Abstract
Interferons (IFN) are an assemblage of signaling proteins made and released by various host cells in response to stimuli, including viruses. Respiratory syncytial virus (RSV), influenza virus, and SARS-CoV-2 are major causes of respiratory disease that induce or antagonize IFN responses depending on various factors. In this review, the role and function of type I, II, and III IFN responses to respiratory virus infections are considered. In addition, the role of the viral proteins in modifying anti-viral immunity is noted, as are the specific IFN responses that underly the correlates of immunity and protection from disease.
Keywords:
respiratory syncytial virus; RSV; influenza; SARS-CoV-2; COVID-19; interferons; IFN; immunity 1. Interferons
Interferons (IFNs) were first discovered during a study on viral interference in egg chorioallantoic membranes. The early study found that a factor was released which had the ability to induce viral interference, hence “interferon” [1]. This soluble IFN was detected within 3 h of infection and was found to confer resistance to viral replication in treated cells before infection [1,2]. There are now three known types of IFNs: type I, II, and III, each with distinct and overlapping functions which signal through different receptors. Type I IFNs signal through the IFN alpha receptor (IFNAR), type II signaling occurs through the IFN gamma receptor (IFNGR), and type III IFN signaling occurs through the IFN lambda receptor (IFNLR) [1,3,4,5]. Type I IFN consists of 14 subtypes of IFNα, INFß, and IFNκ and IFNω [6]. Most cell types are capable of secreting IFNß, but plasmacytoid dendritic cells (pDCs) and other innate immune cells are the primary producers of IFNα [7]. IFNγ is the only type II IFN [8]. Adaptive and innate immune cells, including T helper 1 (Th1) cells, CD8+ cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and innate lymphoid cells (ILCs), are responsible for the majority of IFNγ production [9]. IFNγ expression is linked to cell-mediated immunity against intracellular pathogens/viruses, macrophage activation and polarization towards an M1 phenotype, and IgG class switching [9,10]. Type III IFNs include four subtypes: IFN-λ1 (IL-29), IFN-λ2 and IFN-λ3 (IL-28A and IL28B, respectively), and IFN-λ4 [11]. IFNλ4 is the most recently discovered IFN. Type III IFNs are the first defense against virus replication in epithelial cells and are less likely to cause damaging inflammatory responses compared to the more potent type I IFN response. The IFNLR receptor is found in mucosal sites, inducing less potent inflammatory properties [3,4,5]. The IFN response is crucial in limiting viral replication and dissemination [12].
2. Downstream Signaling
The development of innate and adaptive antiviral immunity is influenced by pattern recognition receptors (PRRs). The activation of innate immunity by respiratory viruses can occur either on the cell surface through host cell receptors or by the interaction of intracellular and cytosolic PRRs. These receptors detect viral components such as RNA or DNA, resulting in the generation of downstream pathways that lead to antiviral immunity [13]. When PRRs recognize pathogen-associated molecular patterns (PAMPs), type I and III IFN responses are initiated which activate IFN regulatory factors (IRFs) in a signaling cascade [14]. Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-1) receptors, and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) are the principal IFN-stimulating PRRs [14,15,16]. TLR recognition of viral PAMPs signals through myeloid differentiation primary response 88 (MYD88) which acts as an adapter connecting proteins that receive signals from outside the cell to the proteins that relay signals inside the cell [17]. RIG-like receptors (RLRs) primarily detect viral genomes [18] and signal through the mitochondrial antiviral signaling protein (MAVS) and TIR domain-containing adaptor inducing INFß (TRIF) [19]. The binding of viral PAMPs ultimately leads to the activation of IRF3 and IRF7, which drive the transcription of type I and type III IFNs [20]. These pathways also activate nuclear factor κ-B (NF-κB) leading to the transcription of other inflammatory cytokines [14,21]. Binding of type I and III IFNs results in the recruitment of signal transducer and activator of transcription (STAT) for phosphorylation by Janus kinases (JAKs) [22]. The JAK/STAT pathway is the major signaling pathway activated by IFNs, leading to the expression of IFN-stimulated genes (ISGs). To reduce IFN effector function, viruses have developed various strategies to antagonize the JAK/STAT pathway [23].
The primary function of ISGs is to detect viral RNA, thus inhibiting viral replication [24]. The activation of the genes responsible for producing IFNs creates a positive feedback loop. ISGs, including transcription factors, have the ability to activate multiple genes. Differential activation is associated with the various subtypes of type I and type III IFNs and is believed to occur due to varying binding affinities of the IFN subtypes and the shared receptor. The outcomes of these activations are still being studied [5,25]. In response to viral DNA and RNA, the IFIT family of ISGs is commonly induced by type I and type III IFNs. IFIT genes suppress viral infection primarily by limiting viral RNA and DNA replication and impairing the entry of enveloped viruses [26]. Oligoadenylate synthetase (OAS1) functions in the activation of RNase L which cleaves viral RNA species [27]. ISG15 is a ubiquitin-like protein that can target viral proteases, preventing replication [28]. ISG20 is a 3′-5′ exonuclease which can cleave viral RNA, thereby reducing replication and triggering PRRs [29]. Viperin functions by disrupting lipid rafts which interferes with viral replication complexes [30]. As multiple ISGs exist, we direct readers to a recently published comprehensive review of ISGs [24].
IFNγ is different from types I and III and has a unique structure and function. It is primarily secreted by activated CD4 T helper and CD8 T cells, natural killer (NK) cells, NK T cells, and professional antigen-presenting cells (APCs) [31]. The production of IFNγ is prompted by IL-12 and IL-18, which are primarily produced by macrophages and dendritic cells [32,33]. While Th1 CD4 cells are major producers of IFNγ, CD8 CTLs also express it [9]. IL-12 drives the Th1 differentiation of naïve CD4 T cells, which is necessary for the expression of IFNγ [34]. The expression of IFNγ relies on transcription factors such as STAT4, Tbet, AP-1, and Eomes [35,36,37]. When IFNγ binds to IFNGR it results in JAK phosphorylation of STAT1 which causes the formation of homodimers that act as transcription factors for γ-activated sites (GAS) [9]. GAS includes a multitude of genes, including transcription factors that drive the expression of other genes, similar to ISGs stimulated by type I and type III IFNs. When macrophages are activated by IFNγ it results in upregulated antigen presentation, increased sensitivity to cytokines and chemokines, increased sensitivity of PRRs, and decreased sensitivity to anti-inflammatory cytokines such as IL-10 [9]. IFNγ is essential in linking T cells and macrophages and cellular immunity to intracellular pathogens so any shortcomings in this response can lead to increased susceptibility [38,39].
3. IFNs in Early Life and Childhood
Infants initially have a Th17/Th2-biased immune response with lower levels of IFNs [40,41,42]. A study of a longitudinal cohort found that levels of type I (and type III) IFNs were reduced in cord blood but increased with age [43]. Interestingly, infants with low IFN levels at birth had a higher risk of developing severe respiratory tract infections and persistent wheezing later in life [43]. When stimulated, cord blood responses showed decreased IFNα expression compared to whole blood from adult donors, while IFNß production in neonatal samples was similar to monocytes and slightly increased to whole blood from adult donors [44]. Another study found that IFNß expression was impaired in cord blood and monocyte-derived dendritic cells in response to TLR4 stimulation, due to defective CREB binding protein (CBP) binding to IRF3 [45]. Similarly, pDC-dependent IFNα response in cord blood plasmacytoid dendritic cells (PDC) has been shown to be deficient in response to TLR9 stimulation due to defective nuclear translocation of IRF7 [46]. Infants, particularly pre-term ones, produce lower levels of IFNs after TLR9 stimulation compared to adults [47]. Researchers continue to investigate the role of type III IFNs in human development. A recent study conducted on primary human airway epithelium cultures (AECs) showed that infant AECs produce more IFNλ1 than young children when exposed to poly I:C stimulation [48]. The study also found that infants hospitalized with respiratory virus infections produce higher levels of IFNλ1 in nasal aspirate than young children [48]. Interestingly, the study also observed that IFNLR1/IL10RB upregulation was specific to certain pathogens in infants aged 1–6 months, and this was associated with more severe bronchiolitis and eosinophilia, implying that IFNλ signaling is connected to disease in certain cases [49].
At birth, the expression of IFNγ and other Th1-associated responses is reduced but increases as age progresses [50,51,52]. This pattern is observed in a mouse model of respiratory syncytial virus (RSV) infection as well. This could be attributed to the fact that the fetal liver gives rise to neonatal T cells, whereas in adults, it is the bone marrow [53]. In a mouse model that involved thymic transfer of fetal derived T cells into adult mice it was observed that fetal-derived T cells produced higher levels of cytokines than adult-derived T cells and were skewed towards a Th2-bias [53,54,55]. This effect maybe partly due to the lack of IL-12 production by dendritic cells (DCs) in neonatal mice, as IL-12 is a cytokine associated with differentiation of Th1 cells [56]. In addition, cord blood and moDCs stimulated with LPS showed impaired CBP and IRF binding compared to adults. This is relevant because IRF3 is a necessary transcription factor for IL-12 expression [45]. The epigenetic profile of neonatal T cells is linked to enhanced IL-4 and IL-13 expression, and the IL-13Rα1, in conjunction with IL4Rα acts as a receptor for IL-4 in a neonatal mouse model and induces apoptosis of Th1 cells further driving the Th2-bias [57,58,59]. GATA3, a Th2-associated transcription factor, was found to be upregulated in neonatal CD4+ T cells [58]. In human cord blood and neonatal adenoid tissue, IL4 receptor expression at an early stage in development decreased with age and may contribute to Th2-biasing [60]. All these studies support the conclusion that age-dependent immune responses to viral infections occur, with infants producing lower IFN levels and subsequent Th2 biasing.
4. Mice as a Model
Mice are often used in translational research due to their easy housing and relatively low cost, as well as the availability of a range of immunology reagents. However, IFN responses vary between strains of mice and between mice and humans. In mice, there is a notable difference in the expression of type III IFN compared to humans. Humans possess up to four functional subtypes of IFNλ, while mice only have functional IFNλ2/3 and a pseudogene for IFNλ1 [61,62]. Studies have shown that age-dependent differences in IFN response also exist in mice, which reflect those observed in humans. For instance, in the context of RSV, neonatal mice exhibit reduced IFNα expression by pDCs and a tendency towards Th2-type cytokines [63,64]. Furthermore, it has been shown that aged mice are more susceptible to severe SARS-CoV-2 disease, a feature linked to an impaired IFN and antibody response [65]. Although mouse models do recapitulate age-related discrepancies in the severity of respiratory virus infection observed in humans, it is essential to consider the biological differences between the two species to interpret the data accurately. Genetic susceptibility to infection is determined by defects in genes that control non-redundant pathways of type I and III IFN responses [66,67].
Certain inbred strains (e.g., C57/BL6 and BALB/c) lack a functional Mx1 gene, which is a dominant antiviral resistance gene known as Mx for ‘myxovirus resistance’ [68,69]. Mice with the Mx1 gene are able to survive infection with mouse-adapted influenza A virus at doses that would be lethal for standard inbred strains [70]. Interestingly, neonatal mice with a functioning Mx1 gene remain just as susceptible to influenza challenge as their Mx1-negative counterparts [71]. However, when treated with exogenous IFN, Mx1-competent mice become resistant, similarly to their adult counterparts, whereas neonates without a functioning Mx gene do not respond to IFN treatment [72,73]. These experiments demonstrate the significance of IFN in the susceptibility of newborns and young children to influenza virus as modeled in neonatal mice [72,73]. Other mammals used in respiratory virus infection studies, such as cotton rats, hamsters, and ferrets, have functional Mx genes [74,75,76]. In humans, the MxA gene has an important role in inhibiting viral replication by interfering with the assembly of viral ribonucleoprotein, although the exact mechanism is yet to be determined [77,78].
5. Respiratory Syncytial Virus
RSV is a virus that can cause serious respiratory tract diseases and death in certain groups of people, including young children (up to 60 months old), the elderly, and those with weakened immune systems [79]. A 2019 review of RSV cases around the world estimated that there were 3.6 million hospital admissions, 26,300 in-hospital deaths, and 101,400 total deaths attributable to RSV [80]. Appallingly, 45,700 of those deaths were infants between the ages of 0–6 months, accounting for 2% of all deaths in that age range [80]. RSV is a single strand of RNA and belongs to the Pneumoviridae family. Its genome is 15.2 kb long and contains 10 genes that produce 11 proteins. One of the genes, M2, actually produces two different proteins, M2.1 and M2.2 [81,82]. RSV’s genome contains two nonstructural (NS) proteins, NS1 and NS2, as well as nucleocapsid proteins N, L, and P, regulatory protein M2, the inner envelope protein M1, and three surface proteins SH, G, and F. The G and F proteins are the main antigenic proteins. RSV has two main lineages, A and B, which are defined by differences in their G protein sequences [81,82]. RSV usually infects ciliated airway epithelial cells [83]. RSV can cause a variety of health problems, including changes in lung structure, decreased lung function, and increased mucosal responses [79]. RSV’s proteins are known to cause a range of immune responses that can suppress the body’s antiviral response and even cause damage to the host’s immune system [82,84,85]. Several studies have shown that RSV can fail to elicit a strong type I IFN response [86,87,88,89].
Research has shown that the RSV NS1 and NS2 proteins are effective in suppressing the IFN response. Removing these proteins results in a weakened response making NS1/NS2 deletion mutants’ potential candidates for a vaccine [86,90,91]. NS1 and NS2 are known to inhibit type I IFNs by blocking STAT2 in human epithelial cells [92]. Although NS2 was originally believed to be redundant to NS1, recent studies have revealed significant differences between them, such as the lack of structural homology in the crystallographic structure of NS2 [93]. NS2 has been shown to inhibit RIG-1 activation by binding to the N-terminal caspase activation and recruitment domain (CARD) of RIG-1, thus preventing downstream interaction with MAVS [94]. In contrast, NS1 binds to MAVS and inhibits its interaction with RIG-1 [93,95]. In mice, alveolar macrophages induced by MAVS coupled with RLRs are the primary source of type I IFNs during RSV infection [96]. Overall, the NS proteins block separate signaling proteins within identical pathways, resulting in a weak IFN response to infection.
The RSV G protein has been found to affect the immune response through a CX3C motif that is present in all RSV strains and is similar to the natural chemokine fractalkine [97]. The G protein inhibits the production of type I IFN by interacting with the CX3CR1 receptor on pDCs and monocytes through its CX3C motif. Blocking this interaction through a CX4C mutant virus or mAb treatment that targets the interaction increased IFNα and pro-inflammatory cytokines such as TNFα [85,98]. A recent study that examined anti-RSV G monoclonal antibodies (mAbs) 3D3 and 2D10 found that treatment improved the types I and III response in BALB/c mice and mouse lung epithelial cells (MLE-15) in a neutralization-independent mechanism, likely through binding the CX3C motif [99]. Neutralizing anti-F protein mAb, Palivizumab, did not improve the IFN response. This is expected since the F protein stimulates the production of IFN [99,100]. Studies that examined the human peripheral blood mononuclear cell response to RSV found that pDCs were the primary mediator of IFNα production in a RIG-1-dependent manner [101,102]. This response was impaired in infants suggesting that pDCs have a role in responding to RSV infection but are less responsive in young children [103].
It has also been shown that the administration of IFNα to mice before RSV challenge can decrease IL-4Rα and Th2 polarization [104]. In a clinical trial, topical administration of IFNα2a improved symptoms, but had no effect on viral shedding suggesting that viral load is not a correlate of disease severity [105]. Although no side effects were noted in this study, IFNα2a has shown dose-dependent side effects in other contexts including influenza-like symptoms, neurotoxicity [106], and pulmonary toxicity [107]. However, IFNα treatment is generally safe with appropriate caution and monitoring of potential toxicity [105,108,109]. The IFN response of the host undoubtedly influences RSV disease, and infants and children show dysfunctions, indicating the role of the impaired IFN response in severe disease in these groups [64,103,110].
The function of type III IFNs in RSV infection is an area that requires further investigation. Research has shown that in an in vitro model of primary human nasal epithelial cells, RSV infection induced IFNλ1 through RIG-1 activation [111]. Pretreatment with IFNλ1 resulted in resistance to RSV replication [112]. Surprisingly, no type I IFNs were expressed following RSV infection. However, in primary human airway epithelium cultures, IFNλ1 pretreatment did not inhibit RSV infection [113]. These conflicting results are likely due to differences in the assays used. In a study of young children with RSV it was found that the mRNA levels of IFNλ1-4 were positively correlated with age in the control group of healthy children. However, no correlation was noted in RSV-infected children, possibly due to RSV’s ability to suppress the IFN response [114]. Another study found that type I, II, and III IFN responses increased with age and were lowest in children under 6 months of age [115]. Higher levels of IFNλ2/3 were associated with a reduced risk of hospitalization. However, a separate study of infants with RSV did not find a correlation of IFNλ2/3 with clinical outcomes [116]. Instead, IFNλ1 was associated with an increased clinical severity index and respiratory rate. RSV activation of epidermal growth factor (EGF) receptor causes suppression of IRF1, antagonizing the IFNλ response, mediated by RSV F protein [117]. These studies suggest that type III interferons have a role in the immune response to RSV infection. RSV viral proteins, NS1, NS2, and G protein suppress these antiviral responses.
When infants are infected with RSV, a Th2 polarization is often linked to RSV immunopathology, and the lack of a strong Th1 response in infants and young children can lead to more severe illness [82,118,119]. RSV activates Rab5a GTPase in cells and mice to suppress IRF1-dependent IFNγ. Knockdown of Rab5a increases IFNγ by mediating IRF1 nuclear translocation [120]. Studies have shown that minimal IFNγ production in RSV infected neonatal mice can result in reduced viral clearance and increased disease severity. However, intranasal administration of IFNγ can improve these outcomes by activating alveolar macrophages [121]. Interestingly, blocking IFNγ or depleting NK and T cells has been associated with an increased antibody response in neonatal mice during RSV infection [122], whereas in adult mice, this depletion impairs the antibody response due to CD4 T cell-dependent antibody production [122]. This is likely due to increased viral load in IFNγ-depleted mice, resulting in more antigen exposure. While viral loads were lower in IFNγ-depleted mice, only one viral gene was measured by qRT-PCR and no other factors were measured to determine pathology. These findings suggest that IFNγ is needed for protecting neonatal mice from RSV, and this protection may be independent of T cell-mediated antibody production.
Type I IFNs can promote IFNγ expression by CD8 T cells via STAT4 in conjunction with TCR activation [104] and in NK cells with IL-12 via STAT1 [102,123]. However, IFNß has been shown to suppress DC production of IL-12, which is linked to Th1 differentiation and IFNγ production [124]. These studies suggest that IFNγ is protective during RSV infection and may contribute to increased susceptibility of neonates and children. Overall, these findings support clinical evaluations of infected neonates and children, which have concluded that RSV does not induce a robust IFNγ response [41,42], due to a combination of viral and host factors.
6. SARS-CoV-2
COVID-19 is caused SARS-CoV-2. This virus belongs to the family Coronaviradae and has a single-stranded, positive-sense RNA enclosed in an envelope [125]. Its genetic material is made up of 29.9 kb and contains 27 ORFs which encode 31 different proteins, including the four structural proteins (S, E, M, N) [125]. The first two-thirds of the genome encodes two large poly proteins (ORF1a and ORF1ab) which are cleaved by viral proteases to form 16 non-structural proteins (nsp1-16) [126,127,128]. These proteins are involved in replication, transcription, and interfering with the host’s innate defenses. The remaining third of the genome encodes 10 accessory proteins and the four structural proteins mentioned earlier [126,127,128]. As of June 2023, COVID-19 has caused 767 million confirmed cases and 6.9 million deaths worldwide since its emergence in 2019 [129].
Similarly to other respiratory viruses, SARS-CoV-2 triggers an IFN response through the recognition of various viral PAMPs by PRRs [130,131]. These include the S protein by TLR4 and TLR2, the E protein by TLR2 [132], and viral ssRNA and dsRNA intermediates by TLR7/8, TLR3, and RLRs [133,134,135,136]. However, similarly to RSV, SARS-CoV-2 has strategies to evade IFN signaling and expression [137,138,139,140]. Several viral proteins including NSP1, ORF6, and NSP13 have been shown to inhibit type I IFNs through different mechanisms including interference with host mRNA translation, nuclear translocation of IRF3 and STAT1, and binding with STAT1 to prevent phosphorylation, respectively [141,142]. ORF6 has also been shown to inhibit type I IFNs by mechanisms that include the inhibition of nuclear translocation of IRF3 and the inhibition of STAT1 nuclear translocation [143,144]. NSP13, which is highly conserved among coronaviruses, has been shown to inhibit type I and II signaling through binding STAT1 and preventing phosphorylation by JAK1 [145].
Several other SARS-CoV-2 viral proteins have been shown to inhibit the IFN response, specifically NSP1, NSP3, NSP5, NSP12, NSP13, NSP14, NSP15, ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, N, and M reported to inhibit IFNß expression by suppression of RLR-mediated signaling [137,138,142,144]. Further studies indicate ORF7a interferes with TANK-binding kinase (TBK1) preventing IRF3 phosphorylation, and ORF9b interferes with the interaction of MAVS and RIG-1 [146]. NSP 6 has been shown to interact with TBK1 to inhibit IRF3 activation and STAT1/2 phosphorylation. ORF 7a, ORF 7b, ORF3a, and the M protein were shown to inhibit IFNß in a luciferase reporter assay, a feature attributed to STAT1 and/or STAT2 activation depending on the viral protein [142]. Moreover, using a luciferase reporter, NSP13, NSP14, NSP15 and ORF6 were found to inhibit IFNß by disrupting nuclear translocation of IRF3 [138].
While SARS-CoV-2 primarily replicates in the nasopharyngeal and type II alveolar epithelial cells, recent evidence has shown that ACE2 expression (the receptor for SARS-CoV-2) and TMPRSS2 (a protease required for viral cell entry) are also present in other cells [147]. A mouse model of SARS-CoV-2 was created by expressing hACE2 in transgenic mice originally developed to study SARS-CoV-1 [148]. A mouse-adapted strain has also been developed, which is attributed to a mutation in the receptor binding domain of the S protein [149]. Interestingly, new variants of SARS-CoV-2 (including B.1.1.7) contain the same substitution and can infect mice [150,151]. Different small animal models, such as ferrets and golden hamsters, which are both naturally susceptible to SARS-CoV-2 have varying outcomes from mild to lethal [151].
In contrast to other respiratory viruses, children and neonates are not more likely than adults to develop severe COVID-19, possibly due to differences in IFN signaling [152,153,154,155,156]. Elderly adults with severe COVID-19 have shown lower levels of type I IFNs, potentially due to IFN autoreactive antibodies [157,158]. The other major finding related to age-dependent differences in the IFN response found reduced IFNα secretion by pDCs in adults with severe COVID-19 [159]. The other major finding related to age-dependent differences in the IFN response found reduced IFNα secretion by pDCs in adults with severe COVID-19 [159], and patients with autoimmune polyendocrine syndrome type-1 (APS-1) which produces autoantibodies against type I IFNs are at increased risk of severe disease [158], and require mechanical ventilation [160]. pDCs and fibroblasts of these patients indicated blunted IFN responses [161]. Age-related discrepancies in the IFN response to SARS-CoV-2 can be attributed to several plausible mechanisms related to suppression of the IFNs. Several studies have shown a role of IFNs in immune-mediated pathology with a deficient early response followed by a persistent and heightened late response associated with severe disease. ScRNASeq analysis of bronchial alveolar lavage (BAL) or lung tissue samples identified a type I IFN-associated inflammatory signature, consistent with findings in peripheral blood, suggesting that an early and regulated IFN response is protective while a latent dysregulated response is pathogenic [162,163].
The findings suggest that the neonatal immune system changes rapidly in early years of life affecting the IFN response. Neonates are less likely to suffer severe outcomes from SARS-CoV-2 infection, but pre-term infants are at higher risk of COVID-19 and pre-term birth due to medically induced birth.
7. Influenza Virus
Influenza virus is an enveloped ssRNA virus belonging to the Orthomyxoviridae family [164]. It has a segmented genome with eight segments encoding various proteins including polymerase subunits, surface glycoproteins, nucleoprotein, matrix protein, membrane protein, nuclear export protein, and nonstructural proteins [164,165,166]. An analysis from 1996 to 2016 estimated a suggested 32.1 million influenza virus episodes and 5.7 million hospitalizations in adults per year globally with those over 65 years of age having the highest rate of hospitalization [167]. In 2018, an analysis of children under 5 years of age estimated 10.1 million cases of influenza-like respiratory illness with 870,000 hospital admissions [168]. Pandemic strains have a much greater impact than seasonal strains with the 1918 pandemic resulting in estimates of 40–50 million deaths globally [169]. Age is an important factor contributing to disease severity, with the young and old particularly susceptible to severe disease. One of many factors contributing to severity by strains of influenza is their ability to suppress the IFN response [170].
Influenza virus activates TLRs 3, 7, and 8, as well as RIG-1/MAVs, inducing the production of type I and III IFNs [171,172]. The influenza NS1 protein suppresses the IFN response through multiple mechanisms [173,174] including blocking host protein translation by interfering with pre-mRNA processing via binding to cleavage and polyadenylation specificity factor 30 (CPSF30), a component of the pre-mRNA processing machinery responsible for 3′ cleavage and polyadenylation [175,176]. NS1 also prevents dsRNA-mediated IFN responses by scavenging dsRNA and blocking IRF3 phosphorylation [177]. Additionally, NS1 inhibits RIG-1-mediated IFN production by preventing TRIM25 multimerization and CARD domain ubiquitination [178]. Influenza PB1 and PA can also counteract the type I IFN response in mice with PA interacting with IRF3 to prevent its activation and PB1-F2 interfering with the MAVs adapter protein [170,179,180]. PB2 interacts with MAVs and IPS-1 to reduce IFNß transcription, and NS1 is the dominant IFN antagonist of influenza virus, with IFN suppression being associated with more severe disease [181,182,183]. IFNß is protective [72,184], but type I IFNs can contribute to mortality [185]. IFNα and IFNλ reduce viral load but only IFNλ controls viral replication in the upper airways [186].
Neonatal mice are more susceptible to influenza virus due to poor IFN responses [187]. Reduced IFNγ response in young mice delays viral clearance and increases mortality [188]. Adoptive transfer of adult CD8 T cells into neonatal mice confers resistance to influenza challenge dependent on IFNγ [189]. A separate study utilizing IFNγ knockout mice found no difference in mortality of WT and IFNγ-/- mice after IAV challenge, while finding increased antigen-specific T cells in IFNγ-/- mice [190]. In mice, type III IFNs were found to have an important role in the course of infection with IFNλ levels found to be higher in the lungs of influenza-infected BALB/c mice than type I IFNs [191]. Children infected with influenza virus had higher levels of IFN in their nasal wash than children infected with RSV, but that IFN was not associated with reduced viral shedding. Children between the ages of 29 and 54 months who were given a live attenuated influenza vaccine (LAIV) had upregulated ISGs and lower viral loads on days 2 and 7, which was attributed to asymptomatic respiratory virus infection prior to the administration of LAIV [192]. Children with an autosomal recessive IRF7/IRF9 deficiency are at greatest risk for severe influenza due to poor IFN responses [66,193,194]. In one report, three children with a rare TLR3 deficiency were found to be susceptible to severe influenza pneumonitis [193]. It was shown that treatment with IFNα2a and IFNλ1 rescued this susceptibility using pulmonary epithelial cells differentiated from patient-derived induced pluripotent stem cells (iPSCs) [193]. Taken together, these studies corroborate the importance of IFN responses to influenza, particularly in infants, children, and the elderly.
8. Conclusions
IFNs have an early and crucial role in the body’s antiviral response. Three major respiratory viruses affecting humans, i.e., RSV, influenza virus, and SARS-CoV-2, have IFN antagonistic features (summarized in Figure 1) [1,102,195,196]. For RSV, the NS1, NS2, and G genes are key effectors in this response [102], while for SARS-CoV-2 the NSP1, ORF6, and NSP13 genes have a particularly potent effect on IFN. Influenza virus has the NS1 gene as the canonical IFN antagonist [137], but antagonism activities are associated with the PA, PB1, PB1-F2, and PB2 genes [170].
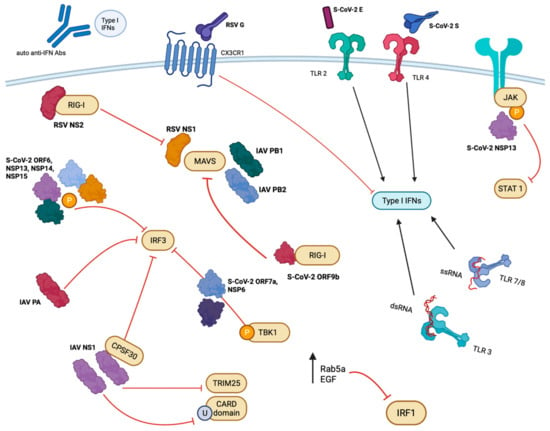
Figure 1.
Overview of viral protein interactions. Viral proteins from RSV, SARS-CoV-2, and IAV antagonize or agonize IFN responses using a variety of mechanisms. An abridged illustration of the viral proteins and host factors involved is pictured summarizing those mentioned throughout the text. RIG-I (retinoic acid-inducible gene 1), EGF (epidermal growth factor), TLR (toll-like receptor), JAK (Janus kinase), MAVS (mitochondrial antiviral-signalizing protein), TRIM25 (tripartite motif-containing protein 25), CARD (caspase recruitment domains), CPSF30 (cleavage and polyadenylation specificity factor subunit 4), IAV (influenza A virus), RSV (respiratory syncytial virus), S-CoV-2 (SARS-CoV-2), NS/NSP (non-structural/non-structural protein), IRF (interferon regulatory transcription factor), TBK1 (TANK-binding kinase), U (ubiquitination), P (phosphorylation). Created with BioRender and Microsoft PowerPoint.
In infants and young children, type I, II, and III IFN responses are lower compared to adults [41,42]. The reduced IFNα producing pDCs and reduced expression of IL-12, (a cytokine that increases production of IFNγ) are principally linked to the diminished IFNγ [44]. Additionally, a general lack of Th1-type responses causes IFNγ to be diminished due to several mechanisms such as a lack of IL-12 production in neonatal moDCs [56], IL-4 induced apoptosis of neonatal Th1 cells and increased expression of GATA3 [58], and increased IL-4 expression [60]. While IFNs are protective against RSV [105,115,121] and influenza [192,193] in neonatal mice and children, they experience more severe disease compared to older counterparts due to blunted IFN responses. However, SARS-CoV-2-infected infants and young children are not at increased risk for severe disease compared to healthy adults [152,153,154,155]. There is evidence that dysregulation of IFNs may contribute to severe disease, but that an early regulated response is protective [197,198].
A beneficial IFN response is associated with lower viral loads, faster viral clearance, and reduced disease severity [66]. IFN treatments have been investigated for viral infections, including approved use of IFNα2a for hepatitis C [199]. However, excessive inflammation has been linked to higher levels of IFNs [198] highlighting the importance of properly regulated anti-viral responses and caution with off-target effects and side effects [199]. Type I and type III IFNs have distinct roles, with type III IFN demonstrating a more targeted role and generally mediating a more diminished inflammatory response [200]. Drugs that boost endogenous IFN have also been investigated as treatments for viral infection [201,202,203]. The lack of a strong interferon response in neonates and children has been associated with susceptibility to influenza and RSV, but this has not been observed in the context of SARS-CoV-2 infection [155]. The protective and pathogenic role of IFNs in respiratory virus infection is likely linked to underlying mechanisms.
Author Contributions
Conceptualization, H.C.B. and R.A.T.; Writing—Original Draft Preparation, H.B, M.R.H., and R.A.T.; Writing—Review & Editing, H.C.B. and R.A.T.; Visualization, H.C.B.; Supervision, R.A.T.; Project Administration, R.A.T.; Funding Acquisition, R.A.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH 1R01AI166066-01A1—Structure-guided engineering to increase respiratory syncytial virus G protein immunogenicity.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Isaacs, A.; Lindenmann, J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957, 147, 258–267. [Google Scholar] [CrossRef]
- Taylor, M.W. Interferons; Springer International Publishing: Berlin/Heidelberg, Germany, 2014; pp. 101–119. [Google Scholar]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Wells, A.I.; Coyne, C.B. Type III Interferons in Antiviral Defenses at Barrier Surfaces. Trends Immunol. 2018, 39, 848–858. [Google Scholar] [CrossRef]
- Pervolaraki, K.; Rastgou Talemi, S.; Albrecht, D.; Bormann, F.; Bamford, C.; Mendoza, J.L.; Garcia, K.C.; McLauchlan, J.; Höfer, T.; Stanifer, M.L.; et al. Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 2018, 14, e1007420. [Google Scholar] [CrossRef]
- Gibbert, K.; Schlaak, J.F.; Yang, D.; Dittmer, U. IFN-alpha subtypes: Distinct biological activities in anti-viral therapy. Br. J. Pharmacol. 2013, 168, 1048–1058. [Google Scholar] [CrossRef]
- Fitzgerald-Bocarsly, P.; Dai, J.; Singh, S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 2008, 19, 3–19. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Ivashkiv, L.B. IFNgamma: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef]
- Mohr, E.; Cunningham, A.F.; Toellner, K.M.; Bobat, S.; Coughlan, R.E.; Bird, R.A.; MacLennan, I.C.; Serre, K. IFN-gamma produced by CD8 T cells induces T-bet-dependent and -independent class switching in B cells in responses to alum-precipitated protein vaccine. Proc. Natl. Acad. Sci. USA 2010, 107, 17292–17297. [Google Scholar] [CrossRef]
- Broggi, A.; Granucci, F.; Zanoni, I. Type III interferons: Balancing tissue tolerance and resistance to pathogen invasion. J. Exp. Med. 2020, 217, e20190295. [Google Scholar] [CrossRef]
- Murira, A.; Lamarre, A. Type-I Interferon Responses: From Friend to Foe in the Battle against Chronic Viral Infection. Front. Immunol. 2016, 7, 609. [Google Scholar] [CrossRef]
- Suresh, R.; Mosser, D.M. Pattern recognition receptors in innate immunity, host defense, and immunopathology. Adv. Physiol. Educ. 2013, 37, 284–291. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, H.K. Innate immune recognition of respiratory syncytial virus infection. BMB Rep. 2014, 47, 184–191. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, L.; Chen, P.; Liang, G. Myeloid Differentiation Primary Response Protein 88 (MyD88): The Central Hub of TLR/IL-1R Signaling. J. Med. Chem. 2020, 63, 13316–13329. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef]
- Ullah, M.O.; Sweet, M.J.; Mansell, A.; Kellie, S.; Kobe, B. TRIF-dependent TLR signaling, its functions in host defense and inflammation, and its potential as a therapeutic target. J. Leukoc. Biol. 2016, 100, 27–45. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host. Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Zhang, Y.J. Interplay between Janus Kinase/Signal Transducer and Activator of Transcription Signaling Activated by Type I Interferons and Viral Antagonism. Front. Immunol. 2017, 8, 1758. [Google Scholar] [CrossRef]
- Ezeonwumelu, I.J.; Garcia-Vidal, E.; Ballana, E. JAK-STAT Pathway: A Novel Target to Tackle Viral Infections. Viruses 2021, 13, 2379. [Google Scholar] [CrossRef]
- Yang, E.; Li, M.M.H. All About the RNA: Interferon-Stimulated Genes That Interfere with Viral RNA Processes. Front. Immunol. 2020, 11, 605024. [Google Scholar] [CrossRef] [PubMed]
- Stanifer, M.L.; Pervolaraki, K.; Boulant, S. Differential Regulation of Type I and Type III Interferon Signaling. Int. J. Mol. Sci. 2019, 20, 1445. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Di, H.; Elbahesh, H.; Brinton, M.A. Characteristics of Human OAS1 Isoform Proteins. Viruses 2020, 12, 152. [Google Scholar] [CrossRef]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, N.; Woodson, S.E.; Dong, Q.; Wang, J.; Liang, Y.; Rijnbrand, R.; Wei, L.; Nichols, J.E.; Guo, J.T.; et al. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 2011, 409, 175–188. [Google Scholar] [CrossRef]
- Fitzgerald, K.A. The interferon inducible gene: Viperin. J. Interferon Cytokine Res. 2011, 31, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Cardoso, A.P.; Goncalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Swain, S.L. Interleukin 18. J. Exp. Med. 2001, 194, F11–F14. [Google Scholar] [CrossRef]
- Okamura, H.; Kashiwamura, S.; Tsutsui, H.; Yoshimoto, T.; Nakanishi, K. Regulation of interferon-gamma production by IL-12 and IL-18. Curr. Opin. Immunol. 1998, 10, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, N.G.; Szabo, S.J.; Weber-Nordt, R.M.; Zhong, Z.; Schreiber, R.D.; Darnell, J.E.; Murphy, K.M. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J. Exp. Med. 1995, 181, 1755–1762. [Google Scholar] [CrossRef]
- Szabo, S.J.; Sullivan, B.M.; Stemmann, C.; Satoskar, A.R.; Sleckman, B.P.; Glimcher, L.H. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 2002, 295, 338–342. [Google Scholar] [CrossRef]
- Lugo-Villarino, G.; Maldonado-López, R.; Possemato, R.; Peñaranda, C.; Glimcher, L.H. T-bet is required for optimal production of IFN-γ and antigen-specific T cell activation by dendritic cells. Proc. Natl. Acad. Sci. USA 2003, 100, 7749–7754. [Google Scholar] [CrossRef] [PubMed]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Dorman, S.E.; Uzel, G.; Roesler, J.; Bradley, J.S.; Bastian, J.; Billman, G.; King, S.; Filie, A.; Schermerhorn, J.; Holland, S.M. Viral infections in interferon-γ receptor deficiency. J. Pediatr. 1999, 135, 640–643. [Google Scholar] [CrossRef]
- Remus, N.; Reichenbach, J.; Picard, C.; Rietschel, C.; Wood, P.; Lammas, D.; Kumararatne, D.S.; Casanova, J.-L. Impaired Interferon Gamma-Mediated Immunity and Susceptibility to Mycobacterial Infection in Childhood. Pediatr. Res. 2001, 50, 8–13. [Google Scholar] [CrossRef]
- Wang, W.; Sung, N.; Gilman-Sachs, A.; Kwak-Kim, J. T Helper (Th) Cell Profiles in Pregnancy and Recurrent Pregnancy Losses: Th1/Th2/Th9/Th17/Th22/Tfh Cells. Front. Immunol. 2020, 11, 2025. [Google Scholar] [CrossRef] [PubMed]
- Tobias; Levy, O.; Ruth; Goriely, S. Innate Immune Function by Toll-like Receptors: Distinct Responses in Newborns and the Elderly. Immunity 2012, 37, 771–783. [Google Scholar] [CrossRef]
- Dowling, D.J.; Levy, O. Ontogeny of early life immunity. Trends Immunol. 2014, 35, 299–310. [Google Scholar] [CrossRef]
- Holt, P.G.; Mok, D.; Panda, D.; Renn, L.; Fabozzi, G.; Deklerk, N.H.; Kusel, M.M.H.; Serralha, M.; Hollams, E.M.; Holt, B.J.; et al. Developmental regulation of type 1 and type 3 interferon production and risk for infant infections and asthma development. J. Allergy Clin. Immunol. 2019, 143, 1176–1182.e5. [Google Scholar] [CrossRef] [PubMed]
- Kollmann, T.R.; Crabtree, J.; Rein-Weston, A.; Blimkie, D.; Thommai, F.; Wang, X.Y.; Lavoie, P.M.; Furlong, J.; Fortuno, E.S., 3rd; Hajjar, A.M.; et al. Neonatal innate TLR-mediated responses are distinct from those of adults. J. Immunol. 2009, 183, 7150–7160. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, E.; Albarani, V.; Nguyen, M.; Laes, J.F.; Ruelle, J.L.; De Wit, D.; Willems, F.; Goldman, M.; Goriely, S. Interferon regulatory factor 3-dependent responses to lipopolysaccharide are selectively blunted in cord blood cells. Blood 2007, 109, 2887–2893. [Google Scholar] [CrossRef]
- Danis, B.; George, T.C.; Goriely, S.; Dutta, B.; Renneson, J.; Gatto, L.; Fitzgerald-Bocarsly, P.; Marchant, A.; Goldman, M.; Willems, F.; et al. Interferon regulatory factor 7-mediated responses are defective in cord blood plasmacytoid dendritic cells. Eur. J. Immunol. 2008, 38, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Bender, G.; Minh Thang, C.; Quang Thanh, L.; Thi Trang Dai, V.; Van Thanh, P.; Thi Hong Nhu, B.; Ngoc Xuan Trang, D.; Thi Phuong Trinh, P.; Vu Thuong, N.; et al. TLR Responses in Preterm and Term Infant Cord Blood Mononuclear Cells. Pathogens 2023, 12, 596. [Google Scholar] [CrossRef]
- Salka, K.; Arroyo, M.; Chorvinsky, E.; Abutaleb, K.; Perez, G.F.; Wolf, S.; Xuchen, X.; Weinstock, J.; Gutierrez, M.J.; Pérez-Losada, M.; et al. Innate IFN-lambda responses to dsRNA in the human infant airway epithelium and clinical regulatory factors during viral respiratory infections in early life. Clin. Exp. Allergy 2020, 50, 1044–1054. [Google Scholar] [CrossRef]
- Pierangeli, A.; Statzu, M.; Nenna, R.; Santinelli, L.; Petrarca, L.; Frassanito, A.; Gentile, M.; Antonelli, G.; Midulla, F.; Scagnolari, C. Interferon lambda receptor 1 (IFNL1R) transcript is highly expressed in rhinovirus bronchiolitis and correlates with disease severity. J. Clin. Virol. 2018, 102, 101–109. [Google Scholar] [CrossRef]
- Wilson, C.B.; Westall, J.; Johnston, L.; Lewis, D.B.; Dower, S.K.; Alpert, A.R. Decreased production of interferon-gamma by human neonatal cells. Intrinsic and regulatory deficiencies. J. Clin. Investig. 1986, 77, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.L.; Kemp, A.S.; Thorburn, J.; Hill, D.J. Reduced interferon-gamma secretion in neonates and subsequent atopy. Lancet 1994, 344, 983–985. [Google Scholar] [CrossRef]
- Forsthuber, T.; Yip, H.C.; Lehmann, P.V. Induction of TH1 and TH2 immunity in neonatal mice. Science 1996, 271, 1728–1730. [Google Scholar] [CrossRef]
- Adkins, B. Peripheral CD4+ lymphocytes derived from fetal versus adult thymic precursors differ phenotypically and functionally. J. Immunol. 2003, 171, 5157–5164. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Hardy, R.R.; Herzenberg, L.A.; Herzenberg, L.A. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J. Exp. Med. 1985, 161, 1554–1568. [Google Scholar] [CrossRef] [PubMed]
- Rudd, B.D. Neonatal T Cells: A Reinterpretation. Annu. Rev. Immunol. 2020, 38, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-H.; Hoeman, C.M.; Hardaway, J.C.; Guloglu, F.B.; Ellis, J.S.; Jain, R.; Divekar, R.; Tartar, D.M.; Haymaker, C.L.; Zaghouani, H. Delayed maturation of an IL-12–producing dendritic cell subset explains the early Th2 bias in neonatal immunity. J. Exp. Med. 2008, 205, 2269–2280. [Google Scholar] [CrossRef]
- Zaghouani, H.; Hoeman, C.M.; Adkins, B. Neonatal immunity: Faulty T-helpers and the shortcomings of dendritic cells. Trends Immunol. 2009, 30, 585–591. [Google Scholar] [CrossRef]
- Li, L.; Lee, H.H.; Bell, J.J.; Gregg, R.K.; Ellis, J.S.; Gessner, A.; Zaghouani, H. IL-4 utilizes an alternative receptor to drive apoptosis of Th1 cells and skews neonatal immunity toward Th2. Immunity 2004, 20, 429–440. [Google Scholar] [CrossRef]
- Rose, S.; Lichtenheld, M.; Foote, M.R.; Adkins, B. Murine neonatal CD4+ cells are poised for rapid Th2 effector-like function. J. Immunol. 2007, 178, 2667–2678. [Google Scholar] [CrossRef]
- Hebel, K.; Weinert, S.; Kuropka, B.; Knolle, J.; Kosak, B.; Jorch, G.; Arens, C.; Krause, E.; Braun-Dullaeus, R.C.; Brunner-Weinzierl, M.C. CD4+ T cells from human neonates and infants are poised spontaneously to run a nonclassical IL-4 program. J. Immunol. 2014, 192, 5160–5170. [Google Scholar] [CrossRef]
- Lasfar, A.; Lewis-Antes, A.; Smirnov, S.V.; Anantha, S.; Abushahba, W.; Tian, B.; Reuhl, K.; Dickensheets, H.; Sheikh, F.; Donnelly, R.P.; et al. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006, 66, 4468–4477. [Google Scholar] [CrossRef]
- Donnelly, R.P.; Kotenko, S.V. Interferon-Lambda: A New Addition to an Old Family. J. Interferon Cytokine Res. 2010, 30, 555–564. [Google Scholar] [CrossRef] [PubMed]
- You, D.; Marr, N.; Saravia, J.; Shrestha, B.; Lee, G.I.; Turvey, S.E.; Brombacher, F.; Herbert, D.R.; Cormier, S.A. IL-4Ralpha on CD4+ T cells plays a pathogenic role in respiratory syncytial virus reinfection in mice infected initially as neonates. J. Leukoc. Biol. 2013, 93, 933–942. [Google Scholar] [CrossRef]
- Cormier, S.A.; Shrestha, B.; Saravia, J.; Lee, G.I.; Shen, L.; Devincenzo, J.P.; Kim, Y.-I.; You, D. Limited Type I Interferons and Plasmacytoid Dendritic Cells during Neonatal Respiratory Syncytial Virus Infection Permit Immunopathogenesis upon Reinfection. J. Virol. 2014, 88, 9350–9360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Liu, F.; Ye, Z.; Song, W.; Lee, A.C.Y.; Shuai, H.; Lu, L.; To, K.K.-W.; Chan, J.F.-W.; et al. Age-associated SARS-CoV-2 breakthrough infection and changes in immune response in a mouse model. Emerg. Microbes Infect. 2022, 11, 368–383. [Google Scholar] [CrossRef]
- Ciancanelli, M.J.; Huang, S.X.; Luthra, P.; Garner, H.; Itan, Y.; Volpi, S.; Lafaille, F.G.; Trouillet, C.; Schmolke, M.; Albrecht, R.A.; et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 2015, 348, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.L.; Abel, L. Human genetics of infectious diseases: Unique insights into immunological redundancy. Semin. Immunol. 2018, 36, 1–12. [Google Scholar] [CrossRef]
- Verhelst, J.; Spitaels, J.; Nurnberger, C.; De Vlieger, D.; Ysenbaert, T.; Staeheli, P.; Fiers, W.; Saelens, X. Functional Comparison of Mx1 from Two Different Mouse Species Reveals the Involvement of Loop L4 in the Antiviral Activity against Influenza A Viruses. J. Virol. 2015, 89, 10879–10890. [Google Scholar] [CrossRef]
- Staeheli, P.; Grob, R.; Meier, E.; Sutcliffe, J.G.; Haller, O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 1988, 8, 4518–4523. [Google Scholar] [CrossRef]
- Haller, O.; Kochs, G. Mx genes: Host determinants controlling influenza virus infection and trans-species transmission. Hum. Genet. 2020, 139, 695–705. [Google Scholar] [CrossRef]
- Shin, D.L.; Hatesuer, B.; Bergmann, S.; Nedelko, T.; Schughart, K. Protection from Severe Influenza Virus Infections in Mice Carrying the Mx1 Influenza Virus Resistance Gene Strongly Depends on Genetic Background. J. Virol. 2015, 89, 9998–10009. [Google Scholar] [CrossRef]
- Lindenmann, J. Inheritance of Resistance to Influenza Virus in Mice. Proc. Soc. Exp. Biol. Med. 1964, 116, 506–509. [Google Scholar] [CrossRef]
- Haller, O.; Arnheiter, H.; Gresser, I.; Lindenmann, J. Virus-specific interferon action. Protection of newborn Mx carriers against lethal infection with influenza virus. J. Exp. Med. 1981, 154, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Bessiere, P.; Wasniewski, M.; Picard-Meyer, E.; Servat, A.; Figueroa, T.; Foret-Lucas, C.; Coggon, A.; Lesellier, S.; Boue, F.; Cebron, N.; et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. PLoS Pathog. 2021, 17, e1009427. [Google Scholar] [CrossRef]
- Pletneva, L.M.; Haller, O.; Porter, D.D.; Prince, G.A.; Blanco, J.C. Interferon-inducible Mx gene expression in cotton rats: Cloning, characterization, and expression during influenza viral infection. J. Interferon Cytokine Res. 2006, 26, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Leon, A.J.; Borisevich, V.; Boroumand, N.; Seymour, R.; Nusbaum, R.; Escaffre, O.; Xu, L.; Kelvin, D.J.; Rockx, B. Host gene expression profiles in ferrets infected with genetically distinct henipavirus strains. PLoS Negl. Trop. Dis. 2018, 12, e0006343. [Google Scholar] [CrossRef]
- Verhelst, J.; Hulpiau, P.; Saelens, X. Mx proteins: Antiviral gatekeepers that restrain the uninvited. Microbiol. Mol. Biol. Rev. 2013, 77, 551–566. [Google Scholar] [CrossRef]
- Verhelst, J.; Parthoens, E.; Schepens, B.; Fiers, W.; Saelens, X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 2012, 86, 13445–13455. [Google Scholar] [CrossRef]
- Carvajal, J.J.; Avellaneda, A.M.; Salazar-Ardiles, C.; Maya, J.E.; Kalergis, A.M.; Lay, M.K. Host Components Contributing to Respiratory Syncytial Virus Pathogenesis. Front. Immunol. 2019, 10, 2152. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Blau, D.M.; Caballero, M.T.; Feikin, D.R.; Gill, C.J.; Madhi, S.A.; Omer, S.B.; Simões, E.A.F.; Campbell, H.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: A systematic analysis. Lancet 2022, 399, 2047–2064. [Google Scholar] [CrossRef] [PubMed]
- Battles, M.B.; McLellan, J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019, 17, 233–245. [Google Scholar] [CrossRef]
- Bergeron, H.C.; Tripp, R.A. Immunopathology of RSV: An Updated Review. Viruses 2021, 13, 2478. [Google Scholar] [CrossRef]
- Zhang, L.; Peeples, M.E.; Boucher, R.C.; Collins, P.L.; Pickles, R.J. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 2002, 76, 5654–5666. [Google Scholar] [CrossRef] [PubMed]
- Oshansky, C.M.; Krunkosky, T.M.; Barber, J.; Jones, L.P.; Tripp, R.A. Respiratory syncytial virus proteins modulate suppressors of cytokine signaling 1 and 3 and the type I interferon response to infection by a toll-like receptor pathway. Viral. Immunol. 2009, 22, 147–161. [Google Scholar] [CrossRef]
- Kauvar, L.M.; Harcourt, J.L.; Haynes, L.M.; Tripp, R.A. Therapeutic targeting of respiratory syncytial virus G-protein. Immunotherapy 2010, 2, 655–661. [Google Scholar] [CrossRef]
- Hall, C.B.; Douglas, R.G., Jr.; Simons, R.L.; Geiman, J.M. Interferon production in children with respiratory syncytial, influenza, and parainfluenza virus infections. J. Pediatr. 1978, 93, 28–32. [Google Scholar] [CrossRef]
- McIntosh, K. Interferon in nasal secretions from infants with viral respiratory tract infections. J. Pediatr. 1978, 93, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, D. Production of interferon in respiratory syncytial virus bronchiolitis. Arch. Dis. Child. 1989, 64, 92–95. [Google Scholar] [CrossRef]
- Taylor, C.E.; Webb, M.S.; Milner, A.D.; Milner, P.D.; Morgan, L.A.; Scott, R.; Stokes, G.M.; Swarbrick, A.S.; Toms, G.L. Interferon alfa, infectious virus, and virus antigen secretion in respiratory syncytial virus infections of graded severity. Arch. Dis. Child. 1989, 64, 1656–1660. [Google Scholar] [CrossRef]
- Teng, M.N.; Whitehead, S.S.; Bermingham, A.; St. Claire, M.; Elkins, W.R.; Murphy, B.R.; Collins, P.L. Recombinant Respiratory Syncytial Virus That Does Not Express the NS1 or M2-2 Protein Is Highly Attenuated and Immunogenic in Chimpanzees. J. Virol. 2000, 74, 9317–9321. [Google Scholar] [CrossRef]
- Spann, K.M.; Tran, K.-C.; Chi, B.; Rabin, R.L.; Collins, P.L. Suppression of the Induction of Alpha, Beta, and Gamma Interferons by the NS1 and NS2 Proteins of Human Respiratory Syncytial Virus in Human Epithelial Cells and Macrophages. J. Virol. 2004, 78, 4363–4369. [Google Scholar] [CrossRef]
- Lo, M.S.; Brazas, R.M.; Holtzman, M.J. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 2005, 79, 9315–9319. [Google Scholar] [CrossRef]
- Pei, J.; Wagner, N.D.; Zou, A.J.; Chatterjee, S.; Borek, D.; Cole, A.R.; Kim, P.J.; Basler, C.F.; Otwinowski, Z.; Gross, M.L.; et al. Structural basis for IFN antagonism by human respiratory syncytial virus nonstructural protein 2. Proc. Natl. Acad. Sci. USA 2021, 118, e2020587118. [Google Scholar] [CrossRef]
- Ling, Z.; Tran, K.C.; Teng, M.N. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 2009, 83, 3734–3742. [Google Scholar] [CrossRef] [PubMed]
- Boyapalle, S.; Wong, T.; Garay, J.; Teng, M.; San Juan-Vergara, H.; Mohapatra, S.; Mohapatra, S. Respiratory Syncytial Virus NS1 Protein Colocalizes with Mitochondrial Antiviral Signaling Protein MAVS following Infection. PLoS ONE 2012, 7, e29386. [Google Scholar] [CrossRef]
- Goritzka, M.; Makris, S.; Kausar, F.; Durant, L.R.; Pereira, C.; Kumagai, Y.; Culley, F.J.; Mack, M.; Akira, S.; Johansson, C. Alveolar macrophage–derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J. Exp. Med. 2015, 212, 699–714. [Google Scholar] [CrossRef]
- Tripp, R.A.; Jones, L.P.; Haynes, L.M.; Zheng, H.; Murphy, P.M.; Anderson, L.J. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2001, 2, 732–738. [Google Scholar] [CrossRef]
- Chirkova, T.; Boyoglu-Barnum, S.; Gaston, K.A.; Malik, F.M.; Trau, S.P.; Oomens, A.G.P.; Anderson, L.J. Respiratory Syncytial Virus G Protein CX3C Motif Impairs Human Airway Epithelial and Immune Cell Responses. J. Virol. 2013, 87, 13466–13479. [Google Scholar] [CrossRef]
- Bergeron, H.C.; Kauvar, L.M.; Tripp, R.A. Anti-G protein antibodies targeting the RSV G protein CX3C chemokine region improve the interferon response. Ther. Adv. Infect. Dis. 2023, 10, 20499361231161157. [Google Scholar] [CrossRef] [PubMed]
- Kurt-Jones, E.A.; Popova, L.; Kwinn, L.; Haynes, L.M.; Jones, L.P.; Tripp, R.A.; Walsh, E.E.; Freeman, M.W.; Golenbock, D.T.; Anderson, L.J.; et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000, 1, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Schijf, M.A.; Lukens, M.V.; Kruijsen, D.; van Uden, N.O.; Garssen, J.; Coenjaerts, F.E.; Van’t Land, B.; van Bleek, G.M. Respiratory syncytial virus induced type I IFN production by pDC is regulated by RSV-infected airway epithelial cells, RSV-exposed monocytes and virus specific antibodies. PLoS ONE 2013, 8, e81695. [Google Scholar] [CrossRef]
- Hijano, D.R.; Vu, L.D.; Kauvar, L.M.; Tripp, R.A.; Polack, F.P.; Cormier, S.A. Role of Type I Interferon (IFN) in the Respiratory Syncytial Virus (RSV) Immune Response and Disease Severity. Front. Immunol. 2019, 10, 566. [Google Scholar] [CrossRef] [PubMed]
- Marr, N.; Wang, T.I.; Kam, S.H.; Hu, Y.S.; Sharma, A.A.; Lam, A.; Markowski, J.; Solimano, A.; Lavoie, P.M.; Turvey, S.E. Attenuation of respiratory syncytial virus-induced and RIG-I-dependent type I IFN responses in human neonates and very young children. J. Immunol. 2014, 192, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.B.; Watford, W.T.; Salomon, R.; Hofmann, S.R.; Pien, G.C.; Morinobu, A.; Gadina, M.; O’Shea, J.J.; Biron, C.A. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 2002, 297, 2063–2066. [Google Scholar] [CrossRef]
- Sung, R.Y.; Yin, J.; Oppenheimer, S.J.; Tam, J.S.; Lau, J. Treatment of respiratory syncytial virus infection with recombinant interferon alfa-2a. Arch. Dis. Child. 1993, 69, 440–442. [Google Scholar] [CrossRef][Green Version]
- Caraceni, A.; Gangeri, L.; Martini, C.; Belli, F.; Brunelli, C.; Baldini, M.; Mascheroni, L.; Lenisa, L.; Cascinelli, N. Neurotoxicity of interferon-? in melanoma therapy. Cancer 1998, 83, 482–489. [Google Scholar] [CrossRef]
- Savale, L.; Sattler, C.; Günther, S.; Montani, D.; Chaumais, M.-C.; Perrin, S.; Jaïs, X.; Seferian, A.; Jovan, R.; Bulifon, S.; et al. Pulmonary arterial hypertension in patients treated with interferon. Eur. Respir. J. 2014, 44, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, J.; Hicks, R.; Pacheco, F.; Olson, L. Pilot study of recombinant interferon alpha-2a for treatment of infants with bronchiolitis induced by respiratory syncytial virus. Antimicrob. Agents Chemother. 1988, 32, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Dubois, J.; Hershon, L.; Carmant, L.; Belanger, S.; Leclerc, J.M.; David, M. Toxicity profile of interferon alfa-2b in children: A prospective evaluation. J. Pediatr. 1999, 135, 782–785. [Google Scholar] [CrossRef]
- Comerlato Scotta, M.; Greff Machado, D.; Goecks Oliveira, S.; De Moura, A.; Rhoden Estorgato, G.; De Souza, A.P.D.; Nery Porto, B.; De Araújo, P.D.; Sarria, E.E.; Pitrez, P.M.; et al. Evaluation of nasal levels of interferon and clinical severity of influenza in children. J. Clin. Virol. 2019, 114, 37–42. [Google Scholar] [CrossRef]
- Okabayashi, T.; Kojima, T.; Masaki, T.; Yokota, S.; Imaizumi, T.; Tsutsumi, H.; Himi, T.; Fujii, N.; Sawada, N. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 2011, 160, 360–366. [Google Scholar] [CrossRef]
- Essaidi-Laziosi, M.; Geiser, J.; Huang, S.; Constant, S.; Kaiser, L.; Tapparel, C. Interferon-Dependent and Respiratory Virus-Specific Interference in Dual Infections of Airway Epithelia. Sci. Rep. 2020, 10, 10246. [Google Scholar] [CrossRef]
- Broadbent, L.; Bamford, C.G.G.; Lopez Campos, G.; Manzoor, S.; Courtney, D.; Ali, A.; Touzelet, O.; McCaughey, C.; Mills, K.; Power, U.F. An endogenously activated antiviral state restricts SARS-CoV-2 infection in differentiated primary airway epithelial cells. PLoS ONE 2022, 17, e0266412. [Google Scholar] [CrossRef]
- Tovo, P.-A.; Garazzino, S.; Savino, F.; Daprà, V.; Pruccoli, G.; Dini, M.; Filisetti, G.; Funiciello, E.; Galliano, I.; Bergallo, M. Expressions of Type I and III Interferons, Endogenous Retroviruses, TRIM28, and SETDB1 in Children with Respiratory Syncytial Virus Bronchiolitis. Curr. Issues Mol. Biol. 2023, 45, 1197–1217. [Google Scholar] [CrossRef] [PubMed]
- Taveras, J.; Garcia-Maurino, C.; Moore-Clingenpeel, M.; Xu, Z.; Mertz, S.; Ye, F.; Chen, P.; Cohen, S.H.; Cohen, D.; Peeples, M.E.; et al. Type III Interferons, Viral Loads, Age, and Disease Severity in Young Children with Respiratory Syncytial Virus Infection. J. Infect. Dis. 2022, 227, 61–70. [Google Scholar] [CrossRef]
- Selvaggi, C.; Pierangeli, A.; Fabiani, M.; Spano, L.; Nicolai, A.; Papoff, P.; Moretti, C.; Midulla, F.; Antonelli, G.; Scagnolari, C. Interferon lambda 1–3 expression in infants hospitalized for RSV or HRV associated bronchiolitis. J. Infect. 2014, 68, 467–477. [Google Scholar] [CrossRef]
- Kalinowski, A.; Galen, B.T.; Ueki, I.F.; Sun, Y.; Mulenos, A.; Osafo-Addo, A.; Clark, B.; Joerns, J.; Liu, W.; Nadel, J.A.; et al. Respiratory syncytial virus activates epidermal growth factor receptor to suppress interferon regulatory factor 1-dependent interferon-lambda and antiviral defense in airway epithelium. Mucosal Immunol. 2018, 11, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Nenna, R.; Fedele, G.; Frassanito, A.; Petrarca, L.; Di Mattia, G.; Pierangeli, A.; Scagnolari, C.; Papoff, P.; Schiavoni, I.; Leone, P.; et al. Increased T-helper Cell 2 Response in Infants with Respiratory Syncytial Virus Bronchiolitis Hospitalized Outside Epidemic Peak. Pediatr. Infect. Dis. J. 2020, 39, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Christiaansen, A.F.; Knudson, C.J.; Weiss, K.A.; Varga, S.M. The CD4 T cell response to respiratory syncytial virus infection. Immunol. Res. 2014, 59, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.; Tang, W.; Xie, J.; Chen, S.; Ren, L.; Zang, N.; Xie, X.; Deng, Y.; Gao, L.; Liu, E. Respiratory syncytial virus activates Rab5a to suppress IRF1-dependent IFN-lambda production, subverting the antiviral defense of airway epithelial cells. J. Virol. 2021, 95, 10-1128. [Google Scholar] [CrossRef]
- Eichinger, K.M.; Egaña, L.; Orend, J.G.; Resetar, E.; Anderson, K.B.; Patel, R.; Empey, K.M. Alveolar macrophages support interferon gamma-mediated viral clearance in RSV-infected neonatal mice. Respir. Res. 2015, 16, 122. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Wang, B.L.; McDonald, J.U.; Yamaguchi, Y.; Harker, J.A.; Goritzka, M.; Johansson, C.; Bukreyev, A.; Collins, P.L.; Openshaw, P.J. Neonatal antibody responses are attenuated by interferon-γ produced by NK and T cells during RSV infection. Proc. Natl. Acad. Sci. USA 2013, 110, 5576–5581. [Google Scholar] [CrossRef]
- Hunter, C.A.; Gabriel, K.E.; Radzanowski, T.; Neyer, L.E.; Remington, J.S. Type I interferons enhance production of IFN-gamma by NK cells. Immunol. Lett. 1997, 59, 1–5. [Google Scholar] [CrossRef]
- Rudd, B.D.; Luker, G.D.; Luker, K.E.; Peebles, R.S.; Lukacs, N.W. Type I Interferon Regulates Respiratory Virus Infected Dendritic Cell Maturation and Cytokine Production. Viral Immunol. 2007, 20, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef] [PubMed]
- Ellis, P.; Somogyvári, F.; Virok, D.P.; Noseda, M.; McLean, G.R. Decoding Covid-19 with the SARS-CoV-2 Genome. Curr. Genet. Med. Rep. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Weekly Epidemiological Update on COVID-19—8 June 2023. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---8-june-2023 (accessed on 6 June 2023).
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife 2021, 10, e68563. [Google Scholar] [CrossRef]
- Zhao, Y.; Kuang, M.; Li, J.; Zhu, L.; Jia, Z.; Guo, X.; Hu, Y.; Kong, J.; Yin, H.; Wang, X.; et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021, 31, 818–820. [Google Scholar] [CrossRef]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Strazzabosco, G.; Fernandez, M.; Caccuri, F.; Caruso, A.; Rizzo, R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms 2021, 9, 1820. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.E.; Lee, H.K. Current Understanding of the Innate Control of Toll-like Receptors in Response to SARS-CoV-2 Infection. Viruses 2021, 13, 2132. [Google Scholar] [CrossRef]
- Sampaio, N.G.; Chauveau, L.; Hertzog, J.; Bridgeman, A.; Fowler, G.; Moonen, J.P.; Dupont, M.; Russell, R.A.; Noerenberg, M.; Rehwinkel, J. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021, 11, 13638. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Reuschl, A.K.; Zuliani-Alvarez, L.; Whelan, M.V.X.; Turner, J.; Noursadeghi, M.; Jolly, C.; Towers, G.J. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021, 40, e107826. [Google Scholar] [CrossRef]
- Znaidia, M.; Demeret, C.; Van Der Werf, S.; Komarova, A.V. Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options. Viruses 2022, 14, 1247. [Google Scholar] [CrossRef]
- Yuen, C.-K.; Lam, J.-Y.; Wong, W.-M.; Mak, L.-F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Liao, C.H.; Wang, Q.; Tan, Y.J.; Luo, R.; Qiu, Y.; Ge, X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Shemesh, M.; Aktepe, T.E.; Deerain, J.M.; McAuley, J.L.; Audsley, M.D.; David, C.T.; Purcell, D.F.J.; Urin, V.; Hartmann, R.; Moseley, G.W.; et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLOS Pathog. 2021, 17, e1009800. [Google Scholar] [CrossRef]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.-A.; Leibundgut, M.; Thiel, V.; Mühlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Z.; Xiao, X.; Tian, Z.; Dong, X.; Wang, C.; Li, L.; Ren, L.; Lei, X.; Xiang, Z.; et al. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell. Mol. Immunol. 2021, 18, 945–953. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Fung, S.-Y.; Siu, K.-L.; Lin, H.; Chan, C.-P.; Yeung, M.L.; Jin, D.-Y. SARS-CoV-2 NSP13 helicase suppresses interferon signaling by perturbing JAK1 phosphorylation of STAT1. Cell Biosci. 2022, 12, 36. [Google Scholar] [CrossRef]
- Kouwaki, T.; Nishimura, T.; Wang, G.; Oshiumi, H. RIG-I-Like Receptor-Mediated Recognition of Viral Genomic RNA of Severe Acute Respiratory Syndrome Coronavirus-2 and Viral Escape from the Host Innate Immune Responses. Front. Immunol. 2021, 12, 700926. [Google Scholar] [CrossRef]
- Gawish, R.; Starkl, P.; Pimenov, L.; Hladik, A.; Lakovits, K.; Oberndorfer, F.; Cronin, S.J.; Ohradanova-Repic, A.; Wirnsberger, G.; Agerer, B.; et al. ACE2 is the critical in vivo receptor for SARS-CoV-2 in a novel COVID-19 mouse model with TNF- and IFNgamma-driven immunopathology. Elife 2022, 11, e74623. [Google Scholar] [CrossRef] [PubMed]
- McCray, P.B.; Pewe, L.; Wohlford-Lenane, C.; Hickey, M.; Manzel, L.; Shi, L.; Netland, J.; Jia, H.P.; Halabi, C.; Sigmund, C.D.; et al. Lethal Infection of K18-hACE2 Mice Infected with Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2007, 81, 813–821. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.-Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.; Yuen, T.T.; Yoon, C.; Hu, J.C.; Wen, L.; Hu, B.; Yang, D.; Wang, Y.; Hou, Y.; et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine 2021, 73, 103643. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.-W.; Yuen, K.-Y. Animal models in SARS-CoV-2 research. Nat. Methods 2022, 19, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Viner, R.M.; Mytton, O.T.; Bonell, C.; Melendez-Torres, G.J.; Ward, J.; Hudson, L.; Waddington, C.; Thomas, J.; Russell, S.; Van Der Klis, F.; et al. Susceptibility to SARS-CoV-2 Infection among Children and Adolescents Compared with Adults. JAMA Pediatr. 2021, 175, 143. [Google Scholar] [CrossRef] [PubMed]
- Götzinger, F.; Santiago-García, B.; Noguera-Julián, A.; Lanaspa, M.; Lancella, L.; Calò Carducci, F.I.; Gabrovska, N.; Velizarova, S.; Prunk, P.; Osterman, V.; et al. COVID-19 in children and adolescents in Europe: A multinational, multicentre cohort study. Lancet Child Adolesc. Health 2020, 4, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, B.K.; Allaire, J.M.; Irvine, M.A.; Lopez, A.A.; Sauve, L.J.; Vallance, B.A.; Jacobson, K. Severe COVID-19 Infection and Pediatric Comorbidities: A Systematic Review and Meta-Analysis. Int. J. Infect. Dis. 2021, 103, 246–256. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y.; et al. SARS-CoV-2 Infection in Children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Thomas, P.G.; Randolph, A.G. Immunology of SARS-CoV-2 infection in children. Nat. Immunol. 2022, 23, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Bartleson, J.M.; Radenkovic, D.; Covarrubias, A.J.; Furman, D.; Winer, D.A.; Verdin, E. SARS-CoV-2, COVID-19 and the Ageing Immune System. Nat. Aging 2021, 1, 769–782. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Bastard, P.; Orlova, E.; Sozaeva, L.; Lévy, R.; James, A.; Schmitt, M.M.; Ochoa, S.; Kareva, M.; Rodina, Y.; Gervais, A.; et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 2021, 218, e20210554. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020, 5, eabd1554. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. S4), D49–D53. [Google Scholar] [CrossRef] [PubMed]
- Uyeki, T.M.; Hui, D.S.; Zambon, M.; Wentworth, D.E.; Monto, A.S. Influenza. Lancet 2022, 400, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Lafond, K.E.; Porter, R.M.; Whaley, M.J.; Suizan, Z.; Ran, Z.; Aleem, M.A.; Thapa, B.; Sar, B.; Proschle, V.S.; Peng, Z.; et al. Global burden of influenza-associated lower respiratory tract infections and hospitalizations among adults: A systematic review and meta-analysis. PLoS Med. 2021, 18, e1003550. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; O’Brien, K.L.; Madhi, S.A.; Widdowson, M.-A.; Byass, P.; Omer, S.B.; Abbas, Q.; Ali, A.; Amu, A.; et al. Global burden of respiratory infections associated with seasonal influenza in children under 5 years in 2018: A systematic review and modelling study. Lancet Glob. Health 2020, 8, e497–e510. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.; Krewski, D. Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef]
- Liedmann, S.; Hrincius, E.R.; Guy, C.; Anhlan, D.; Dierkes, R.; Carter, R.; Wu, G.; Staeheli, P.; Green, D.R.; Wolff, T.; et al. Viral suppressors of the RIG-I-mediated interferon response are pre-packaged in influenza virions. Nat. Commun. 2014, 5, 5645. [Google Scholar] [CrossRef]
- Le Goffic, R.; Pothlichet, J.; Vitour, D.; Fujita, T.; Meurs, E.; Chignard, M.; Si-Tahar, M. Cutting Edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol. 2007, 178, 3368–3372. [Google Scholar] [CrossRef]
- Lee, N.; Wong, C.K.; Hui, D.S.C.; Lee, S.K.W.; Wong, R.Y.K.; Ngai, K.L.K.; Chan, M.C.W.; Chu, Y.J.; Ho, A.W.Y.; Lui, G.C.Y.; et al. Role of human Toll-like receptors in naturally occurring influenza A infections. Influenza Other Respir. Viruses 2013, 7, 666–675. [Google Scholar] [CrossRef]
- Garcia-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef]
- Dauber, B.; Heins, G.; Wolff, T. The Influenza B Virus Nonstructural NS1 Protein Is Essential for Efficient Viral Growth and Antagonizes Beta Interferon Induction. J. Virol. 2004, 78, 1865–1872. [Google Scholar] [CrossRef] [PubMed]
- Nemeroff, M.E.; Barabino, S.M.L.; Li, Y.; Keller, W.; Krug, R.M. Influenza Virus NS1 Protein Interacts with the Cellular 30 kDa Subunit of CPSF and Inhibits 3′ End Formation of Cellular Pre-mRNAs. Mol. Cell 1998, 1, 991–1000. [Google Scholar] [CrossRef]
- Fortes, P.; Beloso, A.; Ortín, J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994, 13, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Donelan, N.R.; Basler, C.F.; GarcíA-Sastre, A. A Recombinant Influenza A Virus Expressing anRNA-Binding-Defective NS1 Protein Induces High Levels of BetaInterferon and Is Attenuated in Mice. J. Virol. 2003, 77, 13257–13266. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Albrecht, R.A.; Urano, T.; Inn, K.-S.; Huang, I.C.; Carnero, E.; Farzan, M.; Inoue, S.; Jung, J.U.; García-Sastre, A. Influenza A Virus NS1 Targets the Ubiquitin Ligase TRIM25 to Evade Recognition by the Host Viral RNA Sensor RIG-I. Cell Host Microbe 2009, 5, 439–449. [Google Scholar] [CrossRef]
- Schreiber, A.; Liedmann, S.; Brunotte, L.; Anhlan, D.; Ehrhardt, C.; Ludwig, S. Type I interferon antagonistic properties of influenza B virus polymerase proteins. Cell. Microbiol. 2020, 22, e13143. [Google Scholar] [CrossRef]
- Yi, C.; Zhao, Z.; Wang, S.; Sun, X.; Zhang, D.; Sun, X.; Zhang, A.; Jin, M. Influenza A Virus PA Antagonizes Interferon-beta by Interacting with Interferon Regulatory Factor 3. Front. Immunol. 2017, 8, 1051. [Google Scholar] [CrossRef]
- Dudek, S.E.; Wixler, L.; Nordhoff, C.; Nordmann, A.; Anhlan, D.; Wixler, V.; Ludwig, S. The influenza virus PB1-F2 protein has interferon antagonistic activity. Biol. Chem. 2011, 392, 1135–1144. [Google Scholar] [CrossRef]
- Varga, Z.T.; Ramos, I.; Hai, R.; Schmolke, M.; García-Sastre, A.; Fernandez-Sesma, A.; Palese, P. The Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon at the Level of the MAVS Adaptor Protein. PLoS Pathog. 2011, 7, e1002067. [Google Scholar] [CrossRef] [PubMed]
- García-Sastre, A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011, 162, 12–18. [Google Scholar] [CrossRef]
- Kumova, O.K.; Fike, A.J.; Thayer, J.L.; Nguyen, L.T.; Mell, J.C.; Pascasio, J.; Stairiker, C.; Leon, L.G.; Katsikis, P.D.; Carey, A.J. Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal Lactobacillus rhamnosus GG. PLOS Pathog. 2019, 15, e1008072. [Google Scholar] [CrossRef] [PubMed]
- Kumova, O.K.; Galani, I.E.; Rao, A.; Johnson, H.; Triantafyllia, V.; Matt, S.M.; Pascasio, J.; Gaskill, P.J.; Andreakos, E.; Katsikis, P.D.; et al. Severity of neonatal influenza infection is driven by type I interferon and oxidative stress. Mucosal. Immunol. 2022, 15, 1309–1320. [Google Scholar] [CrossRef]
- Klinkhammer, J.; Schnepf, D.; Ye, L.; Schwaderlapp, M.; Gad, H.H.; Hartmann, R.; Garcin, D.; Mahlakoiv, T.; Staeheli, P. IFN-lambda prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife 2018, 7, e33354. [Google Scholar] [CrossRef] [PubMed]
- Reuman, P.D.; Ayoub, E.M.; Small, P.A. Influenza Infection in the Infant Mouse. Pediatr. Res. 1983, 17, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, D.; Perry, S.; Pryharski, K. Control of influenza infection is impaired by diminished interferon-gamma secretion by CD4 T cells in the lungs of toddler mice. J. Leukoc. Biol. 2016, 100, 203–212. [Google Scholar] [CrossRef] [PubMed]
- You, D.; Ripple, M.; Balakrishna, S.; Troxclair, D.; Sandquist, D.; Ding, L.; Ahlert, T.A.; Cormier, S.A. Inchoate CD8+ T cell responses in neonatal mice permit influenza-induced persistent pulmonary dysfunction. J. Immunol. 2008, 181, 3486–3494. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.J.; Olivas, E.; Gutierrez, A.; Diaz, G.; Doherty, P.C. Disregulated influenza A virus-specific CD8+ T cell homeostasis in the absence of IFN-gamma signaling. J. Immunol. 2007, 178, 7616–7622. [Google Scholar] [CrossRef] [PubMed]
- Jewell, N.A.; Cline, T.; Mertz, S.E.; Smirnov, S.V.; Flano, E.; Schindler, C.; Grieves, J.L.; Durbin, R.K.; Kotenko, S.V.; Durbin, J.E. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 2010, 84, 11515–11522. [Google Scholar] [CrossRef]
- Costa-Martins, A.G.; Mane, K.; Lindsey, B.B.; Ogava, R.L.T.; Castro, Í.; Jagne, Y.J.; Sallah, H.J.; Armitage, E.P.; Jarju, S.; Ahadzie, B.; et al. Prior upregulation of interferon pathways in the nasopharynx impacts viral shedding following live attenuated influenza vaccine challenge in children. Cell Rep. Med. 2021, 2, 100465. [Google Scholar] [CrossRef]
- Lim, H.K.; Huang, S.X.L.; Chen, J.; Kerner, G.; Gilliaux, O.; Bastard, P.; Dobbs, K.; Hernandez, N.; Goudin, N.; Hasek, M.L.; et al. Severe influenza pneumonitis in children with inherited TLR3 deficiency. J. Exp. Med. 2019, 216, 2038–2056. [Google Scholar] [CrossRef]
- Hernandez, N.; Melki, I.; Jing, H.; Habib, T.; Huang, S.S.Y.; Danielson, J.; Kula, T.; Drutman, S.; Belkaya, S.; Rattina, V.; et al. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 2018, 215, 2567–2585. [Google Scholar] [CrossRef]
- Su, H.C.; Jing, H.; Zhang, Y.; Casanova, J.-L. Interfering with Interferons: A Critical Mechanism for Critical COVID-19 Pneumonia. Annu. Rev. Immunol. 2023, 41, 561–585. [Google Scholar] [CrossRef]
- Killip, M.J.; Fodor, E.; Randall, R.E. Influenza virus activation of the interferon system. Virus Res. 2015, 209, 11–22. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Eskandarian Boroujeni, M.; Sekrecka, A.; Antonczyk, A.; Hassani, S.; Sekrecki, M.; Nowicka, H.; Lopacinska, N.; Olya, A.; Kluzek, K.; Wesoly, J.; et al. Dysregulated Interferon Response and Immune Hyperactivation in Severe COVID-19: Targeting STATs as a Novel Therapeutic Strategy. Front. Immunol. 2022, 13, 888897. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.Y.; Ho, J.X.; Kow, A.S.F.; Liang, G.; Tham, C.L.; Ho, Y.C.; Lee, M.T. Interferon therapy and its association with depressive disorders—A review. Front. Immunol. 2023, 14, 1048592. [Google Scholar] [CrossRef] [PubMed]
- Stanifer, M.L.; Guo, C.; Doldan, P.; Boulant, S. Importance of Type I and III Interferons at Respiratory and Intestinal Barrier Surfaces. Front. Immunol. 2020, 11, 608645. [Google Scholar] [CrossRef]
- Grant, C.R.; Holder, B.S.; Liberal, R.; Heneghan, M.A.; Ma, Y.; Mieli-Vergani, G.; Vergani, D.; Longhi, M.S. Immunosuppressive drugs affect interferon (IFN)-gamma and programmed cell death 1 (PD-1) kinetics in patients with newly diagnosed autoimmune hepatitis. Clin. Exp. Immunol. 2017, 189, 71–82. [Google Scholar] [CrossRef]
- Kopitar-Jerala, N. The Role of Interferons in Inflammation and Inflammasome Activation. Front. Immunol. 2017, 8, 873. [Google Scholar] [CrossRef] [PubMed]
- Goulet, M.-L.; Olagnier, D.; Xu, Z.; Paz, S.; Belgnaoui, S.M.; Lafferty, E.I.; Janelle, V.; Arguello, M.; Paquet, M.; Ghneim, K.; et al. Systems Analysis of a RIG-I Agonist Inducing Broad Spectrum Inhibition of Virus Infectivity. PLoS Pathog. 2013, 9, e1003298. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).