Abstract
Nucleopeptides represent an intriguing class of nucleic acid analogues, in which nucleobases are placed in a peptide structure. The incorporation of D- and/or L-amino acids in nucleopeptide molecules allows the investigation of the role of backbone stereochemistry in determining the formation of DNA and RNA hybrids. Circular Dichroism (CD) spectroscopic studies indicated the nucleopeptide as having fully l-backbone configuration-formed stable hybrid complexes with RNA molecules. Molecular Dynamics (MD) simulations suggested a potential structure of the complex resulting from the interaction between the l-nucleopeptide and RNA strand. From this study, both the backbone (ionics and H-bonds) and nucleobases (pairing and π-stacking) of the chiral nucleopeptide appeared to be involved in the hybrid complex formation, highlighting the key role of the backbone stereochemistry in the formation of the nucleopeptide/RNA complexes.
1. Introduction
Nucleic acid–protein interactions are crucial in numerous cell functions, including transcription, translation and RNA maturation, which in turn modulate numerous physio-pathological processes [1,2]. They regulate genetic information transfer by proper interaction between protein binding domains and specific regions of nucleic acids [3,4,5,6]. The formation of highly stable and specific complexes depends on the strict recognition rules regulating the nucleobase association for nucleic acids and on a wide array of aminoacid–nucleobase interactions, including hydrophobic, ionic and hydrogen bonds for the nucleic acid–protein complexes. In the present day, the use of nucleobases constituting DNA and RNA aptamers is a well-established approach to interfere with both nucleic acid and protein targets for the diagnosis and the therapeutic treatment of numerous diseases, including cancer, inflammation and infections [7]. In spite of their great potential, the full capitalization of these chemotypes as therapeutics is still limited by several drawbacks, such as low chemical variability, binding affinity and metabolic stability. In this regard, DNA aptamers containing pyrimidine bases endowed with amino acid residues (SOMAmer) [8,9] represent an enhancement in chemical diversity providing wide application as diagnostic and therapeutic tools. Alternatively, nucleobases have been added to peptides as recognition units by employing nucleobase amino acids (NBA) [10,11,12,13,14,15]. To date, no study has been reported describing the use of NBA as elements to improve the selective protein recognition of target nucleic acids. Nevertheless, this basic concept has been proposed in several studies [16] involving peptide-based molecules, in order to make novel compounds that bind DNA and RNA specifically, such as Peptide Nucleic Acid (PNA) [17,18,19,20,21]. PNAs are nucleic acid analogues first described by Nielsen and coworkers, which have been largely applied for chemical biology investigations. They are able to efficiently mimic DNA molecules through a pseudopeptide backbone formed by achiral N-(2-aminoethyl) glycine (aeg) moieties bearing nucleobases linked via a methylene carbonyl bridge. Although PNAs selectively recognize DNA and RNA sequences affording very stable hybrids, the absence of chiral centers in its backbone hampers the folding in specific conformations, reducing its ability to discriminate highly structured DNA and RNA molecules. Additionally, PNAs exhibit numerous disadvantages, including low solubility in an aqueous medium, ambiguous directional selectivity of DNA/RNA binding, and low cell permeability.
To address some of the above-mentioned issues, several modifications have been performed on the original PNA structure [22,23,24]. For instance, the incorporation of N-(2-aminoethyl)-D-Lysine residues led to the synthesis of cationic PNAs, which combined the nucleic acid recognition ability of PNA with the cell permeability of cationic cell penetrating peptides (CPP). These new PNA variants displayed advantages in terms of solubility and binding to DNA [23,24]. Crystallographic studies demonstrated that the backbone chirality could conformationally modulate the PNA helical direction, thereby enhancing the recognition specificity of the target [25,26,27]. In line with the results achieved by studying CPPs, the presence of the positive charge of Lys side-chains enhanced their solubility and especially their cell permeability, most likely through an endosomal pathway or direct cell membrane penetration [28]. Similarly, new classes of chiral PNAs (α, and ɣ-GPNA) have been recently designed by merging the recognition ability of PNA with the uptake features of cationic guanidinium groups [29,30]. These studies showed that GPNAs with α-d and/or ɣ-l-backbone configuration exhibited improved cellular uptake, increased binding affinity for DNA and RNA, and enhanced thermal stability of the resulting hybrids. These favorable properties were ascribable to the conformational preorganization of chiral PNAs and the electrostatic interactions between cationic guanidine and phosphate groups. In particular, they demonstrated that the backbone stereochemistry contributed more with respect to the electrostatic forces in stabilizing the hybrid duplex.
In this regard, nucleopeptides represent alternative chemotypes in which nucleobases are combined with a peptide structure [31,32,33]. This fascinating class of molecules are characterized by the presence of amino acids-bearing nucleobases as side chains that promote nucleic acid recognition through both base pairing and stacking interactions. In nucleopeptides, the nucleobase-functionalized amino acids are alternated with underivatized amino acids at defined positions in the peptide backbone. Diederichsen et al. [34] and Mihara et al. [15,35,36] reported the synthesis of α and β-peptides with nucleobase-functionalized side chains and the use of l-α-amino-ɣ-nucleobase-butyric acid to assemble RNA binding molecules and functional peptides, respectively. More recently, we have reported new synthetic strategies to obtain both homo- and hetero-sequences of cationic nucleopeptides that exhibited advantageous properties in terms of binding, specificity, cell permeability and delivery for RNA, DNA and PNA molecules (Figure 1) [37,38]. Although the therapeutic potential of nucleopeptides is less known with the respect to the parent PNA molecules, they are under investigation for their ability to interfere with the Reverse Transcriptase of the Human Immunodeficiency Virus (RT-HIV), which is fundamental for HIV infection [39]. Additionally, nucleopeptides have been successfully employed in combination with Doxorubicin, to improve its efficacy in Multidrug Resistant Cancer (MRD) by sequestering adenosine triphosphate (ATP) [40].
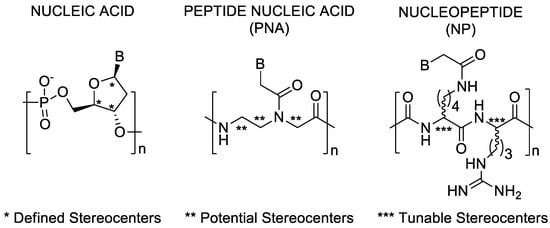
Figure 1.
Stereogenic sites in DNA, PNA, and Arginine-based nucleopeptide building blocks.
Herein, we describe the use of our synthetic approach to introduce specific structural modifications, such as the incorporation of d- and/or l-amino acids, in nucleopeptides with the aim to investigate the role of backbone stereochemistry in determining the formation of DNA and RNA hybrids. CD spectroscopy and UV-thermal stability investigations provided evidence of the impact of the backbone stereochemistry on the ability of nucleopeptides to hybridize DNA and RNA. Finally, the interaction of the best performing chiral nucleopeptide was studied by molecular dynamics (MD) simulations to achieve a new insight on the structure of the resulting complexes with RNA complementary strands.
2. Materials and Methods
All solvents for peptide synthesis, reagent grade HPLC solvents (water and ACN), Fmoc-Rink amide-Am resin, 1-hydroxybenzotriazole hydrate (HOBt) (purity > 97% dry weight, water ≈ 12%), triisopropylsilane (TIS) (purity 98%), acetic anhydride (Ac2O, purity > 98%), anhydrous N,N’-dimethylformamide (DMF), (1H-7-Azabenzotriazol-1-yl-oxy)tris-pyrrolidinophosphonium hexafluorophosphate (PyAOP) (purity 96%), 1-hydroxy-7-azabenzotriazole (HOAt, purity 96%), and anhydrous dichloromethane (DCM) were purchased from commercial sources (Sigma-Aldrich, Milano, Italy) and used without any additional purification. The following reagents were obtained from IRIS Biotech (Marktredwitz, Germany): O-benzotriazole-N,N,N’,N’-tetra-methyl-uroniumhexafluorophosphate (HBTU, purity 99%), N,N-diisopropylethylamine (DIEA, purity 99%), piperidine, trifluoroacetic acid (TFA, purity 99%), and Nα-Fmoc-protected amino acids. from Sigma-Aldrich (Milano, Italy). Nucleopeptides were purified by preparative HPLC (Shimadzu HPLC system) by using a C18-bounded preparative RP-HPLC column (Phenomenex Kinetex 21.2 mm × 150 mm, 5 μM). The analyses were accomplished by analytical HPLC (Shimadzu Prominence analytic HPLC system) on a C18-bounded analytical RP-HPLC column (Phenomenex Kinetex, 4.6 mm × 150 mm, 5 μM), using a gradient elution from 10 to 90% ACN in water (0.1% TFA) over 20 min, a flow rate of 1.0 mL/min and detecting by an UV diode array. Molecular weights of nucleopeptides were assessed by mass spectrometry analyses carried out by matrix assisted laser desorption ionization time of-flight (MALDI-TOF) mass spectrometer implemented with a pulsed nitrogen laser (k = 337 nm).
2.1. Synthesis of Nucleopeptides np-1/4
Rink Amide AM-PS resin (60 mg, 0.033 mmol, 0.55 mmol/g) was suspended for 30 min in DCM/DMF 1:1 and then washed with DCM (3 × 1 min) and DMF (3 × 1 min). The Fmoc deprotection was carried out by treatment with a solution of 20% (v/v) piperidine in DMF (1 × 5 min; 1 × 25 min). The succeeding amide couplings were performed using 4 equivalents (according to the original loading of the resin) of the Fmoc-protected l or d-aminoacids. To a DMF (1.2 mL) solution of Fmoc-l (or d)-Arg(Pbf)–OH (84,5 mg, 0.13 mmol, 4 equivalents), HBTU (50 mg, 0.13 mmol, 4 equivalents) and HOBt (19 mg, 0.13 mmol, 4 equivalents) DIPEA (45 µL, 0.26 mmol, 8 equivalents) was added and the resulting mixture was added to the support. The mixture was shaken for 2 h at room temperature and then washed with DMF (3 × 1 min) and DCM (3 × 1 min). Next, the unreacted free amine groups were acetylated by treatment of the resin with 1 mL of DMF capping solution containing of Ac2O and of DIPEA (2 and 3 equivalents relative to the initial loading of the resin, respectively) and shaken for 5 min. The completion of the peptide sequences was achieved by repeating the cycles of the Fmoc deprotection and coupling reaction with Fmoc-l (or d)-Lys(Alloc)-OH and Fmoc-l (or d)-Arg(Pbf)-OH as above reported. These steps were monitored by Kaiser ninhydrine and TNBS tests and ascertained by quantitative UV spectroscopic measurements of Fmoc chromophore. After the last Fmoc removal, the treatment with a mixture of Ac2O (6.2 μL, 2 equiv) and DIPEA (17.3 μL, 3 equivalents) in DMF (1 mL) for 10 min, allowed N-terminal acetylation of the peptides.
The Alloc groups were removed by treating the supported full-protected peptide twice with a solution of Tetrakis(triphenylphosphine)palladium (0) (11 mg, 0.01 mmol, 5% mol relative to each Alloc group) and DMBA (123 mg, 0.0.79 mmol, 4 equivalents relative to each Alloc protective group) in dry DMF/DCM (1:1) for 1 h. The resulting solution was drained off and the resin was washed (3 × 1 min) with DMF and with DCM. The catalyst traces were eliminated by treating the support with a DMF solution of potassium N,N-diethylcarbamodithioate (0.06 M, 1.5 mL) for 1 h. To obtain the thyminyl-functionalized nucleopeptides, Thymin-1-yl-acetic acid (108,1 mg, 0.59 mmol, 3 equivalents for each amine group), HOAt (87 mg, 0.59 mmol) and PyAOP (308 mg, 0.59 mmol), DIPEA (205 µL, 1.08 mmol, 6 equivalents for each amine group) were dissolved in DMF (2 mL) and then added to resin for reacting over 8 h at room temperature. This latter step was repeated once.
2.2. Cleavage, HPLC Purification and Characterization of Nucleopeptides np-1/4
After washing with DMF (3 × 1 min), DCM (3 × 1 min), and Et2O (3 × 1 min) the supported nucleopeptides np-1/4 were then dried exhaustively. The np-1/4 were released from the solid support by 3 h treatment with TFA/TIS (95:5, 1.5 mL) at room temperature. Then, the resin was filtered off and the crude nucleopeptides were precipitated from cold Et2O (15 mL) and then centrifuged (6000 rpm × 15 min). After removal of supernatant, the precipitate was suspended once in Et2O, centrifuged and the supernatant removed. The resulting wet solid was dried under vacuum, dissolved in water/ACN (9:1) and purified by RP-HPLC (solvent A: water + 0.1% TFA; solvent B: acetonitrile + 0.1% TFA; from 10 to 70% of solvent B over 25 min, flow rate: 10 mL min−1). The collected fractions were evaporated from organic solvents under reduced pressure, frozen and then lyophilized. The final products were analyzed by analytical RP-HPLC (10–90% acetonitrile in water (0.1% TFA) over 20 min, flow rate of 1.0 mL/min) and characterized by MALDI-TOF spectrometry (Table 1). Yield, purity, retention times, and analytical data are reported in Table 1.

Table 1.
Yield, purity, retention times, and analytical data of the described nucleopeptides.
2.3. UV and CD procedures
UV measurements were performed by a spectrophotometer equipped with a Peltier temperature controller. Nucleopeptide stock solutions were prepared using doubly distilled water and RNAse free water. Their concentration was calculated by UV measurements at 260 nm at 90 °C by using the following molar extinction coefficient: Ɛ 260 = 13.7 mL/(μmol × cm) for A, and Ɛ 260 = 8.8 mL/(μmol × cm) for T. CD spectra and melting curves were acquired in 1 cm path-length quartz cell, on a Jasco J-810 Circular Dichroism Spectrometer equipped with a Peltier temperature programmer. Samples were lyophilized for 16 h. Duplexes were formed by mixing equimolar concentrations of np-1/4 (10 μM each base) and single stranded complementary polyA (M.W. average 100–500 kDa) and polydA (M.W. average 85–170 kDa) (10 μM each base) in a 10 mM phosphate buffer, 100 mM NaCl, pH 7.0 solution. The samples were annealed by incubating at 90 °C for 5 min, slowly cooling down at room temperature and then kept at 4 °C overnight. UV and CD melting was monitored at λ = 260 nm (for UV) and at at λ = 267 nm for the polyA containing hybrids and at λ = 247 nm for polydA containing hybrids, with a scan rate of 1 °C min−1 in the 5–85 °C temperature range. Melting temperatures (Tm) were extrapolated by the first derivatives of CD melting curves. CD spectra were performed in the 210–300 nm spectral range, keeping the temperature constant at 20 °C.
2.4. CD Measurements of Apparent Kd Determination of np-1/polyA and np1/polydA
The titration experiment was carried out by the addition of increasing concentrations of np-1 (0, 1.87, 3.76, 5.60, 7.47, 9.33, 11.20, 13.07, 14.93, 16.8, 18.67 10 μM each base) to a solution containing the single stranded complementary polyA (10 μM each base) and polydA (10 μM each base) in a 10 mM phosphate buffer, 100 mM NaCl, pH 7.0 solution. CD spectra were acquired at 20 °C, monitoring the signal decrements at λ = 260 nm for the polyA and at λ = 216 nm for polydA. The detected changes in CD intensity were plotted to generate saturation isotherm curves as a function of added np-1 amounts. Assuming the formation of 1:1 complex for each nucleobase, we calculated the apparent Kd values by using the following equation:
where Θ is the CD intensity of the characteristic hybrid wavelength, Θmin and Θmax are the minimum and maximum value of CD intensity, respectively, and cA and cT are the Adenine and Thymine nucleobase concentrations, respectively.
Θ = Θmin − [(Θmin − Θmax)/(2cA)] × {Kd+cT+cA − [(Kd + cT + cA)2 − 4cAcT]1/2}
2.5. Building of np-1/RNA Complex
Maestro [41,42] was used to build cp-1, the complex structure, using a double-stranded B-DNA hexamer of complementary A-T bases as a template. The phosphofuranose backbone chain of the Thymine-containing single-strand was removed and a nucleopeptide chain, consisting of 6 L-Arg interpolated with six L-Lys residues (np-1), was constructed in its place in an antiparallel fashion, while keeping the A-T base paring fixed. Finally, hydroxyl groups with R configuration were added to the 2’ carbon atoms of the remaining furanose rings, and the C- and N-terminal ends of np-1 was capped with an amino and an acetyl group respectively. The initial structure of cp-1 was constructed base on the experimentally determined 2:1 stoichiometric interaction of two nucleopeptide molecules with a single 12 mer RNA oligonucleotide [37] To finally assemble the structure of cp-1 composed of a single-stranded 12 mer Adenine RNA oligonucleotide (A12 RNA ss) and two molecules of nucleopeptide np-1, the previously obtained cp-1 complex was duplicated and the RNA chains were linked covalently. The union was made by setting one copy of cp-1 on top of the other and connecting the 3’ end of one RNA strands with the 5’ end of the other through a newly added phosphate group.
2.6. Molecular Dynamics Simulation
Cp-1 complex was initially minimized with MacroModel using an OPLS 2005 force field in water and a Polak-Ribiere Conjugate Gradient minimization protocol of 100,000 maximum iterations and a convergence threshold of 0.001. A constrain in the base paring distances of A-T residues was used to fix the double-strand conformation during the process. When satisfactory minimized structures were obtained (i.e. no anomalous bond angles, amide dihedral angles, bond distances, etc.), molecular dynamic (MD) simulations were launched using Desmond as implemented in the Schrodinger suite [43,44,45]. The model system was created using the System Builder tool of Desmond with an explicit TIP3P water solvent model. The positive charge of the complex was neutralized by 1 chloride ion, and additional ions up to a total concentration of 0.15 M KCl were added, all embedded into a rectangular box of 15 Å in each direction. Relaxation of the model system before performing the MD simulation was carried out with the default Desmond parameters, followed by a 200 ns unrestrained MD simulation with an NPT ensemble (300 K, 1.013 bar). An integration time-step of 2 fs was used for the multistep protocol with an interpolating function for electrostatic interactions (cut-off radius of 9.0 Å). Steady temperature and pressure values were maintained using the Nose–Hoover chain thermostat [45].
2.7. RMSD Clustering Analysis
The clustering analysis was carried out using the MD movie Tool in UCSF Chimera 1.10.1 (University of California, San Francisco, CA, USA) [45] as follows: the final MD simulation frames were exported with Maestro (Schrödinger, Cambridge, MA, USA) to separate pdb files and opened with UCSF Chimera MD movie Tool. A cluster analysis was next carried out, setting frame 1 as the starting one, and a step size of 150. The remaining options were set as default. The final cluster-representative structures given by Chimera were minimized with MacroModel (Schrödinger, Cambridge, MA, USA) with an OPLS 2005 force field in a Polak-Ribier Conjugate Gradient method for a maximum of 5000 iterations and a convergence threshold of 0.05 in water.
3. Results and Discussion
3.1. Synthesis
The synthesis of the nucleopeptides set was performed by a general synthetic path, that was developed by using Allyloxycarbonyl [46,47,48] (Alloc) as temporary protection strategy for the amino group connecting the nucleobase. The Alloc approach allowed using commercially available building blocks avoiding the solution phase synthetic steps required for the preparation of the NƐ-protected derivatives or nucleoamino acids needed for the subsequent assembly of nucleopeptide sequences on the solid support. The use of Alloc protecting group is widely employed to preserve amino acid side chains, including Lysine and Ornitine, those are required to be selectively released for direct functionalization on solid support (i.e. FITC, fatty acid etc) or lactam bridged cyclization [45,46]. Interestingly, in spite of the application of an Alloc-protecting strategy for single amino acid being consolidated, few investigations have been reported for the protection of multiple sites. Thus, we aimed to explore its use also for oligomers containing numerous amino groups, those can be simultaneously released for further on-line derivatization, including the introduction of nucleobases in the case of nucleopeptides. As described in Scheme 1, the 12 mer amino acidic backbone of the nucleopeptides consisting of alternation of l-Arg and l-Lys (np-1) or d-Arg and l-Lys (np-2) or l-Arg and d-Lys (np-3) or d-Arg and d-Lys (np-4) was assembled using a Rink-amide-aminomethyl polystyrene (AM-PS) resin. After acetylation of the N-terminal function, all the Lys side chains were concurrently deprotected by a DMF/DCM solution of Pd(PPh3)4 and N,N-Dimethylbarbituric acid (DMBA) as scavenger. The treatment was reiterated three times to assure the complete removal of Alloc protecting groups as confirmed by HPLC-MS analyses. Next, the Thymine-1-acetic acids (T-CH2COOH) were placed by one-pot reaction by a 3 h reaction in the presence of HBTU/HOBt as activating/additive agents. All the nucleopeptides (np-1/4) were eventually released from the resin, purified by RP-HPLC and characterized by MALDI-TOF MS.
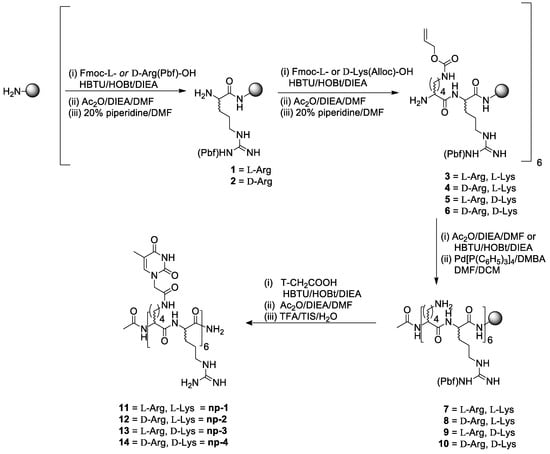
Scheme 1.
Synthetic path to nucleopeptides np-1/4.
3.2. CD Spectroscopic Studies
CD studies were carried out with np-1/4 and with the hybrids obtained by combining the synthesized nucleopeptides np-1/4 with polyA and polydA ss targets. All the CD analyses were performed using 10 μM concentration of np-1/4 in 10 mM phosphate buffer (pH = 7.0) at 20 °C. As depicted in Figure 2, all the single stranded nucleopeptides (np-1/4) exhibited the distinctive CD spectra of linear cationic peptides in an unordered state endowed with a single band below 200 nm.
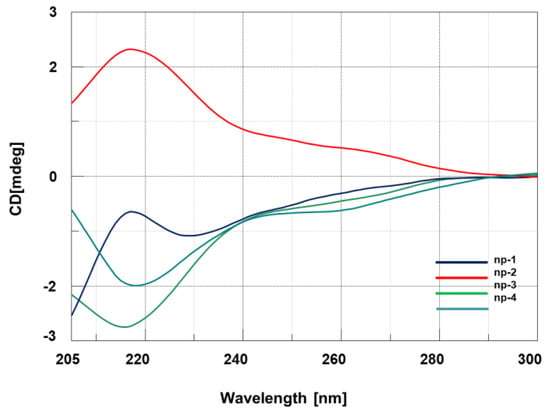
Figure 2.
CD spectra of nucleopeptides np-1/4.
As expected, enantiomeric nucleopeptides containing l-Arg/l-Lys and d-Arg/d-Lys (np-1 and -4) and d-Arg/l-Lys and l-Arg/d-Lys (np-2 and -3) exhibited mirror CD spectra. Next, we investigated the spectroscopic behavior of the hybrids composed of nucleopeptides np-1/4 with PolyA and PolydA (1:1 ratio in nucleobases). Long oligomers, such as polyA and polydA, were chosen for the CD experiments to amplify the resulting binding effects in CD spectra and to take advantage from “the flanking base effect”, that allows the nucleobases next to 5’ and 3’ termini to stabilize the conformation of inner bases even if they do not contribute to the base pairing. Compared to polyA and polydA ss, the CD spectra of np-1/4/polyA (Figure 3a) and np-1/4/polydA (Figure 3b) hybrids exhibited hypochromic effects that could be ascribable to the cationic character of the side chain of Arginine residues.
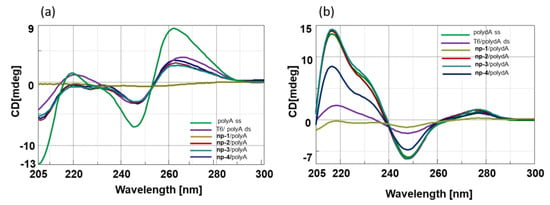
Figure 3.
(a) CD spectra of polyA ss and np-1/4/polyA hybrids; (b) CD spectra of polydA ss and np-1/4/polydA hybrids.
To assess the interaction between nucleopeptides and nucleic acids, CD binding experiments were performed in tandem cells, collecting ‘‘sum’’ CD spectra relative to the separated np-1/4 and ss polyA and polydA, and ‘‘mix’’ CD spectra, acquired after cell mixing and annealing of np-1/4 with polyA and polydA ss. The differences observed comparing the ‘‘sum’’ and ‘‘mix’’ CD spectra strongly suggested the occurrence of the hybridization event in both cases (Figure 4a,b).
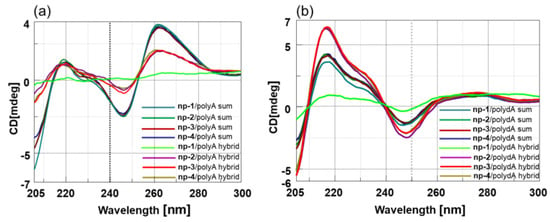
Figure 4.
(a) CD spectra of np-1/4/polyA sum and np-1/4/polyA mix; (b) CD spectra of np-1/4/polydA sum and np-1/4/polydA mix.
In particular, we observed dramatic changes in the CD spectra of hybrids that could be attributed to the loss of nucleic acids helicity resulting from non-specific electrostatic interactions with cationic nucleopeptides. Intriguingly, the strongest hypochromic effect was observed for hybrids containing full L-nucleopeptide, suggesting that besides the electrostatic contribution, the backbone stereochemistry plays a main role in determining the conformation assumed by the hybrid duplexes.
The thermal stability (melting temperature, Tm) of hybrids involving nucleopeptides np-1/4 and both polyA and polydA complements was determined by CD and UV experiments, and compared with Tm of the hexathymine oligodeoxynucleotide (T6)/PolyA and PolydA and ss. (Table 2).

Table 2.
Tm values of np-1/4 polyA and polydA hybrids. Lowercase indicates d-amino acid; uppercase indicates l-amino acid.
Notably, the highest detected thermal stability was observed for np-1/polyA hybrid (Tm = 45 °C), revealing that np-1 represented the most stabilizing RNA binder even if compared to np-3/4/polyA and polydA hybrids. Moreover, the melting profile appeared as a sigmoid shape for np-1/polyA when compared to melting profiles of the any other hybrids (Figure 5). In particular, the melting profiles of polyA hybrids in the presence of l-Arg/d-Lys and d-Arg/d-Lys containing nucleopeptides (np-2 and -3) appeared very complex, suggesting the existence of more than two species and a multistate equilibrium (Figure 5).
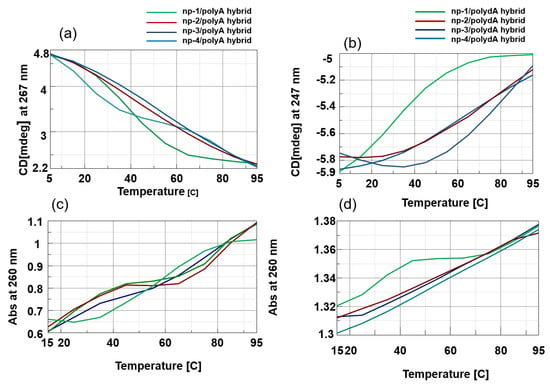
Figure 5.
(a) CD melting profiles of np-1/4/polyA hybrids; (b) CD melting profiles of np-1-np-4/polydA hybrids; (c) UV melting profiles of np-1/4/polyA hybrids; (d) UV melting profiles of np-1/4/polydA hybrids.
The CD melting profiles suggested that the dissociation of np-1/polyA and np-1/polydA was a two-state process. Thus, we were able to estimate the apparent Kd values of np-1 to polyA and polydA by performing CD titration experiment with increasing concentrations of np-1 in the presence of a constant concentration of nucleic acids. As depicted in Figure 6, the apparent Kd values further corroborated the results achieved by us and other groups [37,39], indicating that cationic nucleopeptides are endowed with higher affinity for RNA (Figure 6a) with the respect to DNA molecules (Figure 6b).
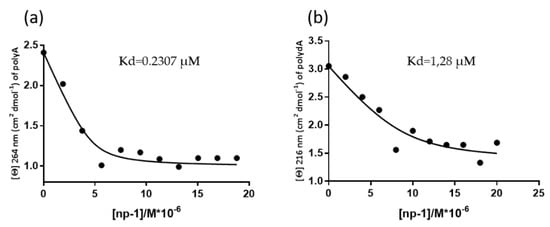
Figure 6.
Binding curve of 10 uM polyA (a) and polydA (b) titrated with increasing concentration of np-1.
3.3. Molecular Dynamics Simulations
To gain structural insights into the binding capabilities of chiral nucleopeptide np-1 toward RNA, we performed molecular dynamics (MD) simulations of the single nucleopeptide consisting of interpolated l-Arg and l-Lys residues covalently linked to Thymine nucleobases. In this case, np-1 contained six l-Arg and six l-Lys residues with the Thymine nucleobases linked to the ε-NH2-group of the Lysine side chain through an acetyl group (Figure 6). This polymer was chosen as a model for the MD simulations due to its interesting spectroscopic behavior toward RNA ss.
The interactions of the molecular complex cp1 (Figure 7) made up by a single-stranded 12 mer Adenine RNA oligonucleotide (A12 RNA ss) and two nucleopeptide np-1 molecules were assessed through MD simulations. The initial simulation structure was constructed based on the experimentally determined 2:1 stoichiometric interaction of np-1 with a complementary single-stranded 12 mer Adenine RNA oligonucleotide, and assuming an initial recognition event between the three macromolecules guided by the base-pairing interactions of the three species [40] (Figure 7).
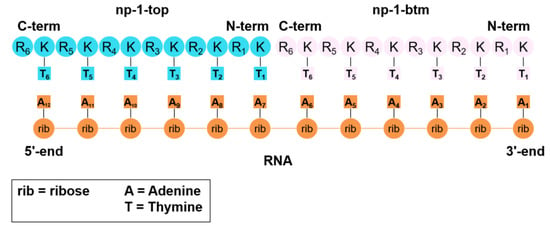
Figure 7.
Schematic representation of complex cp-1 submitted to MD.
After running the 200 ns long MD simulation (see Materials and Methods section), we analyzed the conformational stability of the simulated complex cp-1 in terms of the root-mean-square deviation (RMSD) of all the non-hydrogen atoms in the complex (Figure 8). Overall, the analysis of the MD trajectories demonstrated that after an initial equilibration period the complex reaches a fairly stable conformation with an 8 Å RMSD deviation from the initial input structure. This increase in RMSD is followed by a moderate second change, after approximately 40 ns, to reach a second conformation with an 11 Å RMSD deviation from the initial structure. This last conformation demonstrates to be fairly stable all through the remaining part of the simulation (160 ns).
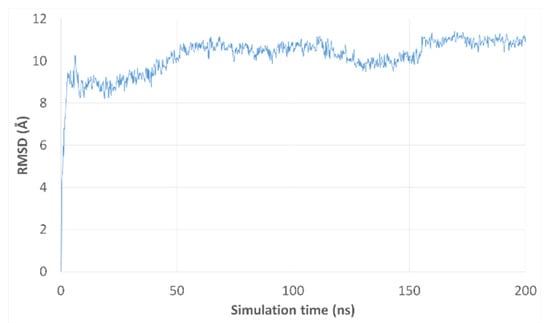
Figure 8.
RMSD plot of MD trajectories from their respective initial frame in complex cp-1.
To better determine the conformational behavior of the simulated complex, we conducted an RMSD-based clustering analysis of the MD 1002 trajectory frames obtained after the MD simulation. A total of three clusters, were obtained accounting for the most representative structures in the MD simulation of cp-1. Of the three clusters, the last one stands for the highest number of frames and includes the structures calculated for the last 160 ns of the simulations. The most representative structure of this cluster is depicted in Figure 9a, corresponding to a wrapped conformation of the complex.
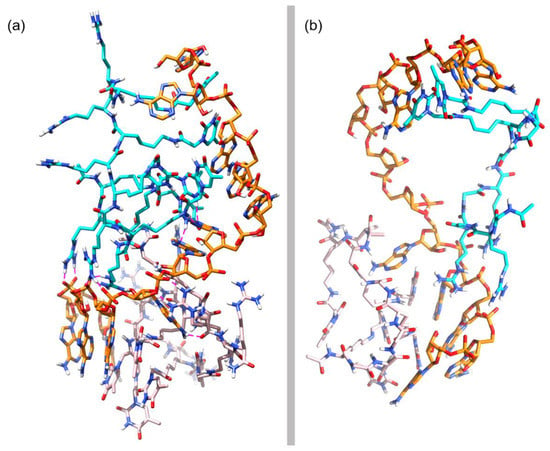
Figure 9.
(a) Representative structure of the most populated cluster in cp-1; (b) Simplified representation of a zenithal view of cp-1.
Next, we analyzed the structure depicted in Figure 9a to gather some information about the way np-1 interacts with the RNA counterpart. As can be observed in Figure 9a, the base-pairing of both np-1 molecules with the RNA chain is almost completely lost, with other chemical interactions taking place in the complex. More importantly, the RNA chain has lost its helical conformation and is adopting an almost linear disposition as can be seen in a zenithal view of the complex (Figure 9b). Responsible for this loss of helicity are the two np-1 molecules. Of these, the np-1 molecule (np-1-top) originally attached to the 5’ end of the complex, uses its three N-terminal Arginines (R1, R2, and R3) to form stable ionic interactions with the RNA phosphate backbone at the 3’ end. Moreover, np-1-top also forms H-bonds between its first and second N-terminal Thymine rings (T1 and T2) with the two middle RNA Adenine (A6 and A7), helping to keep the RNA chain in a near-linear conformation (Figure 10a). Similarly, important is the H-bond formed by the amide group in Thymine T3 arm with Adenine A7, which helps to hold the RNA chain before the turn that takes place between Adenine A7 and A8. The ionic interactions, however, are not the only chemical interactions between np-1-top and RNA. The complex also shows π-stacking interactions among C-terminal Thymine rings (T5 and T6) and Adenine A10 and A12. Both T5 and T6 are sandwiched between A10 and A12, holding the top half of the RNA chain with a successive six nucleobases π-stacking.
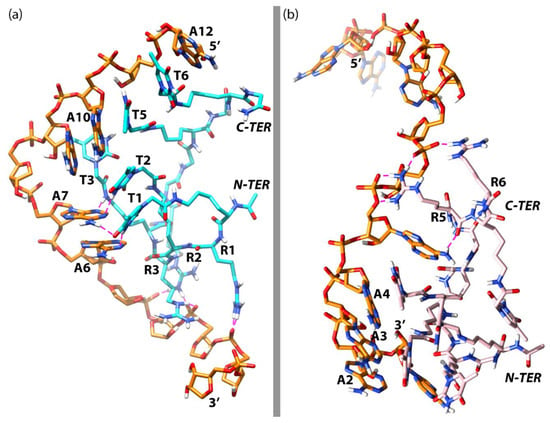
Figure 10.
(a) Simplified representation of the most important interactions between np-1-top and RNA; (b) Simplified representation of the most important interactions between np-1-btm and RNA. RNA chain in orange, np-1-top in cyan blue, np-1-btm in pink, and ionic interactions in purple.
For the second np-1 molecule (np-1-btm) (Figure 10b), only the two C-terminal Arginines R5 and R6 are forming stable ionic interactions with the RNA phosphate backbone in the middle of the chain. The rest of the np-1-btm molecule, however, seems to be interacting exclusively at the 3’ end of the RNA, where the majority of the Thymine rings are grouped in a cluster with Adenine (A2, A3, and A4) by π-stacking interactions, participating in the stabilization of the wrapped RNA conformation.
To account for the importance of the ionic interactions described above, we plotted its distances during the complete MD simulation (Figure 11). The results show that all the interactions are highly conserved during the most part of the simulation. For np-1-top, after an initial equilibration period of approximately 40 ns, a fair stabilization of the H-bond distances occurs for Arginines R1, R2 and R3 (Figure 11a), while for Thymines T1, T2, and T3 the H-bond distances (two for T1, one for T2 and one for T3) are quite stable from the beginning of the simulation, particularly those between T1 and A6, and T2 and A7 (Figure 11b). As for np-1-btm, only Arginine R5 is able to form stable and significant ionic interactions with the phosphate backbone after the initial 40 ns equilibration period, contributing to the stabilization of the extended RNA chain (Figure 11c). These data remark the importance of the Arginine chains in the nucleopeptide np-1 to induce and maintain an extended conformation of the RNA chain, as well as the role of the nucleobase groups, not only for the initial recognition of the RNA chain but also to stabilize such an interaction with RNA.
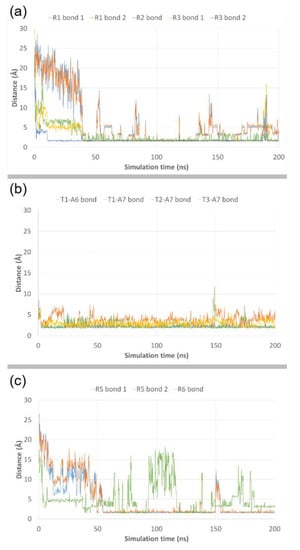
Figure 11.
RMSD plots of bond distances from their respective initial frame for Arginines R1, R2 and R3 (a), Thymine T1, T2 and T3 in np-1-top (b), and Arginines R5 and R6 in np-1-btm (c).
4. Conclusions
In this study, we report the synthesis of Arginine-based chiral nucleopeptides in which stereochemically-defined nucleobase and underivatized amino acids were alternated in the peptide backbone. With the purpose to investigate the role of backbone stereochemistry in determining the formation of DNA and RNA hybrids, we performed spectroscopic studies. CD experiments highlighted that the nucleopeptide having a fully l-backbone configuration and consisting of interpolated l-Arginine and l-Lysine residues covalently linked to Thymine nucleobases, formed the most intriguing and stable hybrid complexes with RNA molecules. Molecular Dynamics (MD) simulations provided insight into potential structure of the resulting complex of the l-nucleopeptide with RNA complementary strands, pointing out detailed chemical interactions involving both the l-backbone (ionics and H-bonds) and nucleobases (pairing and π-stacking) of the chiral nucleopeptide. In conclusion, we provided the first evidence of the effect of the backbone stereochemistry on the chemical interactions involved in the formation of the nucleopeptide/RNA complexes. These findings added a new piece to the complex nucleopeptide/nucleic interaction puzzle that could contribute to the future design of peptide-based molecules acting as antisense agents or diagnostic probes.
Author Contributions
Conceptualization, A.M.and S.D.M.; Data curation, S.T., F.F.M., R.R., A.M. and S.D.M.; Funding acquisition, S.D.M.; Investigation, S.T., F.F.M., A.M. and S.D.M.; Methodology, S.T., A.M. and S.D.M.; Project administration, A.M. and S.D.M.; Writing–original draft, A.M.; Writing–review & editing, E.N., A.M. and S.D.M.
Funding
This research was supported by Scientific Independence of Young Researchers (SIR) 2014 (RBSI142AMA) and University of Campania Luigi Vanvitelli (Valere) to S.D.M.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Caprara, M.G.; Nilsen, T.W. RNA: Versatility in form and function. Nat. Struct. Biol. 2000, 7, 831–833. [Google Scholar] [CrossRef]
- Hudson, W.H.; Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15, 749–760. [Google Scholar] [CrossRef]
- Brennan, R.G.; Matthews, B.W. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989, 264, 1903–1906. [Google Scholar]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Vinson, C.R.; Sigler, P.B.; McKnight, S.L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 1989, 246, 911–916. [Google Scholar] [CrossRef]
- Pavletich, N.P.; Pabo, C.O. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science 1991, 252, 809–817. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Cell-type-specific: Aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 2014, 3, e169. [Google Scholar] [CrossRef]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D.; et al. Chemically Modified DNA Aptamers Bind Interleukin-6 with High Affinity and Inhibit Signaling by Blocking Its Interaction with Interleukin-6 Receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef]
- Matsumura, S.; Takahashi, T.; Ueno, A.; Mihara, H. Complementary nucleobase interaction enhances peptide-peptide recognition and self-replicating catalysis. Chem.-A Eur. J. 2003, 9, 4829–4837. [Google Scholar] [CrossRef]
- Matsumura, S.; Ueno, A.; Mihara, H. Peptides with nucleobase moieties as a stabilizing factor for a two-stranded R-helix. Chem. Commun. 2000, 1615–1616. [Google Scholar] [CrossRef]
- Takahashi, T.; Hamasaki, K.; Kumagai, I.; Ueno, A.; Mihara, H. Design of a nucleobase-conjugated peptide that recognizes HIV-1 RRE IIB RNA with high affinity and specificity. Chem. Commun. 2000, 349–350. [Google Scholar] [CrossRef]
- Takahashi, T.; Hamasaki, K.; Ueno, A.; Mihara, H. Construction of peptides with nucleobase amino acids: Design and synthesis of the nucleobase-conjugated peptides derived from HIV-1 Rev and their binding properties to HIV-1 RRE RNA. Bioorg. Med. Chem. 2001, 9, 991–1000. [Google Scholar] [CrossRef]
- Takahashi, T.; Ueno, A.; Mihara, H. Nucleobase amino acids incorporated into HIV-1 nucleocapsid protein increased the binding affinity and specificity to a hairpin RNA. ChemBioChem. 2002, 3, 543–549. [Google Scholar] [CrossRef]
- Takahashi, T.; Yana, D.; Mihara, H. Utilization of l-a-Nucleobase Amino Acids (NBAs) as Protein Engineering Tools: Construction of NBA-Modified HIV-1 Protease Analogues and Enhancement of Dimerization Induced by Nucleobase Interaction. ChemBioChem. 2006, 7, 729–732. [Google Scholar] [CrossRef]
- Bai, X.; Talukder, P.; Daskalova, S.M.; Roy, B.; Chen, S.; Li, Z.; Dedkova, L.M.; Hecht, S.M. Enhanced Binding Affinity for an i-Motif DNA Substrate Exhibited by a Protein Containing Nucleobase Amino Acids. J. Am. Chem. Soc. 2017, 139, 4611–4614. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef]
- Wittung, P.; Nielsen, P.E.; Buchardt, O.; Egholm, M.; Norden, B. DNA-like double helix formed by peptide nucleic acid. Nature 1994, 368, 561–563. [Google Scholar] [CrossRef]
- Nielsen, P.E. Peptide nucleic acid: A versatile tool in genetic diagnostics and molecular biology. Curr. Opin. Biotechnol. 2001, 12, 16–20. [Google Scholar] [CrossRef]
- Nielsen, P.E. Peptide Nucleic Acid. A Molecule with Two Identities. Acc. Chem. Res. 1999, 32, 624–630. [Google Scholar] [CrossRef]
- Ganesh, K.N.; Nielsen, P.E. Peptide Nucleic Acids Analogs and Derivatives. Curr Org. Chem. 2000, 4, 931–943. [Google Scholar] [CrossRef]
- Kumar, V.A.; Ganesh, K.N. Conformationally Constrained PNA Analogues: Structural Evolution toward DNA/RNA Binding Selectivity. Acc. Chem. Res. 2005, 38, 404–412. [Google Scholar] [CrossRef]
- Kumar, V.A.; Ganesh, K.N. Structure-Editing of Nucleic Acids for Selective Targeting of RNA. Curr. Top. Med. Chem. 2007, 7, 715–726. [Google Scholar] [CrossRef]
- Sforza, S.; Corradini, R.; Ghirardi, S.; Dossena, A.; Marchelli, R. DNA Binding of A D-Lysine-Based Chiral PNA: Direction Control and Mismatch Recognition. Eur. J. Org. Chem. 2000, 2905–2913. [Google Scholar] [CrossRef]
- Tedeschi, T.; Sforza, S.; Dossena, A.; Corradini, R.; Marchelli, R. Lysine-based peptide nucleic acids (PNAs) with strong chiral constraint: Control of helix handedness and DNA binding by chirality. Chirality 2005, 17, 196–204. [Google Scholar] [CrossRef]
- Bose, T.; Banerjee, A.; Nahar, S.; Maiti, S.; Kumar, V.A. β,γ-Bis-substituted PNA with configurational and conformational switch: Preferred binding to cDNA/RNA and cell-uptake studies. Chem. Commun. 2015, 51, 7693–7696. [Google Scholar] [CrossRef]
- Futaki, S.; Nakase, I. Cell-Surface Interactions on Arginine-Rich Cell-Penetrating Peptides Allow for Multiplex Modes of Internalization. Acc. Chem. Res. 2017, 50, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wang, M.; Du, L.; Fisher, G.W.; Waggoner, A.; Ly, D.H. Novel Binding and Efficient Cellular Uptake of Guanidine-Based Peptide Nucleic Acids (GPNA). J. Am. Chem. Soc. 2003, 125, 6878–6879. [Google Scholar] [CrossRef]
- Dragulescu-Andrasi, A.; Zhou, P.; He, G.; Ly, D.H. Cell-permeable GPNA with appropriate backbone stereochemistry and spacing binds sequence-specifically to RNA. Chem. Commun. 2005, 244–246. [Google Scholar] [CrossRef]
- Diederichsen, U. Pairing Properties of Alanyl Peptide Nucleic Acids Containing an Amino Acid Backbone with Alternating Configuration. Angew. Chem. Int. Ed. Engl. 1996, 35, 445–448. [Google Scholar] [CrossRef]
- Diederichsen, U.; Schmitt, H.W. β-Homoalanyl PNAs: Synthesis and Indication of Higher Ordered Structures. Angew. Chem. Int. Ed. 1998, 37, 302–305. [Google Scholar] [CrossRef]
- Hoffmann, M.F.H.; Bruckner, A.M.; Hupp, T.; Engels, B.; Diederichsen, U. Specific Purine-Purine Base Pairing in Linear Alanyl-Peptide Nucleic Acids. Helv. Chim. Acta 2000, 83, 2580–2593. [Google Scholar] [CrossRef]
- Chakraborty, P.; Diederichsen, U. Three-Dimensional Organization of Helices: Design Principles for Nucleobase-Functionalized β-Peptides. Chem. Eur J. 2005, 11, 3207–3216. [Google Scholar] [CrossRef]
- Miyanishi, H.; Takahashi, T.; Mihara, H. De Novo Design of Peptides with l-α-Nucleobase Amino Acids and Their Binding Properties to the P22 boxB RNA and Its Mutants. Bioconjugate Chem. 2004, 15, 694–698. [Google Scholar] [CrossRef]
- Watanabe, S.; Tomizaki, K.; Takahashi, T.; Usui, K.; Kajikawa, K.; Mihara, H. Interactions between peptides containing nucleobase amino acids and T7 phages displaying S. cerevisiae proteins. Peptide Sci. 2007, 88, 131–140. [Google Scholar] [CrossRef]
- Mercurio, M.E.; Tomassi, S.; Gaglione, M.; Russo, R.; Chambery, A.; Lama, S.; Stiuso, P.; Cosconati, S.; Novellino, E.; Di Maro, S.; et al. Switchable Protecting Strategy for Solid Phase Synthesis of DNA and RNA Interacting Nucleopeptides. J. Org. Chem. 2016, 81, 11612–11625. [Google Scholar] [CrossRef]
- Tomassi, S.; Ieranò, C.; Mercurio, M.E.; Nigro, E.; Daniele, A.; Russo, R.; Chambery, A.; Baglivo, I.; Pedone, P.V.; Rea, G.; et al. Cationic nucleopeptides as novel non-covalent carriers for the delivery of peptide nucleic acid (PNA) and RNA oligomers. Bioorg. Med. Chem. 2018, 26, 2539–2550. [Google Scholar] [CrossRef]
- Roviello, G.N.; Gaetano, S.D.; Capasso, D.; Franco, S.; Crescenzo, C.; Bucci, E.M.; Pedone, C. RNA-Binding and Viral Reverse Transcriptase Inhibitory Activity of a Novel Cationic Diamino Acid-Based Peptide. J. Med. Chem. 2011, 54, 2095–2101. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Qin, Y.; Wang, J.; Xu, B. Nucleopeptide Assemblies Selectively Sequester ATP in Cancer Cells to Increase the Efficacy of Doxorubicin. Angew. Chem. Int. Ed. 2018, 130, 5025–5029. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Schrödinger Release 2018-3: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2018. Available online: http://www.schrodinger.com/ (accessed on 17 April 2019).
- Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, USA, 2018. Available online: http://www.schrodinger.com/ (accessed on 17 April 2019).
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kunz, H.; Unverzagt, C. The Allyloxycarbonyl (Aloc) Moiety-Conversion of an Unsuitable into a Valuable Amino Protecting Group for Peptide Synthesis. Angew. Chem. Int. Ed. Engl. 1984, 23, 436–437. [Google Scholar] [CrossRef]
- Di Maro, S.; Zizza, P.; Salvati, E.; De Luca, V.; Capasso, C.; Fotticchia, I.; Pagano, B.; Marinelli, L.; Gilson, E.; Novellino, E.; et al. Shading the TRF2 Recruiting Function: A New Horizon in Drug Development. J. Am. Chem. Soc. 2014, 136, 16708–16711. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, F.S.; Tomassi, S.; Di Maro, S.; Reichart, F.; Notni, J.; Dangi, A.; Marelli, U.K.; Brancaccio, D.; Merlino, F.; Wester, H.J.; et al. From a Helix to a Small Cycle: Metadynamics-Inspired αvβ6 Integrin Selective Ligands. Angew. Chem. Int. Ed. Engl. 2018, 57, 14645–14649. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).