process [12]. However, these byproducts are still considered waste and are burned to generate energy to operate their process [13,14]. Recently, pyrolysis or gasification of these biomass byproducts has been alternatively applied to valorize lignin fractions [31,32].
Although these lignin fractions would be potentially useful resources, lignin depolymerization is a barrier to producing valuable chemicals from lignin due to its heterogeneous and complex structure, the different physical properties of pretreated byproducts, difficulties associated with linkage cleavages in monolignols, repolymerization of depolymerized products, and so on [2,16,25,29]. Nevertheless, there is increasing interest in developing novel biocatalyst methods for lignin valorization by mimicking the natural processes of biodegradation in microorganisms [17,18,19].
4. Lignin Biodegradation and Ligninolytic Enzymes
Lignin is degraded by microorganisms under aerobic conditions, although the degradation rate of lignin is much lower than it is for cellulose and hemicellulose. The intact biomass is attacked first by Basidiomycetes fungi [17,18,19,33]. These fungi initiate degradation processes enzymatically to decompose lignin, cellulose, and hemicellulose. The biodegradation of lignin is a heavily oxidative process by indirectly random mechanisms involving several enzymes in the oxidoreductase family [22,25,33]. Enzymatic biodegradation is associated with oxidizing agents such as superoxide anion (O2−), hydrogen peroxide (H2O2), hydroxyl radicals (–OH), and singlet oxygen (1O2) [19,33]. These reactive compounds break the bonds between the subunit of monolignols and depolymerize lignin gradually within ligninolytic enzyme reactions.
The main four enzymes in fungus species reported as key biocatalysts for breaking C–C or C–O bonds in heterogeneous and complex structures of lignin are laccase and laccase-like multicopper oxidase, lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP). These proteins are secreted from fungi species including mushrooms, penetrate into undecayed wood tissue, and initiate lignin degradation using phenolic compounds as mediators by radical mechanisms [19,33]. Since the lignin degradation activities in white- and brown-rot fungi, such as basidiomycetes, last several decays, many ligninolytic enzymes have been identified and characterized [17,18,19]. In addition to fungi, some bacterial lignin degradation activities have been reported in Arthrobacter, Flavobacterium, Micrococcus, Pseudomonas, Brucella, Ochrobactrum, Sphingobium, and Sphingomonas and other related species belonging to Actinobacteria, α- and γ-Proteobacteria [34,35,36]. Nevertheless, fungal enzymes are most commonly used in lignin degradation. Moreover, the ligninolytic enzymes from mushroom species belonging to phylum Basidiomycota are attractive sources for the application of lignin valorization based on chemoenzymatic approaches [17,19,37].
4.1. Laccase and Laccase-Like Multicopper Oxidase
Laccase (EC 1.10.3.2) is a polyphenol oxidase belonging to the copper-containing oxidase family that is capable of oxidizing aromatic compounds with substrate spectra in diverse organisms including fungi, bacteria, insects, and plants [38,39]. With coupling reactions of radical formation as well as one-electron oxidation, a laccase catalyzes the cleaving of C–C or C–O bonds in lignin and other aromatic compounds. This enzyme can oxidize a wide range of aromatic compounds, aliphatic amines, hydroxylindoles, polysaccharides, and inorganic or organic metals [17,19]. Consequently, it is favored by various industries for oxidation reactions [38,39,40].
In eukaryotes, laccase activity has been reported from most higher fungi in the orders Ascomycetes and Basidiomycetes [17,19,39]. Besides lignin degradation, laccase and laccase-like multicopper oxidase is an essential enzyme playing a role in multiple functions of development and morphogenesis, pathogenesis and detoxification, the biosynthesis of secondary metabolites (e.g., melanin pigments), and polymerization of polyphenolic compounds, etc. [19,38,39]. Interestingly, mushroom laccases and related enzymes were found to be intracellular or extracellular proteins. These localized enzymes may participate in different cellular and physical functions in mushrooms. The extracellular enzymes are secreted proteins containing signal sequences and protein N-glycosylation for cellular localization, which catalyze high molecular weight complex aromatic compounds such as lignin [39,41]. The intracellular enzymes are endogenous cytoplasmic proteins that convert the low molecular weight of intracellular compounds for biosynthesis of secondary metabolites as well as pigment chemicals [25,42].
As summarized in Table 1 [43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89], several species of mushroom, including Agaricus bisporus, Lentinula edodes, Pleurotus ostreatus, and Trametes versicolor, produce more than one laccase isoenzyme, each with different biochemical features. Although the biological functions of some of these isoenzymes remain unclear, they display differences in terms of their substrate specificities and expression patterns [74,79,86]. In higher eukaryote mushrooms, alternative RNA splicing in the transcription process results in multiple transcript variations for isoenzymes of laccase and laccase-like multicopper oxidase with highly homologous sequences in one species [90,91,92]. The number of isoforms varies depending on the mushroom species, presence or lack of an inducer, growth and stress condition, and other environmental conditions [92,93,94,95,96]. However, the regulatory mechanisms of isoenzymes and the cellular functions have not yet been characterized. Although the biological functions of these laccase isoenzymes are still unclear, most mushroom laccases are secreted extracellular enzymes with different substrate specificities toward aromatic compounds. Therefore, it has been suggested that they play an important role in the degradation of lignin in nutrient uptake to support the growth of mushroom fruit bodies. L. edodes H600 harbors extracellular and intracellular laccases, Lcc1 and Lcc2, within different substrate specificities [59,60]. The activities of both enzymes for typical laccase substrates such as ABTS, p-phenylenediamine, 2,6-dimethoxyphenol, ferulic acid, and guaiacol were similar, but those of intracellular laccase for catechol and L-DOPA were noticeably lower than the extracellular enzyme [60]. The expression of these two laccase-coding genes was high in the caps of the fruiting body, and high enzyme activities were also detected when the pigment of the fruit bodies changed during post-harvesting preservation. Thus, the intracellular laccase could play a role in pigment synthesis by oxidation of phenolic compounds, such as dihydroxyphenols, to making the fruiting bodies turn brown [60]. However, the oxidized chemical structures of dihydroxyphenol precursors are not yet known for the synthesis of melanin in vivo.

Table 1.
Physicochemical and biochemical properties of mushroom laccases.
4.2. Lignin Peroxidase (LiP)
Lignin peroxidase (LiP: EC 1.11.1.14) was the first reported lignin-degrading oxidase. This oxidase is an extracellular heme glycoprotein with approximately 35–50 kDa and pI of 3.0–4.0 belonging to the heme oxidase II family [97,98]. Since LiP activity was identified in a white-rot fungus P. chrysosporium, most laccase-produced mushroom species, including C. cinereus, L. edodes, P. eryngii, P. ostreatus, P. sajor-caju, and T. versicolor, can produce LiP and/or other oxidase enzymes together [43,98]. The number of LiP coding genes for isoenzymes, their expression level, and their productivity depend on the fungal growth condition, inducers, growth stage, medium composition, etc. These LiPs play an active role in lignin depolymerization, similar to other extracellular oxidases including laccase, manganese peroxidases (MnP), and versatile peroxidases (VPs) [99,100,101,102,103]. In lignin depolymerization, LiPs attack Cα–Cβ linkages in phenylpropanoid units and β–O–4 ether linkages between the side chain and the next subunit to open aromatic rings [17]. Since the mechanism of lignin degradation by these extracellular class II peroxidase has been determined by using protein structure models with biochemical characterizations [17,98], the basic reaction is identified as a heme oxidase involving H2O2. In the depolymerization of lignin, this oxido-reduction reaction is initiated when H2O2 accesses the heme prosthetic group via the heme channel, acts as an electron acceptor, and then reduces to water (Figure 2a). Interestingly, LiP has a higher redox potential (E0′ ≈ 1.2 V versus Standard hydrogen electrode, SHE) than other oxidases and therefore can oxidize both phenolic and non-phenolic compounds in the absence of mediators [17,104]. This advantage of LiPs displayed the broad substrate specificities for a variety of applications to degrade recalcitrant petroleum-based chemicals [105,106,107].
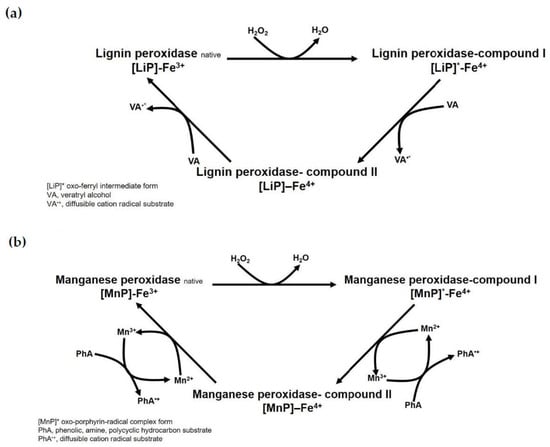
Figure 2.
Comparison of reaction mechanisms for lignin peroxidase (a) and manganese peroxidase (b).
4.3. Manganese Peroxidase (MnP)
Manganese peroxidase (MnP: EC 1.11.1.13) is a heme glycoprotein belonging to the extracellular oxidase II family, similar to LiPs [17,97,98]. The physicochemical properties of these oxidases display secreted enzymes with approximately 40–55 kDa and pI of 3.5–4.5. Even though LiPs were sometimes not observed in the culture media for mushroom growth [43,108,109], MnPs are abundantly secreted proteins in most wood-decayed Basidiomycetes fungi. Recently, the productivity of MnP was shown to be more efficient in terms of H2O2 consuming capacity than a cofactor metal ion and carbon sources in the solid-state medium of A. bisporus [110]. H2O2, when used as a cofactor for peroxidase, is considered a limiting factor for the ligninolytic activity and productivity of lignin degradation. Compared to the catalytic reaction of LiP, MnPs also require H2O2 as an electron acceptor for the oxidation reaction, but this enzyme needs Mn2+ as an electron donor, generating Mn3+. The Mn3+ released from the enzyme is reacted with a carboxyl acid moiety-contained substance, and the diffusible chelated complex plays the role of a redox mediator oxidizing phenolic compounds (Figure 2b) [17,19,98]. Therefore, MnP oxidizes phenolic compounds indirectly via the oxidation of Mn ions and secretion of Mn3+ to the extracellular environment [111]. The diffusible Mn3+ is advantageous due to its elimination of the need for complicated heterogeneous catalytic kinetics to broaden the substrate scope [17,111,112]. Nevertheless, MnPs have lower redox potential than LiPs. Thus, MnP can oxidize phenolic compounds, 2,6-dimethoxyphenol, guaiacol, 4-methoxyphenol, and the phenolic moiety in lignin, but is unable to convert nonphenolic compounds and veratryl alcohol [17,112].
4.4. Versatile Peroxidase (VP)
Versatile peroxidase (VP: EC 1.11.1.16) is the third member of the extracellular oxidase II family used for lignin degradation [17,97,98]. The VP activity has been reported from Pleurotus eryngii [113], but a limited number of VPs have been identified and characterized, mainly from the white fungal Basidiomycetes Pleurotus, Bjerkandera, and a few other fungi genera [99,100,113,114,115,116]. Recently, the comparative genomics for wood-decayed fungi showed that other fungi could harbor VP coding genes [98,117,118]. Indeed, fungal VPs may have evolutionarily split from a common ancestral peroxidase origin and then acquired the ability to catalyze nonphenolic compounds [117]. VPs have similar biochemical characteristics to MnPs. They are secreted glycoproteins with approximately 37–55 kDa in size and with a pI of 3.5–4.5. VPs display multifunctional properties typical of both LiP and MnP: they have a catalytic function similar to LiPs toward nonphenolic substances, such as veratryl alcohol, but these oxidases are capable of oxidizing Mn2+ to Mn3+ via a mechanism similar to that of MnPs [119,120,121]. The versatility of VPs allows it to directly oxidize low- and high-redox potential substrates because it contains two additional active sites [116,122]: one site is exposed to the heme edge to provide VPs with the capability to oxidize substrates directly without Mn2+; and the other site is the exposed tryptophanyl radical (Trp164 residue in VPs of Pleurotus strains), which is exposed on the surface of VPs, and is involved in the direct oxidation of low- and high-redox potential substrates in the presence of redox mediators [119,120,121,122]. On the basis of the Trp residue in the active site, VP has a higher redox potential (E0′ > 1.4V versus SHE at pH 3) to catalyze nonphenolic compounds without a mediator like LiPs. Therefore, this high-redox potential peroxidase could be used for catalyzing the degradation of xenobiotics, depolymerization, or polymerization of functional monomers or macromolecules, through enzymatic oxidoreduction [105,123,124,125].
4.5. Other Ligninolytic Enzymes in Mushroom
In addition to the lignin degradation by three main peroxidases described above, fungal genera also produce other ligninolytic enzymes [17,18,19], such as dye-decolorizing peroxidases (DyPs: EC 1.11.1.19), which are among the extracellular peroxidases. These peroxidases are not evolutionarily related to the oxidase II family for lignin [126,127]. A fungal DyP was identified as an extracellular enzyme from a Basidomycete fungus, Geotrichum candidum (now Bjerkandera adusta) [128]. This fungal peroxidase did not have a homology with another peroxidase, named DyP, which can catalyze the decolorization activity of a wide range of dyes, including poor degrading anthraquinone families such as Reactive blue 5 [129]. Comparative studies of DyPs derived from other wood- and litter-degrading fungi revealed that all fungal DyPs efficiently oxidized recalcitrant dyes as well as the phenolic substrate [129]. These peroxidases showed higher activity than LiP and VP toward Reactive Blue 5, Reactive Black 5, and 2,6-DMP. On the other hand, these enzymes displayed lower activity than other fungal peroxidases toward a recalcitrant heterocyclic dye, Azure B, and a nonphenolic compound (veratryl alcohol). Since fungal DyPs have been reported, the similar heme structure protein, YcdB, as a substrate of the twin-arginine translocation system of Escherichia coli, was shown to be bacterial DyP [130]. An ever-increasing number of prokaryotic genome data is being gathered, and now several thousand bacterial DyP homologous sequences have been identified and their proteins have been characterized [126,127,131]. The protein structures of DyPs revealed the presence of a dimeric α/β ferredoxin-like fold, which is different from any other peroxidase superfamily, which consists of an α-helical fold domain [126]. Indeed, a phylogenetical analysis of the fungal DyPs displayed that the fungal enzymes are distinct from the bacterial DyPs [126]. Comparative studies of the enzyme activities for both fungal and bacterial DyPs toward a variety of lignin-derived substrates have been performed, and a comparison of the kinetic parameters for a general oxidoreductase substrate, ABTS, showed that the fungal enzyme has enhanced oxidation activity compared with the bacterial enzymes [126].
Other minor enzymes involved are phenoxy radical-reducing enzymes, such as glyoxal oxidase (EC 1.2.3.5), aryl alcohol oxidase (EC 1.1.3.7), pyranose 2-oxidase (EC1.1.3.10), and cellobiose dehydrogenase (EC 1.1.99.18) within a cofactor heme or flavin adenine dinucleotide (FAD) to generate extracellular hydrogen peroxide [17,132]. Although these enzymes catalyze different substrates, such as dicarbonyl and methylglyoxal compounds, phenolic and nonphenolic alcohols, and cellobiose in lignin depolymerization, their oxidation reactions commonly produce hydrogen peroxide via the reduction of O2 to H2O2. These secreted hydrogen peroxides could regulate peroxidase activity in vitro [132]. In addition, heme-thiolate haloperoxidases and FDA-dependent glucose-dehydrogenases have also been known as minor oxidoreductases that catalyze the hydroxylation of organic and aromatic substances and scavenge phenolic radicals and quinone compounds [17,133,134].
5. Structural Properties of Mushroom Laccases
For lignin depolymerization, most mushroom laccases are secreted dimeric or tetrameric glycoproteins that contain signal sequences, which are cleaved during a protein secretion process via the productive folding of glycoproteins in the endoplasmic reticulum (ER) [135]. The molecular weights of monomeric fungal laccases range from 30 to 130 kDa (Table 1). The apparent sizes of the proteins are usually greater than predicted theoretical molecular weights based on the calculation of their protein sequences due to protein N-glycosylation.
The overall sequence homologies of fungi laccase proteins are less than 30%. Nevertheless, the copper-binding motifs and the overall structures are well-conserved and share several residues close to the metal-binding site (Figure 3a). Laccases have four copper-binding motifs conserved: NH2-His-x-His-Gly-CO3- for Cu motif I, NH2-His-x-His-CO3− for Cu motif II, NH2-His-x-x-His-x-His-CO3− for Cu motif III, and NH2-His-Cys-His-x-x-x-His-x-x-x-x-Met/Leu/Phe-CO3- for Cu motif IV (where x represents any amino acid). A protein structural analysis of mushroom laccase revealed that the active sites consist of four copper centers with four different catalytic forms (T1, T2, T3, and T2/T3), which are arranged with the identified residues of the motifs for the interaction of copper atoms [136,137] (Figure 3b). The T1 copper atom is bound by Cys, Met, and His residues in the motif IV and another His residue in motif III, with a distance of 2.05–3.27 Å. This T1 copper-binding protein or domain is called cupredoxin. The T2 copper atom of the trinuclear center is coordinated by two His-residues in motifs I and III, with a distance of less than 2.0 Å. The other two T3 copper atoms have interactions with three His-residues in motifs I―IV, with a distance of about 2.0 Å. In addition, the three T2/T3 copper atoms are arranged in a triangular fashion. Six His-residues in the copper-binding motifs coordinate the T3 copper pair, while T2 is coordinated by the other two His residues in the motifs. The four residue-bound T1 copper atoms are distant from the T3 copper atom consisting of the trinuclear center with a distance of around 13 Å. These copper ions binding in the active sites are classified as T1, T2, and T3 according to their different spectroscopic characteristics with the signals of electronic paramagnetic resonance (EPR) [136].
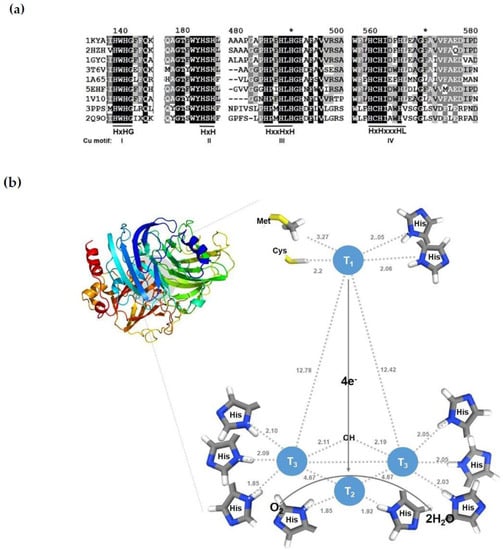
Figure 3.
Sequence alignment (a) and putative structural model (b) of fungal laccases. (a) Partial sequence alignment of Trametes versicolor laccase and its homologs. PDB accession number: 1KYA, Trametes versicolor laccase; 2HZH, T. ochracea laccase; 1GYC, T. versicolor laccase 2; 3T6V, Steccherinum ochraceum laccase; 1A65, Coprinus cinereus laccase; 5EHF, Antrodiella faginea laccase; 1V10, Rigidoporus microporus laccase; 3PPS, Canariomyces arenarius laccase; 2Q9O, Melanocarpus albomyces laccase-1. Four copper (Cu) binding motifs were marked. (b) Overall structure of Hericium erinaceus laccase 1 and a closer view of the active site for putative copper binding in a laccase with modification from the reference [137]. The mushroom laccase structure was predicted by PHYRE2 based on the homologous laccase 3-D structure in Pdb data files.
The oxidation mechanism of laccase occurs in its four copper centers via three main steps: the enzyme initially oxidizes a substrate at a mononuclear copper center T1 or blue copper center. Then, electrons are transferred internally from the T1 copper atom at a distance of around 13 Å though a Cys-His pathway to the tri-nuclear copper center, a normal type (T2), and a binuclear type (T3), where dioxygen is reduced by four electrons, generating two water molecules. In the laccase reaction, the monoelectronic oxidation of substrates generates free radicals. The initial product is unstable and then propagates a second oxidation by enzymatic catalysis or a non-enzymatic reaction [136,137,138].
Although laccase has lower redox potential (≤0.8 V) compared to other peroxidases for oxidation of a phenolic moiety in lignin degradation, a variety of low molecular substrates, intermediators, and products (e.g., reactive and unstable cationic radicals) oxidized by the enzymes can participate as redox mediators to enhance its oxidoreduction [137]. These laccase mediators can expand the catalytic ability toward non-aromatic and aromatic compounds, more than 100 mediators known as synthetic and natural compounds [38,139,140,141,142]. Synthetic mediators include 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 1-hydroxybenotriazole (HBT), N-hydroxyphtalimide (HPI), and 2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO). Natural mediators include acetosyringone (AS), catechol, p-coumaric acid, 4-hydroxybenzoic acid (4-HBA), 4-hydroxybenzilic alcohol, syringaldehyde (SA), vallinin, veratryl alcohol (VA), and violuric acid (VLA). The co-utilization of these mediators and a laccase, known as laccase-mediator systems (LMS), has been intensively studied to apply a variety of the enzymatic processes to pulp and paper bleaching, bioremediation, dye-decolorization, and the degradation of antibiotics and xenobiotics [38,39,40,63,68,134]. Moreover, the dual or triple mediators (ABTS and others) used with laccase for degradation of polyaromatic carbons showed synergistic effects of enhanced oxidative activities compared with a single mediator for the enzyme reaction, due to the recycling of ABTS radical formation [143]. The redox potential of mediators differs depending on their chemical structure and the biochemical activity of an individual laccase [144]. Based on many case studies for LMS, an effective mediator should be a favorable substrate because it has high oxidation potential, which means the oxidized radical form has a long half-life and can propagate more oxidation of the remaining substrate. Nevertheless, synthetic mediators have a limitation because they are unable to be used in a wide variety of industrial applications due to their cost, potential toxicity, and inhibition of laccases [145].
In contrast, natural mediators, such as oxidized lignin units, AS, p-coumaric acid, 4-HBA, SA, vanillin, VA, and VLA, derived from lignin degradation could be potential substances for use instead of synthetic compounds. These naturally-derived phenolic compounds were used as laccases mediators and worked effectively in the application of paper pulp delignification and the degradation of recalcitrant chemicals such as xenobiotic compounds, antibiotics, pesticides, dyes, and polycyclic aromatic hydrocarbons petroleum derivatives [140,142].
6. Bioconversion of Lignin Derivative into Bio-Based Chemicals and Materials
The ligninolytic enzyme-based bioconversion of lignin and its derivatives would be an attractive green technology for reducing wastes and energy consumption. Most ligninolytic enzymes for lignin degradation could be useful biocatalysts for biotechnological applications, due to their oxidation capacity toward both aromatic and non-aromatic compounds [37,38,39,40]. Among these ligninolytic oxidases, laccases are most broadly applicable due to their powerful oxidizing capability in the fields of high demand for highly oxidized agents such as the degradation of xenobiotics, hormones, biocides, and drug chemicals; decolorization of dyes; and detection of redox potential as biosensors [37,38,39,40] (Table 1).
Laccase can oxidize lignin through the abstraction of a single electron from a monolignol subunit, which causes activated radicals to react to functionalization on the lignin surface (Figure 4a). Activation of functionalized structures in lignin could propagate C–C bond and alkyl–aryl linkage cleavages, functional group modification, and radical coupling (Figure 4b). These oxidation mechanisms of laccase are applicable for biomass pretreatment as well as the detoxification of biomass hydrolysate in biorefinery processes [37,38,39,40]. In the biomass pretreatment, physicochemical processes generally use harsh reaction conditions with intensive energy consumption for the breakdown of biomass [1,2,3,4,5]. On the other hand, the biological process using white-rot fungi to produce a ligninolytic enzymes cocktail with laccase could reduce energy consumption and decrease reduce toxic residual compounds [38,39]. Moreover, the enzymatic process with laccase has already been used for typical delignification and bio-bleaching in the pulp and paper industries instead of chemical oxidation agents [146].
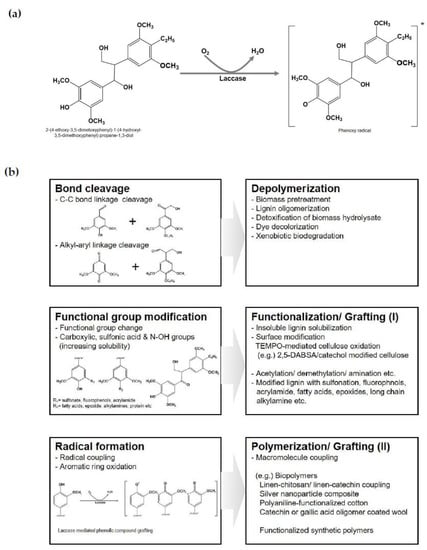
Figure 4.
Summary of the potential applications of various lignin derivatives and other biomolecules by laccase catalytic activity for lignin valorization. (a) Schematic representation of the laccase catalytic mechanism (b) Biotechnological applications for depolymerization, grafting, and polymerization based on laccase catalytic activities.
The oxidation of laccase was also used to degrade inhibitory compounds, phenolic and furanic substances, and carboxylic acids, generated in biomass pretreatment processes due to thermal lignin or sugar decompositions [147]. This detoxification could enhance the fermentation productivity or enzymatic conversion yield of biomass hydrolysate for the biorefinery process. However, the synthetic mediators in LMS-based pretreatment increase the overall process cost. Moreover, laccase-based biomass pretreatment combined with enzymatic hydrolysis showed low sugar generation yields due to laccase mediators inhibiting cellulases and lytic polysaccharide monooxygenases [37,148,149]. Thus, these two processes should be separated to enhance their benefits to biomass utilization.
Another unique application of laccase is grafting, which is the covalent bond formation between a macromolecule, e.g., lignin, and a small molecule, soluble functional groups, carboxylic and sulfonic acid, and hydrophilic N–OH [144,150]. Laccase-mediated grafting would be advantageous to converting hydrophobic lignin into soluble lignin by changing the functional groups on the lignin surface to enhance their reactivity to make enzymes easily accessible in the aqueous reaction phase. Grafting of sulfanilic acid or p-aminobenzoic acid by laccase showed that lignin is soluble in water under acidic or neutral pH conditions [151].
The bioconversion of lignin or lignin derivative directly to valuable chemicals and materials is still challenging with the existing techniques due to lignin’s properties for monomerization via depolymerization and the limitation of suitable enzymes for enzymatic processes. A recent review summarized lignin depolymerization for renewable chemicals based on chemical catalyst processes [29]. Interestingly, at least 37 chemicals could potentially be produced as lignin-based monomers obtained via different homogeneous and heterogeneous catalytic processes. Although the monomer content in lignin is heterogeneous in the origin of lignocellulose biomass, the procedure follows two steps of fractionation and catalytic depolymerization or a single step with reductive catalytic fractionation for platform chemicals. Bulky monomers are then converted into value-added chemicals by functionalization or fuel additives by defunctionalization. Ideally, these fractionation and depolymerization processes are performed by a chemical catalyst under high temperatures (100–300 °C) in acidic, alkaline, or solvent conditions [29]. Most chemical catalyst strategies are not adaptable for bioconversion processes due to the strict thermal and chemical reaction conditions.
Only two monomers among the 37 putative chemicals derived from lignin are biologically producible in temperature below 50 °C and under mild reaction conditions: hexa-2,4-dieneioic acid (muconic acid) and medium-chain-length polyhydroxyalkanoate. These two chemicals were biosynthesized by a wild type or engineered Pseudomonas strain, which is the most dominant soil bacteria capable of mineralizing a broad range of recalcitrant xenobiotics [152,153]. Various Pseudomonas strains and other bacteria can degrade aromatic compounds to either catechol or protocatechuic acid [154,155]. Through subsequent oxidative reactions, these compounds are converted to acetyl-CoA and succinate or pyruvate and acetaldehyde, which are common metabolites in the main metabolisms of most organisms (Figure 5) Depolymerized heterogeneous monomers from lignin consisting of syringyl-, gualacyl-, and hydroxyphenyl-aromatic polymers can also be metabolized to reconstruct various bio-building blocks by microorganisms engineered through metabolic engineering or synthetic biology technology [150,155,156,157]. Since the reconstruction of a bio-building block through bacterial metabolisms of lignin monomers has been proposed, various microorganisms have been engineered to convert lignin and lignin-derived aromatics into a variety of value-added compounds: cis,cis-muconic acid, catechol, protocatechuate, benzoate diol, p-hyroxybenzoate, guaiacol, pyrogallol, syringate vaillin, vanillate, vanillyl alcohol, coniferyl aldehyde, pyruvate, lactate, succinate, pyridine-related organic acids, polyhydroxyalkanoates (PHAs), and lipids [150,155,156,157,158]. Recently, itaconic acid, a chemical building block listed among the top 12 value-added chemicals, was also produced from p-coumaric acid by the engineered Pseudomonas strain [159,160]. In addition, the engineered Pseudomonas strain accumulated and converted p-coumaric/ ferulic acid into 4-vinylphenol/4-vinylguaiacol, which were further applied to polymerize 4-vinylguaiacol and 4-vinylcatechol styrene by the enzymatic reaction of T. versicolor laccase [161].
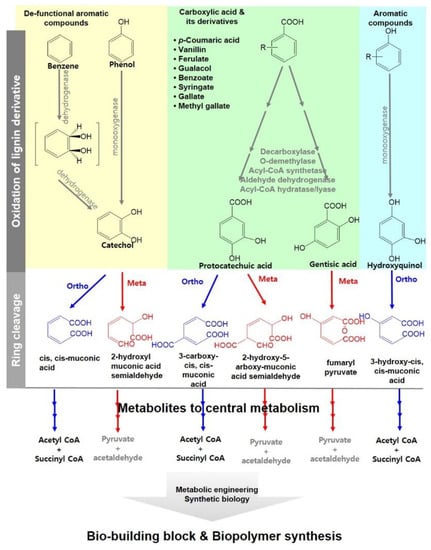
Figure 5.
Metabolic pathways for biodegradation of aromatic compounds derived from depolymerized lignin and its derivative for lignin valorization.
These results showed that bioconversion processes for lignin valorization could be potential cell factories for the production of a variety of renewable chemicals and materials through metabolic engineering and synthetic biology technology. However, the biological processes still have lower productivities and yields than chemical processes due to the physiochemical properties of lignin and its derivatives and their toxicities.
The alternative utilization of lignin as a functional material source without depolymerization has been intensively studied. Although the physicochemical properties of lignin are relatively reactive, immiscibility, strong inter- and intra-hydrogen bonding in complex aromatic compounds, and laccase-mediated grafting can modify functional groups and convert their characteristics by introducing functional molecules (Figure 4b). Enzyme grafting has demonstrated the coupling of various functional molecules such as sulfonation, fluorophnols, acrylamide, fatty acids, epoxides, long-chain alkylamine, etc., onto hydroxyl group moieties in lignin or lignin-derived structures [150]. Among these functionalized lignins, the conjugation with sulfanilic acid or p-aminobenzoic acid onto lignin also showed enhanced miscibility for the application of concrete dispersing agents [151].
In addition to small-molecule conjugation, macromolecule grafting with proteins, polysaccharides, and oligomeric compounds can also be broadly applied to synthesize functionalized novel biopolymers or synthetic polymers exhibiting mechanical strength, antibacterial activity, and antioxidant activity via laccase-mediator systems with ABTS, TEMPO, HOBt, and HPI [161]. Examples of such materials include linen–chitosan and linen–catechin coupling, silver nanoparticle composite, polyaniline-functionalized cotton, and catechin or gallic acid oligomer-coated wool. Additionally, laccase-assisted copolymerization enables the conversion of kraft lignin with methyl-hydroquinone and a trithiol to lignin-core hyperbranched copolymers [162,163]. In the future, the bioconversion of lignin into value-added materials and chemicals via laccase-assisted copolymerization could be expanded to develop novel biomaterials as well as green products based on renewable biomass resources.
7. Concluding Remarks and Future Perspective
Even though the chemical properties of lignin and its derivatives, its biodegradation, and related enzymes have been studied intensively, their application in biorefinery-derived platform chemicals and materials is still in the proof-of-concept phase. To date, the individual processes or techniques for lignin utilization have been studied at the tube-, flask-, and bench-scales. Nevertheless, emerging technologies for the scale-up and industrial application face bottlenecks of bio-based processes for lignin valorization from the raw lignin resource to final products for chemicals or materials. The following prospective could be considered for developing the techniques for lignin valorization in the production of platform chemicals and materials within a biorefinery:
(1) Lignin is a macromolecule consisting of heterogeneously aromatic polymer compounds, containing syringyl-, gualacryl-, and hydroxyphenyl-subunits. Although the predominant subunit composition ratios are different depending on the plant species, these aromatic compounds are potential substances for renewable chemicals, which can be obtained through lignin depolymerization with thermal or catalytic deconstruction. Depolymerized lignin subunits could fractionate based on their chemical properties for further manipulation processes. In addition, these depolymerization processes would need high-energy consumption steps such as the pretreatment of lignocellulose. Mushroom ligninolytic enzymes could be potential biocatalysts for the initial depolymerization step, combined with mild condition extraction by organosolv or ionic liquid from lignocellulose [164,165]. Mushroom ligninolytic enzymes are secreted proteins including laccases, LiPs, MnPs, VPs, and other oxidases, in the solid-state fermentation [37]. The enzymatic processes with secreted oxidase proteomes for lignin depolymerization from the solid phase of lignocellulosic biomass could reduce energy consumption and byproducts. Moreover, the residual biomass after these treatments could be also utilized further for biorefinery products.
(2) Although biocatalyst processes are an eco-friendly technology, the production of ligninolytic enzymes incurs higher financial costs in the scale-up process in lignin valorization rather than chemical catalysts. Since effective and powerful oxidases are produced from higher eukaryotic microorganisms, such as fungi, most researchers have developed a yeast or fungus expression system, such as P. pastoris, K. lactis, S. cerevisiae, A. niger, and A. oryzae, for recombinant protein production considering the secretion and glycosylation of an active enzyme [47,49,57,62,71,75,81]. These heterologous expression systems for the recombinant ligninolytic enzymes should enhance protein productivity to reduce the production costs of renewable chemicals. In addition, the secreted proteins in the solid-state fermentation after mushroom harvesting could be useful resources to obtain native enzymes through further formulation processes. This topic should be further studied to identify proteomes for ligninolysis and establish a novel protocol for the formulation of crude proteins extracted from the solid media.
(3) The bioconversion or biocatalytic process for lignin valorization faces many challenges due to the characteristics of biological molecules such as instability, toxicity, and solubility of lignin-derivative molecules, as well as the strict reaction conditions. Most aromatic compounds derived from depolymerized and fractionated lignin subunits are also hydrophobic, insoluble, and toxic properties for microorganisms. Thus, the high concentration of aromatic derivatives cannot be transformed into products by biological processes. Nevertheless, bioprocesses would still be attractive methods for eco-friendly sophisticated reactions that are simple, low-energy consumption processes without byproducts that can convert lignin monomers to usable chemicals and materials. The disadvantages associated with biological conversion may be overcome by the development of novel chemoenzymatic or hydride processes with the help of chemical catalysts or by improved and optimized unit process designs. Novel emerging technologies should be researched to combine biological and chemical processes.
(4) To date, most research on lignin valorization has been performed at small scales. Utilizing lignin and its derivatives for commercialized products is clearly still in the early phase. To evaluate the economic feasibility of lignin conversion processes, a pilot scheme at a large scale should be established to estimate various aspects of the process design to achieve a commercial scale. These approaches might help to determine whether current technologies can be used to produce lignin-based chemicals or materials, identify the advantages and drawbacks in the process, and assess the modifications or new technologies required to help develop this green technology process.
(5) The last point of view we consider here is the life cycle assessment (LCA) of lignin valorization. Ideally, lignin conversion would reduce environmental impacts by saving energy in comparison with conventional petroleum-based products. Recently, an LCA for the production of lignin-derived adipic acid and catechol showed that it produces less CO2 emissions and byproducts [166,167,168]. Most LCAs performed to data have concluded that lignin-based products offer better environmental performance than petroleum-based products due to their reduced impact in terms of climate change. Nevertheless, over recent decades, the chemicals and materials derived from petroleum oil have been produced under optimized processes to minimize their negative effects on the environment to survive in the competitive market with bio-based products. However, it is not yet known whether lignin valorization would be a more environmentally friendly process than oil refinery products because the procedures used to create the final products have not yet been optimized, and the market processes of these products are uncertain [169,170,171]. Further research on lignin valorization should focus on the environmental impact and cost of processes based on both a techno-economic analysis and a life cycle analysis, and further designs should be developed to optimize the processes involved.
Funding
This work was supported by the basic science research program (NRF-2021R1A2C1005811) through the National Research Foundation of Korea (NRF) and partially by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (IPET), funded by Ministry of Agriculture, Food, and Rural Affairs (1545021966) and the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program grant (KGM5482113).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author declares no conflict of interest.
References
- Brethauer, S.; Studer, M.H. Biochemical conversion processes of lignocellulosic biomass to fuels and chemicals—A review. Chimia 2015, 69, 572–581. [Google Scholar] [CrossRef]
- Capolupo, L.; Faraco, V. Green methods of lignocellulose pretreatment for biorefinery development. Appl. Microbiol. Biotechnol. 2016, 100, 9451–9467. [Google Scholar] [CrossRef]
- Silveira, M.H.; Morais, A.R.; da Costa Lopes, A.M.; Olekszyszen, D.N.; Bogel-Łukasik, R.; Andreaus, J.; Pereira, L.R. Current pretreatment technologies for the development of cellulosic ethanol and biorefineries. ChemSusChem 2015, 8, 3366–3390. [Google Scholar] [CrossRef]
- Shen, Y.; Jarboe, L.; Brown, R.; Wen, Z. A thermochemical-biochemical hybrid processing of lignocellulosic biomass for producing fuels and chemicals. Biotechnol. Adv. 2015, 33, 1799–1813. [Google Scholar] [CrossRef]
- Yoo, C.G.; Meng, X.; Pu, Y.; Ragauska, A.J. The critical role of lignin in lignocellulosic biomass conversion and recent pretreatment strategies: A comprehensive review. Bioresour Technol. 2020, 301, 122784. [Google Scholar] [CrossRef]
- Cantero, D.; Jara, R.; Navarrete, A.; Pelaz, L.; Queiroz, J.; Rodríguez-Rojo, S.; Cocero, M.J. Pretreatment processes of biomass for biorefineries: Current status and prospects. Annu. Rev. Chem. Biomol. Eng. 2019, 10, 289–310. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.M.; Seo, J.W.; Kim, C.H. Sequential acid-/alkali-pretreatment of empty palm fruit bunch fiber. Bioresour. Technol. 2012, 109, 229–233. [Google Scholar] [CrossRef]
- Kim, S. Xylitol production from byproducts generated during sequential acid-/alkali-pretreatment of empty palm fruit bunch fiber by an adapted Candida tropicalis. Front. Energy Res. 2019, 7, 72. [Google Scholar] [CrossRef]
- Kim, S.; Kim, C.H. Bioethanol production using the sequential acid/alkali-pretreated empty palm fruit bunch fiber. Renew. Energ. 2013, 54, 150–155. [Google Scholar] [CrossRef]
- Kim, S. Enhancing bioethanol productivity using alkali-pretreated empty palm fruit bunch fiber hydrolysate. BioMed Res. Int. 2018, 2018, 5272935. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Sung, B.H. Isolation and characterization of the stress-tolerant Candida tropicalis YHJ1 and evaluation of its xylose reductase for xylitol production from acid pre-treatment wastewater. Front. Bioeng. Biotechnol. 2019, 7, 138. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.-D.; Sohn, S.Y. Evaluation of the wastewater generated during alkaline pretreatment of biomass for feasibility of recycling and reusing. Renew. Energ. 2020, 155, 1156–1164. [Google Scholar] [CrossRef]
- Humbird, D.; Davis, R.; Tao, L.; Kinchin, C.; Hsu, D.; Aden, A.; Schoen, P.; Lukas, J.; Olthof, B.; Worley, M.; et al. Process Design and Economics for Biochemical Conversion of Lignocellulosic Biomass to Ethanol. Dilute-Acid Pretreatment and Enzymatic Hydrolysis of Corn Stover, NREL/TP-5100-47764. 2011. Available online: https://www.nrel.gov/docs/fy11osti/47764.pdf (accessed on 30 June 2021).
- Liu, W.; Wang, J.; Richard, T.L.; Hartley, D.S.; Spatari, S.; Volk, T.A. Economic and life cycle assessments of biomass utilization for bioenergy products. Biofuel Bioprod. Biorefin. 2017, 11, 633–647. [Google Scholar] [CrossRef]
- Zakzeski, J.; Bruijnincx, P.C.; Jongerius, A.L.; Weckhuysen, B.M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010, 110, 3552–3599. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, P.; Keller, M.; et al. Lignin valorization: Improving lignin processing in the biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef]
- Pollegioni, L.; Tonin, F.; Rosini, E. Lignin-degrading enzymes. FEBS J. 2015, 282, 1190–1213. [Google Scholar] [CrossRef]
- Plácido, J.; Capareda, S. Ligniolytic enzymes: A biotechnological alternative for bioethanol production. Bioresour. Bioprocess 2015, 2, 23. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Swiderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczynski, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Alexander, M. Nonbiodegradable and other recalcitrant molecules. Biotechnol. Bioeng. 1973, 15, 611–647. [Google Scholar] [CrossRef]
- Brunow, G.; Lundquist, K. Functional groups and bonding patterns in lignin (including the lignin-carbohydrate complexes). In Lignin and Lignan: Advances in Chemistry; CRC Press, Taylor Francis Group: New York, NY, USA, 2010; pp. 267–299. [Google Scholar]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant. Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Scalbert, A.; Monties, B.; Lallemand, J.-Y.; Guittet, E.; Rolando, C. Ether linkage between phenolic acids and lignin fractions from wheat straw. Phytochemistry 1985, 24, 1359–1362. [Google Scholar] [CrossRef]
- Muller-Harvey, I.; Hartley, R.D. Linkage of p-coumaroyl and feruloyl groups to cell-wall polysaccharides of barley straw. Carbohydr. Res. 1986, 148, 71–85. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Ralph, J.; Peng, J.P.; Lu, F.C.; Hatfield, R.D.; Helm, R.F. Are lignins optically active? J. Agric. Food Chem. 1999, 47, 2991–2996. [Google Scholar] [CrossRef]
- Zhu, X.; Akiyama, T.; Yokoyama, T.; Matsumoto, Y. Lignin-biosynthetic study: Reactivity of quinone methides in the diastereopreferential formation of p-hydroxyphenyl- and guaiacyl-type β-O-4 structures. J. Agric. Food Chem. 2019, 67, 2139–2147. [Google Scholar] [CrossRef]
- Novaes, E.; Kirst, M.; Chiang, V.; Winter-Sederoff, H.; Sederoff, R. Lignin and biomass: A negative correlation for wood formation and lignin content in trees. Plant. Physiol. 2010, 154, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Fridrich, B.; de Santi, A.; Elangovan, S.; Barta, K. Bright side of lignin depolymerization: Toward new platform chemicals. Chem. Rev. 2018, 118, 614–678. [Google Scholar] [CrossRef]
- Strassberger, Z.; Tanase, S.; Rothenberg, G. The pros and cons of lignin valorization in an integrated biorefinery. RSC Adv. 2014, 4, 25310–25318. [Google Scholar] [CrossRef]
- Collard, F.-X.; Blin, J. A review on pyrolysis of biomass constituents: Mechanisms and composition of the products obtained from the conversion of cellulose, hemicelluloses and lignin. Renew. Sustain. Energy Rev. 2014, 38, 594–608. [Google Scholar] [CrossRef]
- Kirubakaran, V.; Sivaramakrishnan, V.; Nalini, R.; Sekar, T.; Premalatha, M.; Subramanian, P. A review on gasification of biomass. Renew. Sustain. Energy Rev. 2009, 13, 179–186. [Google Scholar] [CrossRef]
- Ten Have, R.; Teunissen, P.J.M. Oxidative mechanisms involved in lignin degradation by white-rot fungi. Chem. Rev. 2001, 101, 3397–3414. [Google Scholar] [CrossRef]
- Atlas, R.M.; Bartha, R. Biogeochemical cycling: Carbon, hydrogen, and oxygen. In Microbial Ecology: Fundamentals and Applications, 3rd ed.; The Benjamin/Cummings Publishing Co. Inc.: Redwood City, CA, USA, 1993; pp. 289–313. [Google Scholar]
- Tian, J.-H.; Pourcher, A.-M.; Bochez, T.; Gelhaye, E.; Peu, P. Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Appl. Microbiol. Biotechnol. 2014, 98, 9527–9544. [Google Scholar] [CrossRef]
- Bugg, T.D.H.; Rahmanpour, R. Enzymatic conversion of lignin into renewable chemicals. Curr. Opin. Chem. Biol. 2015, 29, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Erjavec, J.; Kos, J.; Ravnikar, M.; Dreo, T.; Sabotič, J. Proteins of higher fungi-from forest to application. Trends Biotechnol. 2012, 30, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.; Spiess, A.C. Laccases for biorefinery applications: A critical review on challenges and perspectives. Bioprocess Bioyst. Eng. 2015, 38, 2285–2313. [Google Scholar] [CrossRef] [PubMed]
- Senthivelan, T.; Kanagaraj, J.; Panda, R.C. Recent trends in fungal laccase for various industrial applications: An eco-friendly approach—A review. Biotechnol. Bioprocess Eng. 2016, 21, 19–38. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; de Los Santos, M.H.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Fact. 2019, 18, 200. [Google Scholar] [CrossRef]
- Maestre-Reyna, M.; Liu, W.C.; Jeng, W.Y.; Lee, C.C.; Hsu, C.A.; Wen, T.N.; Wang, A.H.; Shyur, L.F. Structural and functional roles of glycosylation in fungal laccase from Lentinus sp. PLoS ONE 2015, 10, e0120601. [Google Scholar] [CrossRef]
- Taborda, C.P.; da Silva, M.B.; Nosanchuk, J.D.; Travassos, L.R. Melanin as a virulence factor of Paracoccidioides brasiliensis and other dimorphic pathogenic fungi: A minireview. Mycopathologia 2008, 165, 331–339. [Google Scholar] [CrossRef]
- Bonnen, A.M.; Anton, L.H.; Orth, A.B. Lignin-degrading enzymes of the commercial button mushroom, Agaricus bisporus. Appl. Environ. Microbiol. 1994, 60, 960–965. [Google Scholar] [CrossRef]
- Othman, A.M.; Elsayed, M.A.; Elshafei, A.M.; Hassan, M.M. Purification and biochemical characterization of two isolated laccase isoforms from Agaricus bisporus CU13 and their potency in dye decolorization. Int. J. Biol. Macromol. 2018, 113, 1142–1148. [Google Scholar] [CrossRef]
- Ullrich, R.; Huong, L.M.; Dung, N.L.; Hofrichter, M. Laccase from the medicinal mushroom Agaricus blazei: Production, purification and characterization. Appl. Microbiol. Biotechnol. 2005, 67, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto-Akanuma, A.; Akanuma, S.; Motoi, M.; Yamagishi, A.; Ohno, N. Cloning and characterization of laccase DNA from the royal sun medicinal mushroom, Agaricus brasiliensis (higher Basidiomycetes). Int. J. Med. Mushrooms 2014, 16, 375–393. [Google Scholar] [CrossRef] [PubMed]
- Hamuro, Y.; Tajima, K.; Matsumoto-Akanuma, A.; Sakamoto, S.; Furukawa, R.; Yamagishi, A.; Ohno, N.; Akanuma, S. Characterization of a thermostable mutant of Agaricus brasiliensis laccase created by phylogeny-based design. J. Biosci. Bioeng. 2017, 124, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Caspersen, M.B.; Mondorf, K.; Halkier, T.; Skov, L.K.; Østergaard, P.R.; Brown, K.M.; Brown, S.H.; Xu, F. Characterization of a Coprinus cinereus laccase. Enzym. Microbiol. Technol. 1999, 25, 502–508. [Google Scholar] [CrossRef]
- Yaver, D.S.; Overjero, M.D.; Xu, F.; Nelson, B.A.; Brown, K.M.; Halkier, T.; Bernauer, S.; Brown, S.H.; Kauppinen, S. Molecular characterization of laccase genes from the basidiomycete Coprinus cinereus and heterologous expression of the laccase lcc1. Appl. Environ. Microbiol. 1999, 65, 4943–4948. [Google Scholar] [CrossRef]
- Ducros, V.; Brzozowski, A.; Wilson, K.; Brown, S.H.; Østergaard, P.; Schneider, P.; Yaver, D.S.; Pedersen, A.H.; Davies, G.J. Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nat. Struct. Mol. Biol. 1998, 5, 310–316. [Google Scholar] [CrossRef]
- Bao, S.; Teng, Z.; Ding, S. Heterologous expression and characterization of a novel laccase isoenzyme with dyes decolorization potential from Coprinus comatus. Mol. Biol. Rep. 2013, 40, 1927–1936. [Google Scholar] [CrossRef]
- Mishra, S.; Bisaria, V.S. Production and characterization of laccase from Cyathus bulleri and its use in decolorization of recalcitrant textile dyes. Appl. Microbiol. Biotechnol. 2006, 71, 646–653. [Google Scholar]
- Garg, N.; Bieler, N.; Kenzom, T.; Chhabra, M.; Ansorge-Schumacher, M.; Mishra, S. Cloning, sequence analysis, expression of Cyathus bulleri laccase in Pichia pastoris and characterization of recombinant laccase. BMC Biotechnol. 2012, 12, 75. [Google Scholar] [CrossRef]
- Perie, F.H.; Reddy, G.V.; Blackburn, N.J.; Gold, M.H. Purification and characterization of laccases from the white-rot basidiomycete Dichomitus squalens. Arch. Biochem. Biophys. 1998, 353, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Liu, X.; Chen, L.; Shen, Y.; Zhang, X.; Fang, W.; Wang, X.; Bao, X.; Xiao, Y. Identification of a laccase Glac15 from Ganoderma lucidum 77002 and its application in bioethanol production. Biotechnol. Biofuels 2015, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Nitheranont, T.; Watanabe, A.; Asada, Y. Extracellular laccase produced by an edible basidiomycetous mushroom, Grifola frondosa: Purification and characterization. Biosci. Biotechnol. Biochem. 2011, 75, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Nitheranont, T.; Watanabe, A.; Asada, Y. Heterologous expression of two minor laccase isozyme cDNAs from the edible mushroom Grifola frondosa. Biosci. Biotechnol. Biochem. 2017, 81, 2367–2369. [Google Scholar] [CrossRef]
- Zou, Y.J.; Wang, H.X.; Ng, T.B.; Huang, C.Y.; Zhang, J.X. Purification and characterization of a novel laccase from the edible mushroom Hericium coralloides. J. Microbiol. 2012, 50, 72–78. [Google Scholar] [CrossRef]
- Nagai, M.; Sato, T.; Watanabe, H.; Saito, K.; Kawata, M.; Enei, H. Purification and characterization of an extracellular laccase from the edible mushroom Lentinula edodes, and decolorization of chemically different dyes. Appl. Microbiol. Biotechnol. 2002, 60, 327–335. [Google Scholar] [PubMed]
- Nagai, M.; Kawata, M.; Watanabe, H.; Ogawa, M.; Saito, K.; Takesawa, T.; Kanda, K.; Sato, T. Important role of fungal intracellular laccase for melanin synthesis: Purification and characterization of an intracellular laccase from Lentinula edodes fruit bodies. Microbiology 2003, 149, 2455–2462. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Nakade, K.; Yano, A.; Nakagawa, Y.; Hirano, T.; Irie, T.; Watanabe, H.; Nagai, M.; Sato, T. Heterologous expression of lcc1 from Lentinula edodes in tobacco BY-2 cells results in the production an active, secreted form of fungal laccase. Appl. Microbiol. Biotechnol. 2008, 79, 971–980. [Google Scholar] [CrossRef]
- Wong, K.S.; Huang, Q.; Au, C.H.; Wang, J.; Kwan, H.S. Biodegradation of dyes and polyaromatic hydrocarbons by two allelic forms of Lentinula edodes laccase expressed from Pichia pastoris. Bioresour. Technol. 2012, 104, 157–164. [Google Scholar] [CrossRef]
- Wong, K.S.; Cheung, M.K.; Au, C.H.; Kwan, H.S. A novel Lentinula edodes laccase and its comparative enzymology suggest guaiacol-based laccase engineering for bioremediation. PLoS ONE 2013, 8, e66426. [Google Scholar] [CrossRef]
- Schückel, J.; Matura, A.; van Pée, K.H. One-copper laccase-related enzyme from Marasmius sp.: Purification, characterization and bleaching of textile dyes. Enzyme Microb. Technol. 2011, 48, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.J.; Lim, S.J. Purification and characterization of the laccase involved in dye decolorization by the white-rot fungus Marasmius scorodonius. J. Microbiol. Biotechnol. 2017, 27, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Dedeyan, B.; Klonowska, A.; Tagger, S.; Tron, T.; Iacazio, G.; Gil, G.; Le Petit, J. Biochemical and molecular characterization of a laccase from Marasmius quercophilus. Appl. Environ. Microbiol. 2000, 66, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Quaratino, D.; Federici, F.; Petruccioli, M.; Fenice, M.; D’Annibale, A. Production, purification and partial characterisation of a novel laccase from the white-rot fungus Panus tigrinus CBS 577.79. Antonie Leeuwenhoek 2007, 91, 57–69. [Google Scholar] [CrossRef]
- Ben Younes, S.; Mechichi, T.; Sayadi, S. Purification and characterization of the laccase secreted by the white rot fungus Perenniporia tephropora and its role in the decolourization of synthetic dyes. J. Appl. Microbiol. 2007, 102, 1033–1042. [Google Scholar] [CrossRef]
- Srinivasan, C.; Dsouza, T.M.; Boominathan, K.; Reddy, C.A. Demonstration of laccase in the white rot basidiomycete Phanerochaete chrysosporium BKM-F1767. Appl. Environ. Microbiol. 1995, 61, 4274–4277. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, C.; Guillén, F.; Martínez, A.T.; Martínez, M.J. Laccase isoenzymes of Pleurotus eryngii: Characterization, catalytic properties and participation in activation of molecular oxygen and Mn2+ oxidation. Appl. Environ. Microbiol. 1997, 63, 2166–2174. [Google Scholar] [CrossRef]
- Rodríguez, E.; Ruiz-Dueñas, F.J.; Kooistra, R.; Ram, A.; Martínez, A.T.; Martínez, M.J. Isolation of two laccase genes from the white-rot fungus Pleurotus eryngii and heterologous expression of the pel3 encoded protein. J. Biotechnol. 2008, 134, 9–19. [Google Scholar] [CrossRef]
- Ueda, M.; Shintani, K.; Nakanishi-Anjyuin, A.; Nakazawa, M.; Kusuda, M.; Nakatani, F.; Kawaguchi, T.; Tsujiyama, S.; Kawanishi, M.; Yagi, T.; et al. A protein from Pleurotus eryngii var. tuoliensis C.J. Mou with strong removal activity against the natural steroid hormone, estriol: Purification, characterization, and identification as a laccase. Enzyme Microb. Technol. 2012, 51, 402–407. [Google Scholar] [CrossRef]
- Das, N.; Chakraborty, T.K.; Mukherjee, M. Purification and characterization of a growth-regulating laccase from Pleurotus florida. J. Basic Microbiol. 2001, 41, 261–267. [Google Scholar] [CrossRef]
- Yuan, X.; Tian, G.; Zhao, Y.; Zhao, L.; Wang, H.; Ng, T.B. Biochemical characteristics of three laccase isoforms from the basidiomycete Pleurotus nebrodensis. Molecules 2016, 21, 203. [Google Scholar] [CrossRef]
- Piscitelli, A.; Giardina, P.; Mazzoni, C.; Sannia, G. Recombinant expression of Pleurotus ostreatus laccases in Kluyveromyces lactis and Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2005, 69, 428–439. [Google Scholar] [CrossRef]
- Palmieri, G.; Cennamo, G.; Sannia, G. Remazol Brilliant Blue R decolourisation by the fungus Pleurotus ostreatus and its oxidative enzymatic system. Enzym. Microb. Technol. 2005, 36, 17–24. [Google Scholar] [CrossRef]
- Giardina, P.; Autore, F.; Faraco, V.; Festa, G.; Palmieri, G.; Piscitelli, A.; Sannia, G. Structural characterization of heterodimeric laccases from Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 2007, 75, 1293–1300. [Google Scholar] [CrossRef]
- Faraco, V.; Ercole, C.; Festa, G.; Giardina, P.; Piscitelli, A.; Sannia, G. Heterologous expression of heterodimeric laccase from Pleurotus ostreatus in Kluyveromyces lactis. Appl. Microbiol. Biotechnol. 2008, 77, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Marques De Souza, C.G.; Peralta, R.M. Purification and characterization of the main laccase produced by the white-rot fungus Pleurotus pulmonarius on wheat bran solid state medium. J. Basic Microbiol. 2003, 43, 278–286. [Google Scholar] [CrossRef]
- Linke, D.; Bouws, H.; Peters, T.; Nimtz, M.; Berger, R.G.; Zorn, H. Laccases of Pleurotus sapidus: Characterization and cloning. J. Agric. Food Chem. 2005, 53, 9498–9505. [Google Scholar] [CrossRef] [PubMed]
- Soden, D.M.; O’Callaghan, J.; Dobson, A.D.W. Molecular cloning of a laccase isozyme gene from Pleurotus sajor-caju and expression in the heterologous Pichia pastoris host. Microbiology 2002, 148, 4003–4014. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.E. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: Purification and characterization of the laccase. Appl. Environ. Microbiol. 1996, 62, 1151–1158. [Google Scholar] [CrossRef]
- Ryan, S.; Schnitzhofer, W.; Tzanov, T.; Cavaco-Paulo, A.; Gübitz, G.M. An acid-stable laccase from Sclerotium rolfsii with potential for wool dye decolourization. Enzym. Microb. Technol. 2003, 33, 766–774. [Google Scholar] [CrossRef]
- Daroch, M.; Houghton, C.A.; Moore, J.K.; Wilkinson, M.C.; Carnell, A.J.; Bates, A.D.; Iwanejko, L.A. Glycosylated yellow laccases of the basidiomycete Stropharia aeruginosa. Enzym. Microb. Technol. 2014, 10, 58–59. [Google Scholar] [CrossRef] [PubMed]
- Zouari-Mechichi, H.; Mechichi, T.; Dhouib, A.; Sayadi, S.; Martínez, A.T.; Martínez, M.J. Laccase purification and characterization from Trametes trogii isolated in Tunisia: Decolorization of textile dyes by the purified enzyme. Enzym. Microb. Technol. 2006, 39, 141–148. [Google Scholar] [CrossRef]
- Bertrand, B.; Martinez-Morales, F.; Tinoco, R.; Rojas-Trejo, S.; Serrano-Carreon, L.; Trejo-Hernandez, M.R. Induction of laccases in Trametes versicolor by aqueous wood extracts. World J. Microbiol. Biotechnol. 2014, 30, 135–142. [Google Scholar] [CrossRef]
- Zheng, F.; An, Q.; Meng, G.; Wu, X.J.; Dai, Y.C.; Si, J.; Cui, B.K. A novel laccase from white rot fungus Trametes orientalis: Purification, characterization, and application. Int. J. Biol. Macromol. 2017, 102, 758–770. [Google Scholar] [CrossRef]
- Rudakiya, D.M.; Patel, D.H.; Gupte, A. Exploiting the potential of metal and solvent tolerant laccase from Tricholoma giganteum AGDR1 for the removal of pesticides. Int. J. Biol. Macromol. 2020, 144, 586–595. [Google Scholar] [CrossRef]
- Chen, S.; Ge, W.; Buswell, J.A. Biochemical and molecular characterization of a laccase from the edible straw mushroom, Volvariella volvacea. Eur. J. Biochem. 2004, 271, 318–328. [Google Scholar] [CrossRef]
- Palmieri, G.; Giardina, P.; Bianco, C.; Fontanella, B.; Sannia, G. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 2000, 66, 920–924. [Google Scholar] [CrossRef]
- Velázquez-Cedeño, M.; Farnet, A.M.; Billette, C.; Mata, G.; Savoie, J.M. Interspecific interactions with Trichoderma longibrachiatum induce Pleurotus ostreatus defense reactions based on the production of laccase isozymes. Biotechnol. Lett. 2007, 29, 1583–1590. [Google Scholar] [CrossRef]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar] [CrossRef]
- Courty, P.E.; Hoegger, P.J.; Kilaru, S.; Kohler, A.; Buée, M.; Garbaye, J.; Martin, F.; Kües, U. Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 2009, 182, 736–750. [Google Scholar] [CrossRef]
- Pezzella, C.; Lettera, V.; Piscitelli, A.; Giardina, P.; Sannia, G. Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl. Microbiol. Biotechnol. 2013, 97, 705–717. [Google Scholar] [CrossRef]
- Fernández-Alejandre, K.I.; Flores, N.; Tinoco-Valencia, R.; Caro, M.; Flores, C.; Galindo, E.; Serrano-Carreón, L. Diffusional and transcriptional mechanisms involved in laccases production by Pleurotus ostreatus CP50. J. Biotechnol. 2016, 223, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.D.H.; Costa, P.C.; Fontana, R.C.; de Siqueira, F.G.; Dillon, A.J.P.; Camassola, M. Upscale and characterization of lignin-modifying enzymes from Marasmiellus palmivorus VE111 in a bioreactor under parameter optimization and the effect of inducers. J. Biotechnol. 2019, 295, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Welinder, K.G. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 1992, 2, 388–393. [Google Scholar] [CrossRef]
- Morgenstern, I.; Klopman, S.; Hibbett, D.S. Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. J. Mol. Evol. 2008, 66, 243–257. [Google Scholar] [CrossRef]
- Coconi-Linares, N.; Magaña-Ortíz, D.; Guzmán-Ortiz, D.A.; Fernández, F.; Loske, A.M.; Gómez-Lim, M.A. High-yield production of manganese peroxidase, lignin peroxidase, and versatile peroxidase in Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 2014, 98, 9283–9294. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Dueñas, F.J.; Martínez, M.J.; Martínez, A.T. Molecular characterization of a novel peroxidase isolated from the ligninolytic fungus Pleurotus eryngii. Mol. Microbiol. 1999, 31, 223–235. [Google Scholar] [CrossRef]
- Fernández-Fueyo, E.; Castanera, R.; Ruiz-Dueñas, F.J.; López-Lucendo, M.F.; Ramírez, L.; Pisabarro, A.G.; Martínez, A.T. Ligninolytic peroxidase gene expression by Pleurotus ostreatus: Differential regulation in lignocellulose medium and effect of temperature and pH. Fungal Genet. Biol. 2014, 72, 150–161. [Google Scholar] [CrossRef]
- Singh, R.; Ahlawat, O.P.; Rajor, A. Identification of the potential of microbial combinations obtained from spent mushroom cultivation substrates for use in textile effluent decolorization. Bioresour. Technol. 2012, 125, 217–225. [Google Scholar] [CrossRef]
- Barr, D.P.; Shah, M.M.; Aust, S.D. Veratryl alcohol-dependent production of molecular oxygen by lignin peroxidase. J. Biol. Chem. 1993, 268, 241–244. [Google Scholar] [CrossRef]
- Bissaro, B.; Várnai, A.; Røhr, Å.K.; Eijsink, V.G.H. Oxidoreductases and reactive oxygen species in conversion of lignocellulosic biomass. Microbiol. Mol. Biol. Rev. 2018, 82, e00029-18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chandra, R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef] [PubMed]
- Biko, O.D.V.; Viljoen-Bloom, M.; van Zyl, W.H. Microbial lignin peroxidases: Applications, production challenges and future perspectives. Enzym. Microb. Technol. 2020, 141, 109669. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Bilal, M.; Iqbal, H.M.N.; Raj, A. Lignin peroxidase in focus for catalytic elimination of contaminants—A critical review on recent progress and perspectives. Int. J. Biol. Macromol. 2021, 177, 58–82. [Google Scholar] [CrossRef] [PubMed]
- Hiscox, J.; Baldrian, P.; Rogers, H.J.; Boddy, L. Changes in oxidative enzyme activity during interspecific mycelial interactions involving the white-rot fungus Trametes versicolor. Fungal Genet. Biol. 2010, 47, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Hildén, K.; Mäkelä, M.R.; Lankinen, P.; Lundell, T. Agaricus bisporus and related Agaricus species on lignocellulose: Production of manganese peroxidase and multicopper oxidases. Fungal Genet. Biol. 2013, 55, 32–41. [Google Scholar] [CrossRef]
- Vos, A.M.; Jurak, E.; Pelkmans, J.F.; Herman, K.; Pels, G.; Baars, J.J.; Hendrix, E.; Kabel, M.A.; Lugones, L.G.; Wösten, H.A.B. H2O2 as a candidate bottleneck for MnP activity during cultivation of Agaricus bisporus in compost. AMB Express 2017, 7, 124. [Google Scholar] [CrossRef]
- Wariishi, H.; Valli, K.; Gold, M.H. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992, 267, 23688–23695. [Google Scholar] [CrossRef]
- Hofrichter, M. Review: Lignin conversion by manganese peroxidase (MnP). Enzym. Microb. Technol. 2002, 30, 454–466. [Google Scholar] [CrossRef]
- Camarero, S.; Sarkar, S.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Martínez, A.T. Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J. Biol. Chem. 1999, 274, 10324–10330. [Google Scholar] [CrossRef]
- Heinfling, A.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Bergbauer, M.; Szewzyk, U.; Martínez, A.T. A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett. 1998, 428, 141–146. [Google Scholar] [CrossRef]
- Kamitsuji, H.; Watanabe, T.; Honda, Y.; Kuwahara, M. Direct oxidation of polymeric substrates by multifunctional manganese peroxidase isoenzyme from Pleurotus ostreatus without redox mediators. Biochem. J. 2005, 386, 387–393. [Google Scholar] [CrossRef]
- Knop, D.; Ben-Ari, J.; Salame, T.M.; Levinson, D.; Yarden, O.; Hadar, Y. Mn²-deficiency reveals a key role for the Pleurotus ostreatus versatile peroxidase (VP4) in oxidation of aromatic compounds. Appl. Microbiol. Biotechnol. 2014, 98, 6795–6804. [Google Scholar] [CrossRef] [PubMed]
- Ayuso-Fernández, I.; Ruiz-Dueñas, F.J.; Martínez, A.T. Evolutionary convergence in lignin-degrading enzymes. Proc. Natl. Acad. Sci. USA 2018, 115, 6428–6433. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Dueñas, F.J.; Barrasa, J.M.; Sánchez-García, M.; Camarero, S.; Miyauchi, S.; Serrano, A.; Linde, D.; Babiker, R.; Drula, E.; Ayuso-Fernández, I.; et al. Genomic analysis enlightens Agaricales lifestyle evolution and increasing peroxidase diversity. Mol. Biol. Evol. 2021, 38, 1428–1446. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Boada, M.; Ruiz-Dueñas, F.J.; Pogni, R.; Basosi, R.; Choinowski, T.; Martínez, M.J.; Piontek, K.; Martínez, A.T. Versatile peroxidase oxidation of high redox potential aromatic compounds: Site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J. Mol. Biol. 2005, 354, 385–402. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Dueñas, F.J.; Morales, M.; Pérez-Boada, M.; Choinowski, T.; Martínez, M.J.; Piontek, K.; Martínez, A.T. Manganese oxidation site in Pleurotus eryngii versatile peroxidase: A site-directed mutagenesis, kinetic, and crystallographic study. Biochemistry 2007, 46, 66–77. [Google Scholar] [CrossRef]
- Ruiz-Dueñas, F.J.; Morales, M.; Mate, M.J.; Romero, A.; Martínez, M.J.; Smith, A.T.; Martínez, A.T. Site-directed mutagenesis of the catalytic tryptophan environment in Pleurotus eryngii versatile peroxidase. Biochemistry 2008, 47, 1685–1695. [Google Scholar] [CrossRef]
- Knop, D.; Yarden, O.; Hadar, Y. The ligninolytic peroxidases in the genus Pleurotus: Divergence in activities, expression, and potential applications. Appl. Microbiol. Biotechnol. 2015, 99, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Knop, D.; Levinson, D.; Makovitzki, A.; Agami, A.; Lerer, E.; Mimran, A.; Yarden, O.; Hadar, Y. Limits of versatility of versatile peroxidase. Appl. Environ. Microbiol. 2016, 82, 4070–4080. [Google Scholar] [CrossRef]
- Salvachúa, D.; Prieto, A.; Mattinen, M.L.; Tamminen, T.; Liitiä, T.; Lille, M.; Willför, S.; Martínez, A.T.; Martínez, M.J.; Faulds, C.B. Versatile peroxidase as a valuable tool for generating new biomolecules by homogeneous and heterogeneous cross-linking. Enzym. Microb. Technol. 2013, 52, 303–311. [Google Scholar] [CrossRef]
- Wang, X.; Yao, B.; Su, X. Linking enzymatic oxidative degradation of lignin to organics detoxification. Int. J. Mol. Sci. 2018, 19, 3373. [Google Scholar] [CrossRef]
- Singh, R.; Eltis, L.D. The multihued palette of dye-decolorizing peroxidases. Arch. Biochem. Biophys. 2015, 574, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Linde, D.; Ruiz-Dueñas, F.J.; Fernández-Fueyo, E.; Guallar, V.; Hammel, K.E.; Pogni, R.; Martínez, A.T. Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance. Arch. Biochem. Biophys. 2015, 574, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Shoda, M. Purification and characterization of a novel peroxidase from Geotrichum candidum dec 1 involved in decolorization of dyes. Appl. Environ. Microbiol. 1999, 65, 1029–1035. [Google Scholar] [CrossRef]
- Liers, C.; Pecyna, M.J.; Kellner, H.; Worrich, A.; Zorn, H.; Steffen, K.T.; Hofrichter, M.; Ullrich, R. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl. Microbiol. Biotechnol. 2013, 97, 5839–5849. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A.; Schierhorn, A.; Lindenstrauss, U.; Lilie, H.; Brüser, T. YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J. Biol. Chem. 2006, 281, 13972–13978. [Google Scholar] [CrossRef]
- Catucci, G.; Valetti, F.; Sadeghi, S.J.; Gilardi, G. Biochemical features of dye-decolorizing peroxidases: Current impact on lignin degradation. Biotechnol. Appl. Biochem. 2020, 67, 751–759. [Google Scholar] [CrossRef]
- Ander, P.; Marzullo, L. Sugar oxidoreductases and veratryl alcohol oxidase as related to lignin degradation. J. Biotechnol. 1997, 53, 115–131. [Google Scholar] [CrossRef]
- Daou, M.; Faulds, C.B. Glyoxal oxidases: Their nature and properties. World J. Microbiol. Biotechnol. 2017, 33, 87. [Google Scholar] [CrossRef]
- Martínez, A.T.; Ruiz-Dueñas, F.J.; Camarero, S.; Serrano, A.; Linde, D.; Lund, H.; Vind, J.; Tovborg, M.; Herold-Majumdar, O.M.; Hofrichter, M.; et al. Oxidoreductases on their way to industrial biotransformations. Biotechnol. Adv. 2017, 35, 815–831. [Google Scholar] [CrossRef] [PubMed]
- Ninagawa, S.; George, G.; Mori, K. Mechanisms of productive folding and endoplasmic reticulum-associated degradation of glycoproteins and non-glycoproteins. Biochim. Biophys. Acta. 2021, 1865, 129812. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Hoyos, C.; Morales-Álvarez, E.D.; Poutou-Piñales, R.A.; Pedroza-Rodríguez, A.M.; Rodríguez-Vázquez, R.; Delgado-Boada, J.M. Fungal laccases. Fungal Biol. Rev. 2013, 27, 67–82. [Google Scholar] [CrossRef]
- Reiss, R.; Ihssen, J.; Richter, M.; Eichhorn, E.; Schilling, B.; Thöny-Meyer, L. Laccase versus laccase-like multi-copper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PLoS ONE 2013, 8, e65633. [Google Scholar] [CrossRef]
- Bourbonnais, R.; Paice, M.G. Oxidation of non-phenolic substrates-an expanded role for laccase in lignin biodegradation. FEBS Lett. 1990, 127, 99–102. [Google Scholar] [CrossRef]
- Díaz-González, M.; Vidal, T.; Tzanov, T. Phenolic compounds as enhancers in enzymatic and electrochemical oxidation of veratryl alcohol and lignins. Appl. Microbiol. Biotechnol. 2011, 89, 1693–1700. [Google Scholar] [CrossRef]
- Mehra, R.; Muschiol, J.; Meyer, A.S.; Kepp, K.P. A structural-chemical explanation of fungal laccase activity. Sci. Rep. 2018, 8, 17285. [Google Scholar] [CrossRef]
- Song, Y.; Jiang, J.; Qin, W.; Li, J.; Zhou, Y.; Gao, Y. Enhanced transformation of organic pollutants by mild oxidants in the presence of synthetic or natural redox mediators: A review. Water Res. 2021, 189, 116667. [Google Scholar] [CrossRef]
- Jeon, J.R.; Murugesan, K.; Kim, Y.M.; Kim, E.J.; Chang, Y.S. Synergistic effect of laccase mediators on pentachlorophenol removal by Ganoderma lucidum laccase. Appl. Microbiol. Biotechnol. 2008, 81, 783–790. [Google Scholar] [CrossRef]
- Munk, L.; Sitarz, A.K.; Kalyani, D.C.; Mikkelsen, D.; Meyer, A.S. Can laccases catalyze bond cleavage in lignin? Biotechnol. Adv. 2015, 33, 13–24. [Google Scholar] [CrossRef]
- Ashe, B.; Nguyen, L.N.; Hai, F.I.; Lee, D.-J.; Van de Merwe, J.P.; Leusch, F.D.L.; Price, W.E.; Nghiem, L.D. Impacts of redox-mediator type on trace organic contaminants degradation by laccase: Degradation efficiency, laccase stability and effluent toxicity. Int. Biodeterior. Biodegrad. 2016, 113, 169–176. [Google Scholar] [CrossRef]
- Singh, G.; Arya, S.K. Utility of laccase in pulp and paper industry: A progressive step towards the green technology. Int. J. Biol. Macromol. 2019, 134, 1070–1084. [Google Scholar] [CrossRef]
- Tramontina, R.; Brenelli, L.B.; Sodré, V.; Franco Cairo, J.P.; Travália, B.M.; Egawa, V.Y.; Goldbeck, R.; Squina, F.M. Enzymatic removal of inhibitory compounds from lignocellulosic hydrolysates for biomass to bioproducts applications. World J. Microbiol. Biotechnol. 2020, 36, 166. [Google Scholar] [CrossRef]
- Palonen, H.; Viikari, L. Role of oxidative enzymatic treatments on enzymatic hydrolysis of softwood. Biotechnol. Bioeng. 2004, 86, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Brenelli, L.; Squina, F.M.; Felby, C.; Cannella, D. Laccase-derived lignin compounds boost cellulose oxidative enzymes AA9. Biotechnol. Biofuels 2018, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.; Guebitz, G.M.; Pellis, A.; Nyanhongo, G.S. Harnessing the power of enzymes for tailoring and valorizing lignin. Trends Biotechnol. 2020, 38, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, D.; Heck, T.; Schubert, M.; Yerlikaya, A.; Weymuth, C.; Rentsch, D.; Schober, I.; Richter, M. Enzymatic synthesis of lignin-based concrete dispersing agents. Chembiochem 2018, 19, 1365–1369. [Google Scholar] [CrossRef]
- Linger, J.G.; Vardon, D.R.; Guarnieri, M.T.; Karp, E.M.; Hunsinger, G.B.; Franden, M.A.; Johnson, C.W.; Chupka, G.; Strathmann, T.J.; Pienkos, P.T.; et al. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. USA 2014, 111, 12013–12018. [Google Scholar] [CrossRef]
- Sonoki, T.; Morooka, M.; Sakamoto, K.; Otsuka, Y.; Nakamura, M.; Jellison, J.; Goodell, B. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J. Biotechnol. 2014, 192, 71–77. [Google Scholar] [CrossRef]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. A field of dreams: Lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 2019, 37, 107360. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, Y. Biotransformation of lignin: Mechanisms, applications and future work. Biotechnol. Prog. 2020, 36, e2922. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; You, S.; Beiyuan, J.; Luo, G.; Gupta, J.; Kumar, S.; Singh, L.; Zhang, S.; Tsang, D.C.W. Lignin valorization by bacterial genus Pseudomonas: State-of-the-art review and prospects. Bioresour. Technol. 2021, 320, 124412. [Google Scholar] [CrossRef]
- Martău, G.A.; Călinoiu, L.-F.; Vodnar, D.C. Bio-vanillin: Towards a sustainable industrial production. Trends Food Sci. Technol. 2021, 109, 579–592. [Google Scholar] [CrossRef]
- Elmore, J.R.; Dexter, G.N.; Salvachúa, D.; Martinez-Baird, J.; Hatmaker, E.A.; Huenemann, J.D.; Klingeman, D.M.; Peabody, G.L.; Peterson, D.J.; Singer, C.; et al. Production of itaconic acid from alkali pretreated lignin by dynamic two stage bioconversion. Nat. Commun. 2021, 12, 2261. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.C. Emerging biotechnologies for production of itaconic acid and its applications as a platform chemical. J. Ind. Microbiol. Biotechnol. 2017, 44, 303–315. [Google Scholar] [CrossRef]
- Williamson, J.J.; Bahrin, N.; Hardiman, E.M.; Bugg, T.D.H. Production of substituted styrene bioproducts from lignin and lignocellulose using engineered Pseudomonas putida KT2440. Biotechnol. J. 2020, 15, e1900571. [Google Scholar] [CrossRef]
- Slagman, S.; Zuilhof, H.; Franssen, M.C.R. Laccase-mediated grafting on biopolymers and synthetic polymers: A critical review. Chembiochem. 2018, 19, 288–311. [Google Scholar] [CrossRef]
- Cannatelli, M.D.; Ragauskas, A.J. Laccase-mediated synthesis of lignin-core hyperbranched copolymers. Appl. Microbiol. Biotechnol. 2017, 101, 6343–6353. [Google Scholar] [CrossRef]
- Cannatelli, M.D.; Ragauskas, A.J. Conversion of lignin into value-added materials and chemicals via laccase-assisted copolymerization. Appl. Microbiol. Biotechnol. 2016, 100, 8685–8691. [Google Scholar] [CrossRef]
- Thoresen, P.P.; Matsakas, L.; Rova, U.; Christakopoulos, P. Recent advances in organosolv fractionation: Towards biomass fractionation technology of the future. Bioresour. Technol. 2020, 306, 123189. [Google Scholar] [CrossRef]
- Corona, A.; Biddy, M.J.; Vardon, D.R.; Birkved, M.; Hauschild, M.Z.; Beckham, G.T. Life cycle assessment of adipic acid production from lignin. Green Chem. 2018, 20, 3857–3866. [Google Scholar] [CrossRef]
- Van Duuren, J.B.J.H.; de Wild, P.J.; Starck, S.; Bradtmöller, C.; Selzer, M.; Mehlmann, K.; Schneider, R.; Kohlstedt, M.; Poblete-Castro, I.; Stolzenberger, J.; et al. Limited life cycle and cost assessment for the bioconversion of lignin-derived aromatics into adipic acid. Biotechnol. Bioeng. 2020, 117, 1381–1393. [Google Scholar] [CrossRef]
- Montazeri, M.; Eckelman, M.J. Life cycle assessment of catechols from lignin depolymerization. ACS Sustain. Chem. Eng. 2016, 4, 708–718. [Google Scholar] [CrossRef]
- Bajwa, D.S.; Pourhashem, G.; Ullah, A.H.; Bajwa, S.G. A concise review of current lignin production, applications, products and their environmental impact. Ind. Crops Prod. 2019, 139, 111526. [Google Scholar] [CrossRef]
- Hermansson, F.; Janssen, M.; Svanström, M. Allocation in life cycle assessment of lignin. Int. J. Life Cycle Assess. 2020, 25, 1620–1632. [Google Scholar] [CrossRef]
- Moretti, C.; Corona, B.; Hoefnagels, R.; Vural-Gürsel, I.; Gosselink, R.; Junginger, M. Review of life cycle assessments of lignin and derived products: Lessons learned. Sci. Total Environ. 2021, 770, 144. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).